Abstract
Cryptosporidium spp., Enterocytozoon bieneusi, and Giardia duodenalis are common intestinal pathogens that infect humans and animals. To date, research regarding these three protozoa in the Ningxia Hui Autonomous Region (Ningxia) has mostly been limited to a single pathogen, and comprehensive data on mixed infections are unavailable. This study aimed to evaluate the zoonotic potential of these three protozoa. In this study, small subunit ribosomal RNA (SSU rRNA) and 60 kDa glycoprotein (gp60) genes of Cryptosporidium; internal transcribed spacer (ITS) gene of E. bieneusi; and SSU rRNA, glutamate dehydrogenase (gdh), triosephosphate isomerase (tpi), and beta-giardin (bg) genes of G. duodenalis were examined. DNA extraction, polymerase chain reaction, and sequence analysis were performed on fecal samples collected from 320 dairy cattle at three intensive dairy farms in Ningxia in 2021 to determine the prevalence and genetic characteristics of these three protozoa. The findings revealed that 61.56% (197/320) of the samples were infected with at least one protozoan. The overall prevalence of Cryptosporidium was 19.38% (62/320), E. bieneusi was 41.56% (133/320), and G. duodenalis was 29.38% (94/320). This study identified four Cryptosporidium species (C. bovis, C. andersoni, C. ryanae, and C. parvum) and the presence of mixed infections with two or three Cryptosporidium species. C. bovis was the dominant species in this study, while the dominant C. parvum subtypes were IIdA15G1 and IIdA20G1. The genotypes of E. bieneusis were J, BEB4, and I alongside the novel genotypes NX1–NX8, all belonging to group 2, with genotype J being dominant. G. duodenalis assemblages were identified as assemblages E, A, and B, and a mixed infection involving assemblages A + E was identified, with assemblage E being the dominant one. Concurrently, 11 isolates formed 10 different assemblage E multilocus genotypes (MLGs) and 1 assemblage A MLG and assemblage E MLGs formed 5 subgroups. To the best of our knowledge, this is the first report on mixed infection with two or three Cryptosporidium species in cattle in Ningxia and on the presence of the C. parvum subtype IIdA20G1 in this part of China. This study also discovered nine genotypes of E. bieneusis and novel features of G. duodenalis assemblages in Ningxia. This study indicates that dairy cattle in this region may play a significant role in the zoonotic transmission of Cryptosporidium spp., E. bieneusi, and G. duodenalis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cryptosporidium spp., Enterocytozoon bieneusi, and Giardia duodenalis are three important intestinal protozoa (Yang et al. 2022) that can cause clinical symptoms such as weight loss, malnutrition, and diarrhea in various hosts (Wang et al. 2022a). The infections caused by these parasites are self-limiting in healthy individuals; however, the infection period may be prolonged or even life threatening in immunocompromised individuals (Yang et al. 2022). These pathogens spread primarily through the fecal–oral route (Fan et al. 2021).
To date, at least 45 valid Cryptosporidium species and approximately 120 genotypes have been reported worldwide (Wang et al. 2022a), among which, 19 Cryptosporidium species and 4 genotypes have been reported in humans (Ryan et al. 2021) and 12 Cryptosporidium species have been reported in cattle (Gong et al. 2017), with C. parvum, C. bovis, C. ryanae, and C. andersoni being the most dominant species (Feng et al. 2018; Gong et al. 2017; Hatam-Nahavandi et al. 2019; Santin 2020; Zahedi and Ryan 2020; Zhang et al. 2022). Zoonotic cryptosporidiosis is mainly caused by C. parvum (Feng et al. 2018; Guo et al. 2022; Zhang et al. 2022). In E. bieneusi, > 685 distinct genotypes have been reported (Qin et al. 2022) and classified into 11 distinct groups (groups 1–11) via phylogenetic analysis, with > 90% of the genotypes belonging to group 1 or 2 (Li et al. 2019b; Wang et al. 2022a; Yang et al. 2022). Group 1 is a zoonotic evolutionary group (Lichtmannsperger et al. 2023; Wang et al. 2022b) in which the genotypes A, D, EbpC, and IV are the most common (Santín and Fayer 2011; Wang et al. 2022b). Some genotypes from groups 2, 5, and 6 can also infect humans (Dong et al. 2022). BEB4, BEB6, I, and J in group 2 constitute the main genotypes that infect humans and animals (Li et al. 2019b), whereas genotypes BEB4, I, and J are most commonly reported in cattle globally (Lichtmannsperger et al. 2023). G. duodenalis is currently classified into eight genetically distinct but morphologically identical lineages, namely assemblages A–H (Qi et al. 2016; Ryan et al. 2019; Xiao and Feng 2017). Assemblages A and B exhibit higher zoonotic risk (Kiani-Salmi et al. 2019; Taghipour et al. 2022b; Zhang et al. 2021), while assemblages C–F have also been identified in human patients (Kiani-Salmi et al. 2019). Assemblage E is the most common genotype in cattle globally (Cai et al. 2021; Feng and Xiao 2011; Minetti et al. 2014; Santin 2020; Taghipour et al. 2022b; Zahedi and Ryan 2020; Zhang et al. 2021), followed by assemblages A and B (Feng and Xiao 2011; Qi et al. 2016; Zhang et al. 2021). Assemblages C, D (Ryan and Cacciò 2013), and F have also been reported in cattle (Cardona et al. 2015; Ryan and Cacciò 2013).
Investigation of the three protozoa in cattle via a molecular method revealed that the global pooled prevalence of Cryptosporidium was 29.1% (Hatam-Nahavandi et al. 2019) and the Chinese pooled prevalence of Cryptosporidium was 17.0% (Cai et al. 2019). Furthermore, the global pooled prevalence of E. bieneusi has been reported to be approximately 14% (Taghipour et al. 2022b), 12.0% (Qin et al. 2022), or 16.6% (Ruan et al. 2021), while its Chinese pooled prevalence has been reported to be 14.2% (Taghipour et al. 2022b), 11.2% (Qin et al. 2022), 20.0% (Qiu et al. 2019), or 14.0% (Wang et al. 2018). Furthermore, the global pooled prevalence of G. duodenalis was approximately 22% and its Chinese pooled prevalence was 14.1% (Taghipour et al. 2022b). In recent years, Cryptosporidium (Cui et al. 2014; Huang et al. 2014; Wang et al. 2023; Zhang et al. 2015), E. bieneusi (Dong et al. 2022; Li et al. 2016), and G. duodenalis (Huang et al. 2014; Wang et al. 2023; Zhang et al. 2016) have been studied in cattle from Ningxia. However, most of these studies were limited to a single pathogen, and no comprehensive data regarding mixed infection are available. Thus, to evaluate the zoonotic potential of Cryptosporidium, E. bieneusi, and G. duodenalis, we investigated the molecular prevalence and genetic characteristics of these pathogens infecting dairy cattle in Ningxia, China.
Materials and methods
Study areas and sample collection
From April to May 2021, fresh fecal samples were randomly collected from 320 calves at three intensive dairy farms near Yinchuan and Zhongwei (105°46'E–106°60'E, 36°59'N–38°58'N) in northern Ningxia, China. The samples were collected from 70 preweaned calves (0–60 days), 70 postweaned calves (61–180 days), 70 young cattle (181–360 days), and 110 adult cattle (≥ 361 days). The fecal samples were collected through rectal sampling or from the inner top layer of fresh feces. Detailed information regarding whether the sampled animals had diarrhea was recorded, and the samples were stored at 4℃ before being transported to the laboratory for DNA extraction.
DNA extraction and polymerase chain reaction (PCR)
DNA was extracted from the collected 320 fecal samples using E.Z.N.A.® Stool DNA Kit (Omega Biotek Inc., Norcross, GA, USA) according to the manufacturer’s protocol and stored at − 20 °C. Using the extracted DNA as a template, Premix Taq™ (TaKaRa Taq™ Version 2.0 plus dye) (TaKaRa, Beijing, China) was used to amplify the SSU rRNA gene of Cryptosporidium (Alves et al. 2003) and G. duodenalis (Appelbee et al. 2003). The ITS gene of E. bieneusi (Buckholt et al. 2002) was amplified via nested PCR.
The sequencing of the SSU rRNA gene of Cryptosporidium-positive PCR products was outsourced to Sangon Biotech (Shanghai, China). The results of sequencing analysis were combined with those of the RFLP analysis of the SSU rRNA gene of Cryptosporidium-positive PCR products using restriction enzymes (Ssp I and Mbo II; TaKaRa, Beijing, China) (Feng et al. 2007). Upon confirming the presence of C. parvum DNA, the nested PCR of gp60 (Alves et al. 2003) was subsequently performed, and gp60 sequencing results were used for C. parvum subtype identification (Sulaiman et al. 2005). The SSU rRNA genes of G. duodenalis-positive DNA were further amplified via nested PCR for bg (Lalle et al. 2005), gdh (Cacciò et al. 2008), and tpi (Sulaiman et al. 2003). The ITS gene of E. bieneusi and four genes of G. duodenalis-positive PCR products were also sequenced.
Sequence analysis
The sequences were aligned with reference sequences downloaded from GenBank (http://www.ncbi.nlm.nih.gov) using MEGA 7.0 software (http://www.megasoftware.net/). The sequencing results were analyzed using the BLAST online platform. To comprehensively investigate the relationship among different isolates, phylogenetic analyses were performed using a concatenated dataset of the ITS gene of E. bieneusi and the bg, gdh, and tpi sequences of G. duodenalis. Phylogenetic trees were constructed using the neighbor-joining algorithm based on a matrix of evolutionary distances calculated using the Kimura two-parameter model via MEGA 7.0 software. Bootstrap analysis was performed to assess the robustness of clusters using 1000 replicates.
Statistical analysis
Using chi-squared test in SPSS Statistics 21.0 (IBM Corp., New York, NY, USA) with a 95% confidence interval, the infection rates of Cryptosporidium spp., E. bieneusi, and G. duodenalis were compared among different farms, age groups, and groups with and without diarrhea. A two-tailed p-value of < 0.05 was considered to indicate a statistically significant difference.
Results
Cryptosporidium, E. bieneusis, and G. duodenalis infection status
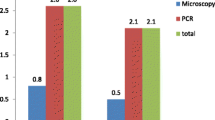
Among the 320 samples collected in this study, 197 (61.56%) were infected with at least one of the following pathogens: Cryptosporidium, E. bieneusis, and G. duodenalis. Furthermore, 10 samples (3.13%) were simultaneously infected with Cryptosporidium, G. duodenalis, and E. bieneusis. In addition, 26 samples (8.13%) exhibited mixed infection with Cryptosporidium and E. bieneusis, 39 samples (12.19%) with E. bieneusis and G. duodenalis, and 7 samples (2.19%) with Cryptosporidium and G. duodenalis (Table 1).
Cryptosporidium infection status
Based on the identification of the SSU rRNA gene of Cryptosporidium, the overall prevalence rate of Cryptosporidium was 19.38% (62/320). The overall prevalence rates of Cryptosporidium in samples from animals with and without diarrhea were 16.13% (5/31) and 19.72% (57/289), respectively; the difference was not significant. The overall prevalence rates of Cryptosporidium in samples from the four age groups (in chronological order) were 24.29% (17/70), 28.57% (20/70), 22.86% (16/70), and 8.18% (9/110). The overall infection rates in the preweaned calves and young cattle were significantly higher than those in adult cattle. The overall infection rate in the postweaned calves was also significantly higher than that in adult cattle. The overall infection rates among samples from the three intensive dairy farms were 35.83% (43/120), 7% (7/100), and 12% (12/100), respectively. The overall infection rate at farm 1 was significantly higher than that at farm 2 (Table 2).
E. bieneusis infection status
Based on the PCR amplification of the ITS gene of E. bieneusi, infection was identified in 133 of the 320 samples (41.56%). The overall infection rates of E. bieneusi in samples collected from animals with and without diarrhea were 48.39% (15/31) and 40.83% (118/289), respectively. The overall infection rates of E. bieneusi in the four age groups (in chronological order) were 40% (28/70), 54.29% (38/70), 61.43% (43/70), and 21.82% (24/110), respectively. The overall infection rates in the postweaned calves and young cattle were significantly higher than those in adult cattle. The overall infection rate in the preweaned calves was significantly higher than that in adult cattle. The overall infection rates of E. bieneusi at the three intensive dairy farms were 52.5% (63/120), 38% (38/100), and 32% (32/100), respectively. The overall infection rate at farm 1 was significantly higher than that at farm 3 (Table 2).
G. duodenalis infection status
Based on the PCR amplification of the SSU rRNA gene of G. duodenalis, infection was identified in 94 of the 320 samples (29.38%). The overall infection rates of G. duodenalis in samples from animals with and without diarrhea were 22.58% (7/31) and 30.10% (87/289), respectively; the difference was not significant. The overall infection rates of G. duodenalis in the four age groups (in chronological order) were 20% (14/70), 28.57% (20/70), 35.71% (25/70), and 31.82% (35/110). A statistically significant difference in the overall infection rate was observed between young cattle and preweaned calves. The overall infection rates of G. duodenalis in samples from the three intensive dairy farms were 20% (24/120), 38% (38/100), and 32% (32/100), respectively. The overall infection rate at farm 2 was significantly higher than that at farm 1. In addition, the overall infection rate between farm 1 and farm 3 was statistically significantly different (Table 2).
Identification of species, genotypes and assemblages and sequence analysis of Cryptosporidium, E. bieneusis, and G. duodenalis
Species identification and sequence analysis of Cryptosporidium
Through RFLP analysis of the PCR amplification products from the 62 SSU rRNA genes and combining them with the 61 obtained sequences, 4 Cryptosporidium species, namely, C. bovis (59.68%, 37/62), C. andersoni (9.68%, 6/62), C. ryanae (33.87%, 21/62), and C. parvum (20.97%, 13/62), were identified. In addition, mixed infections with two or three Cryptosporidium species were identified (Table 2). Infections with these four Cryptosporidium species were identified in all three intensive dairy farms. C. bovis was the dominant species in this survey and was identified in all four age groups, primarily in the postweaned calves (48.65%, 18/37). The preweaned calves were mainly infected with C. parvum (12/17) and the postweaned calves were mainly infected with C. bovis (18/20). In young cattle, C. bovis infection (10/16) was the most common, whereas in adult cattle, C. andersoni (4/9) and C. bovis (5/9) infections were the most common. Three and four of the abovementioned Cryptosporidium spp. were detected in samples from animals with and without diarrhea, respectively. C. bovis was mainly found in the samples from the groups with and without diarrhea (4/5 and 34/57, respectively) (Table 2).
The 10 gp60 sequences obtained in this study were analyzed, and the subtypes of the C. parvum family IId were successfully identified as IIdA15G1 (9) and IIdA20G1 (1) (Table 2).
Genotype identification and sequence analysis of E. bieneusis
Based on the ITS gene of E. bieneusis, 133 credible sequences were obtained, which were used for the sequence alignment of 243-bp ITS region and comparison with the reference sequences of different genotypes. Differences in no more than two bases were identified. The sequences identified in this study were classified into the genotypes J (66.17%, 88/133), BEB4 (24.81%, 33/133), and I (3.01%, 4/133) along with new genotypes NX1–NX8 (0.75%, 1/133) (Table 2).
Phylogenetic analysis classified the genotypes J, BEB4, I, and NX1–NX8 along with the multiple genotypes previously widely reported in humans and animals into group 2 (Fig. 1).
Phylogenetic relationship among Enterocytozoon bieneusi in a neighbor-joining tree based on the ITS gene. The isolates marked with black triangles (▲) and white triangles (△) represent Enterocytozoon bieneusi identified as having genotypes J, BEB4, and I and novel genotypes from cattle in this study, respectively
Assemblage identification and MLG analysis of G. duodenalis
Overall, 94 samples positive for the PCR products of the SSU rRNA gene were obtained, and 92 sequences were evaluated as reliable. Among these, 75 (81.52%, 75/92) were classified into assemblage E, 15 (16.30%, 15/92) into assemblage A, and 2 (2.17%, 2/92) into assemblage B. Assemblage E was identified in all four age groups of cattle, whereas only assemblage E was present in the preweaned calves (Table 2).
Overall, 94 positive samples were obtained via PCR and sequencing of gdh, tpi, and bg from G. duodenalis, yielding 18, 26, and 40 credible sequences, respectively. Overall, 18 isolates of gdh were identified, with 1 being identified as an assemblage A sequence (A1) and 17 identified as 11 assemblage E sequences with the following distribution: E1 (n = 5), E2 (n = 2), E3 (n = 1), E4 (n = 1), E5 (n = 2), and E6–E11 (each n = 1). Among the 26 isolates of tpi, 5 were identified as assemblage A sequences (A1–A5, each n = 1) and 21 were identified as 20 assemblage E sequences (E1–E3 and E5–E20, each n = 1, and E4 [n = 2]). In addition, there were 40 isolates of bg, with 2 being identified as a single assemblage A sequence (A1, n = 2) and 38 as 16 assemblage E sequences [E1 (n = 4), E2 (n = 3), E3 (n = 4), E4 (n = 8), E5 (n = 1), E6 (n = 1), E7 (n = 8), and E8–E16 (each n = 1)] (Table 2). Through the sequence alignment of the above four genes, the genotyping results of the two samples at different gene sites differed and mixed infection was identified (Assemblages A + E).
In a phylogenetic tree constructed based on the gdh (Fig. 2), tpi (Fig. 3), and bg (Fig. 4) sequences of G. duodenalis, these three genes were divided into two branches (assemblages A and E). The sequences of gdh, tpi, and bg were successfully obtained from 11 isolates. These three genes of the 11 isolates were combined for genotyping, resulting in 10 different assemblage E MLGs and 1 assemblage A MLG. Phylogenetic analysis revealed that the assemblage E MLGs formed five subgroups (Fig. 5).
Phylogenetic tree of Giardia duodenalis based on gdh sequences. The phylogenetic tree was inferred via neighbor-joining analysis of genetic distances calculated using the Kimura two-parameter model. Bootstrap values > 50% from 1000 replicates are shown to the left of the nodes. The isolates marked with black triangles (▲) and black squares (■) represent assemblages E and A identified from cattle in this study, respectively
Phylogenetic tree of Giardia duodenalis based on tpi sequences. The phylogenetic tree was inferred via neighbor-joining analysis of genetic distances calculated using the Kimura two-parameter model. Bootstrap values > 50% from 1000 replicates are shown to the left of the nodes. Assemblages E and A isolates identified from cattle in this study are marked with black triangles (▲) and black squares (■), respectively
Phylogenetic tree of Giardia duodenalis based on bg sequences. The phylogenetic tree was inferred via neighbor-joining analysis of genetic distances calculated via the Kimura two-parameter model. Bootstrap values > 50% from 1000 replicates are shown to the left of the nodes. The black triangles (▲) and black squares (■) represent assemblages E and A identified from cattle in this study, respectively
Phylogenetic relationship among Giardia duodenalis assemblage E MLGs. The phylogenetic tree was constructed using a concatenated dataset of bg, tpi, and gdh sequences, and analysis via the neighbor-joining method resulted in identical topologies. Black triangles (▲) and black squares (■) represent assemblages MLGE and MLGA identified from cattle in this study, respectively
Discussion
Cryptosporidium spp., E. bieneusi, and G. duodenalis are three opportunistic pathogens that infect humans and animals (Wang et al. 2022b). To date, numerous studies worldwide have reported Cryptosporidium (Zhang et al. 2022), E. bieneusi (Ruan et al. 2021; Taghipour et al. 2022a), and G. duodenalis (Taghipour et al. 2022b) infections in cattle. In China, high incidences of cryptosporidiosis, E. bieneusi infection, and giardiasis have been reported in dairy cattle (Qiu et al. 2019; Ryan and Cacciò 2013; Santin 2020; Wang et al. 2017a, 2018). Cattle infected with these three protozoa are potential sources through which humans can acquire protozoiasis (Dong et al. 2022; Kiani-Salmi et al. 2019; Ruan et al. 2021; Ryan and Cacciò 2013; Zhang et al. 2015). In the present study, a molecular epidemiological survey of these three protozoa was performed on 320 fecal samples from dairy cattle at three intensive dairy farms in Ningxia, China. The results reconfirmed that dairy cattle in Ningxia are infected with these three protozoa (Cui et al. 2014; Huang et al. 2014; Li et al. 2016; Wang et al. 2023; Zhang et al. 2015, 2016) and further identified new protozoon genotypes. Previously, in Ningxia, only mixed infections involving Cryptosporidium and G. duodenalis were reported (Wang et al. 2023). However, the present study also found mixed infections involving Cryptosporidium, G. duodenalis, and E. bieneusis; Cryptosporidium and E. bieneusis; E. bieneusis and G. duodenalis; and Cryptosporidium and G. duodenalis as well as mixed infections involving different Cryptosporidium species and G. duodenalis assemblages A and E. In addition, mixed infections involving pairs of taxa alongside G. duodenalis assemblage mixed infections have been reported in various animals in other regions, and it is not uncommon both within China and abroad (Jiang et al. 2023; Wang et al. 2022a, 2022b; Yang et al. 2022). However, this phenomenon should be seriously considered and the mutual influence of the infectious agents and the impact of mixed infection on animals should be further studied.
Here, the overall infection rates of Cryptosporidium, E. bieneusi, and G. duodenalis in dairy cattle from Ningxia were 19.38% (62/320), 41.56% (133/320), and 29.38% (94/320), respectively. The infection rate of Cryptosporidium was lower than the previously reported global pooled infection rate (Hatam-Nahavandi et al. 2019) and higher than the Chinese pooled infection rate (Cai et al. 2019). Moreover, the infection rates of E. bieneusi (Qin et al. 2022; Qiu et al. 2019; Ruan et al. 2021; Taghipour et al. 2022a; Wang et al. 2018) and G. duodenalis (Taghipour et al. 2022b) were higher than the global and Chinese pooled infection rates. The infection rates of Cryptosporidium were higher than those previously mentioned in two reports on Ningxia dairy cattle, which were 5.45% (92/1688) (Zhang et al. 2015) and 1.68% (23/1366) (Huang et al. 2014); however, they were lower than those reported in two other studies on Ningxia cattle, which were 21% (53/252) (Cui et al. 2014) and 50.46% (111/220) (Wang et al. 2023). The infection rate of E. bieneusi was lower than that reported in a previous study on Ningxia dairy cattle, which was 46.8% (51/109) (Li et al. 2016). Furthermore, the infection rates of G. duodenalis were higher than those reported in three previous reports on Ningxia cattle, which were 2.12% (29/1366) (Huang et al. 2014), 4.38% (74/1688) (Zhang et al. 2016), and 13.64% (30/220) (Wang et al. 2023). However, it is difficult to compare the infection rate owing to the influence of various factors, including study design, diagnostic method, geographical conditions, climate, sanitation conditions, rearing conditions, total number of samples, age of animals, and sampling season (Gong et al. 2017; Qi et al. 2016; Hatam-Nahavandi et al. 2019). Thus, to prevent and control protozoa in Ningxia, there is an urgent need to clarify the infection rates among local animals.
Here, no significant differences were identified in the overall infection rates of the three protozoa between the groups with and without diarrhea. Moreover, all three protozoa-infected young cattle were present in the group without diarrhea and the Cryptosporidium-infected preweaned calves were found in the group without diarrhea. Infections with Cryptosporidium at farm 2 and Cryptosporidium and G. duodenalis at farm 3 also appeared exclusively in the group without diarrhea. The most common clinical manifestations of infection with these three intestinal protozoa include diarrhea and wasting (Qin et al. 2022; Wang et al. 2022a). In addition, some studies have reported a correlation among these three intestinal protozoa and cattle suffering from diarrhea in cattle (Cai et al. 2019; Qin et al. 2022; Taghipour et al. 2022b; Wang et al. 2017a). However, diarrhea is a common clinical symptom of various diseases, with causative agents including bacteria, viruses, and parasites, or other possible factors (Ichikawa-Seki et al. 2015); hence, it is difficult to evaluate the impact of the three protozoa on diarrhea.
In addition, the overall infection rate of Cryptosporidium and E. bieneusi was significantly higher in the preweaned calves, postweaned calves, and young cattle than in adult cattle. G. duodenalis exhibited the highest overall prevalence in young cattle, which differed significantly compared with that in the preweaned calves. The results of this study show that with increasing age, the infection rate of the three protozoa first increased and then decreased, with the inflection point occurring at the postweaned calf or young cattle stage. Several studies have reported that the infection rate of the three protozoa decreases with increasing age (Qin et al. 2022; Wang et al. 2017a, 2018, 2019). Some studies have also reported a high infection rate in the postweaned calves (Fan et al. 2021; Gillhuber et al. 2014; Hamnes et al. 2006; Liu et al. 2009; Ma et al. 2021; Qi et al. 2015, 2016; Trout et al. 2004, 2005; Zhao et al. 2013); however, many reports have described a high infection rate in the preweaned calves (Feng et al. 2007; Geurden et al. 2012; Hamnes et al. 2006; Heyworth 2016; Jia et al. 2022; Ma et al. 2015; Mravcová et al. 2020; Shrivastava et al. 2017; Sulaiman et al. 2005; Trout et al. 2004, 2005; Wang et al. 2018; Yasouri et al. 2020; Zahedi et al. 2017; Zhao et al. 2013). Moreover, here, the three intensive dairy farms demonstrated different infection rates of the three protozoa. As mentioned earlier, several factors affect the infection rate, regional and age distribution, and occurrence of diarrhea in samples infected with the three different protozoa. Thus, there is a need for a more comprehensive understanding of such factors.
Herein, four Cryptosporidium species were identified at all three intensive dairy farms, consistent with the four dominant species in cattle reported within China and (Li et al. 2022a) abroad (Feng et al. 2018; Gong et al. 2017; Hatam-Nahavandi et al. 2019; Santin 2020; Wang et al. 2017a; Zahedi and Ryan 2020; Zhang et al. 2022). These four Cryptosporidium species were previously isolated from dairy cattle in Ningxia (Zhang et al. 2015). The dominant species in this study was C. bovis; however, this is inconsistent with the finding that C. andersoni was the most common species in China (Gong et al. 2017). In this study, C. bovis infection was primarily detected in postweaned calves; however, other reports have described that C. bovis infection is dominant in preweaned calves in China (Feng and Xiao 2017; Gong et al. 2017; Guo et al. 2022; Wang et al. 2017a). In addition, other previous reports have described that C. bovis infection is very common in postweaned calves (Li et al. 2019a). In the present study, the preweaned calves were mainly infected with C. parvum, consistent with previous findings (Gong et al. 2017; Li et al. 2022a). In addition, young cattle were most commonly infected with C. bovis, and the majority of adult cattle were infected with C. andersoni and C. bovis. This is inconsistent with the finding that C. andersoni was the dominant species among postweaned, juvenile, and adult cattle (Gong et al. 2017).
To the best of our knowledge, this study is the first to report mixed infection with two or three Cryptosporidium species in dairy cattle in Ningxia. The infection type is consistent with the appearance of C. bovis + C. ryanae in Guangdong (Liang et al. 2019), Shanghai (Cai et al. 2017), Xinjiang (Alves et al. 2003; Qi et al. 2020), and Henan (Wang et al. 2010) in China, as well as C. parvum + C. ryanae in Xinjiang (Wu et al. 2020) and Henan (Wang et al. 2010), and also shows C. parvum + C. bovis + C. andersoni. Multiple mixed infections involving Cryptosporidium spp. have also been detected in cattle from other countries (Cai et al. 2019; Gong et al. 2017; Hatam-Nahavandi et al. 2019; Wang et al. 2017a), and humans have also been reported to exhibit mixed infection involving the protozoan species (Helmy et al. 2013). C. parvum is reportedly the chief causative agent of zoonotic cryptosporidiosis (Feng et al. 2018; Guo et al. 2022; Zhang et al. 2022), and IIa and IId C. parvum gp60 subtype families are known to be zoonotically transmitted (Feng et al. 2018; Jia et al. 2022; Li et al. 2022a; Mravcová et al. 2020). Herein, the main C. parvum subtype was found to be IIdA15G1; the IIdA20G1 subtype was also identified. Reportedly, in China, C. parvum infections are entirely caused by IId subtypes in cattle, with IIdA15G1 and IIdA19G1 being the most common subtypes (Guo et al. 2022; Li et al. 2022a; Zahedi et al. 2017; Zhang et al. 2022). In addition, IIdA15G1 (Cui et al. 2014; Huang et al. 2014; Zhang et al. 2015) was previously reported in dairy cattle in Ningxia; however, IIdA20G1 is reported here for the first time in Ningxia to the best of our knowledge. This subtype was previously only reported in Xinjiang (Wu et al. 2020) and Heilongjiang (Tao et al. 2018) in China.
Regarding E. bieneusis, the genotypes identified in this study were J (66.17%), BEB4 (24.81%), and I (3.01%) along with new genotypes NX1–NX8 (all 0.75%), which belong to E. bieneusis group 2. Reportedly, the genotypes BEB4, J, and I are most common in E. bieneusis globally (Lichtmannsperger et al. 2023; Taghipour et al. 2022b). In China, the most common genotypes of cattle are I and J (Wang et al. 2018). Furthermore, BEB4, BEB6, I, and J in group 2 were described as the main genotypes associated with infections in humans and animals (Qin et al. 2022; Wang et al. 2022b). In addition, E. bieneusis infection has been reported in at least 42 countries globally (Qin et al. 2022; Ruan et al. 2021). Cattle are an important vector for the transmission of E. bieneusis (Qin et al. 2022). A previous study reported the infection of dairy cattle with E. bieneusis group 2 (genotypes J and I) and group 1 (genotypes EbpA, CM8, O, CHC5, CHC4, and CHC3) in Ningxia (Alves et al. 2003). Group 1 is a zoonotic evolutionary group (Li et al. 2022b; Lichtmannsperger et al. 2023; Wang et al. 2022b). Herein, BEB4 and NX1–NX8 were the genotypes that were newly discovered in Ningxia and have not been reported in other animals in this region (Dong et al. 2022; Peng et al. 2019, 2020; Yang et al. 2018; Zhang et al. 2020). The obtained findings reveal more and more genotypes of E. bieneusis infecting cattle in Ningxia, which is associated with an increased risk of dairy cattle transmitting E. bieneusis to humans in this region.
Through the SSU rRNA gene analysis, we found that dairy cattle were infected with G. duodenalis assemblages E (81.52%), A (16.30%), and B (2.17%) in Ningxia. As reported in the literature, assemblage E is the most common genotype in dairy cattle worldwide (Cai et al. 2021; Feng and Xiao 2011; Minetti et al. 2014; Qi et al. 2016; Taghipour et al. 2022b; Zhang et al. 2021) followed by assemblages A and B (Feng and Xiao 2011; Qi et al. 2016; Zhang et al. 2021). Combined sequence analysis of the gdh, tpi and bg genes in G. duodenalis revealed the genetic diversity of these isolates and the presence of mixed infection with assemblages A + E, (Qi et al. 2016; Scheuerle et al. 2013).
Assemblages A and B are reportedly associated with a higher risk of zoonotic diseases (Kiani-Salmi et al. 2019; Taghipour et al. 2022b; Zhang et al. 2021), with assemblage E also being reported in human cases in Bangladesh (Li et al. 2023), Egypt (Abdel-Moein and Saeed 2016), Brazil (Fantinatti et al. 2016), and Australia (Zahedi et al. 2017). In the literature, there are reports regarding dairy cattle being infected with G. duodenalis assemblages A, B, and E in Ningxia (Huang et al. 2014; Zhang et al. 2016). This highlights the risk of G. duodenalis spreading to humans from dairy cattle in Ningxia. Indeed, dairy cattle are considered a significant reservoir of human giardiasis in China (Feng and Xiao 2017). Herein, assemblage E was identified in all four cattle age groups, while only assemblage E was present in the preweaned calves. This is inconsistent with the findings of previous studies, which reported that the infection rates of the calves with assemblages A and B were higher than those with assemblage E (Dixon et al. 2011; Geurden et al. 2008; Huang et al. 2014; Ng et al. 2011; Qi et al. 2016). However, our findings are consistent with those of a report from Sichuan, China, reporting that only assemblage E was detected in the preweaned calves (Zhong et al. 2018). The characteristics of the individual loci of G. duodenalis often lead to inconsistent genotyping results (Feng and Xiao 2011). Based on multilocus genotyping, the MLG model can be used to obtain a better understanding of the diversity of G. duodenalis in humans and animals in different geographic regions, which can help reveal the potential and dynamic transmission of zoonotic giardiasis (Huey et al. 2013). Herein, the gdh, tpi, and bg genes obtained from 11 isolates were combined to form 10 different assemblage E MLGs (5 subgroups) and 1 assemblage A MLG. Consistent with the findings of previous studies, G. duodenalis isolates classified into the same assemblage may be classified as distinct MLGs (Qi et al. 2016; Wang et al. 2017b; Zhang et al. 2016; Zhao et al. 2015).
Conclusions
This study reconfirmed that dairy cattle are infected with Cryptosporidium, G. duodenalis and E. bieneusis. Furthermore, to the best of our knowledge, this study is the first to report mixed infections involving two or three Cryptosporidium species in dairy cattle. The subtype IIdA20G1 of C. parvum was also reported here for the first time in Ningxia, and nine genotypes of E. bieneusis and new features of G. duodenalis assemblages were also discovered. Zoonotic potential and diverse genotypes and assemblages of the three protozoa significantly elevate the risk of cryptosporidiosis, microsporidiosis, and giardiasis among local people and animals in Ningxia. Cattle are important reservoirs of these three protozoa, and in countries with a large population of cattle, more attention should be paid to preventing and controlling these pathogens.
Data availability
No datasets were generated or analysed during the current study.
Abbreviations
- SSU rRNA :
-
Small subunit ribosomal RNA
- ITS :
-
Internal transcribed spacer
- gp60:
-
60 KDa glycoprotein
- bg :
-
Beta-giardin
- gdh :
-
Glutamate dehydrogenase
- tpi :
-
Triosephosphate isomerase
- PCR :
-
Polymerase chain reaction
- RFLP :
-
Restriction fragmentlength polymorphism
- MLG :
-
Multilocus genotype
References
Abdel-Moein KA, Saeed H (2016) The zoonotic potential of Giardia intestinalis assemblage E in rural settings. Parasitol Res 115:3197–3202
Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F (2003) Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J Clin Microbiol 41(6):2744–2747
Appelbee A, Frederick L, Heitman T, Olson M (2003) Prevalence and genotyping of Giardia duodenalis from beef calves in Alberta, Canada. Vet Parasitol 112(4):289–294
Buckholt MA, Lee JH, Tzipori S (2002) Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl Environ Microbiol 68(5):2595–2599
Cacciò S, Beck R, Lalle M, Marinculic A, Pozio E (2008) Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int J Parasitol 38(13):1523–1531
Cai M, Guo Y, Pan B, Li N, Wang X, Tang C, Feng Y, Xiao L (2017) Longitudinal monitoring of Cryptosporidium species in pre-weaned dairy calves on five farms in Shanghai, China. Vet Parasitol 241:14–19
Cai Y, Zhang N, Gong Q, Zhao Q, Zhang X (2019) Prevalence of Cryptosporidium in dairy cattle in China during 2008–2018: A systematic review and meta-analysis. Microb Pathog 132:193–200
Cai W, Ryan U, Xiao L, Feng Y (2021) Zoonotic giardiasis: an update. Parasitol Res 120(12):4199–4218
Cardona GA, de Lucio A, Bailo B, Cano L, de Fuentes I, Carmena D (2015) Unexpected finding of feline-specific Giardia duodenalis assemblage F and Cryptosporidium felis in asymptomatic adult cattle in Northern Spain. Vet Parasitol 209(3–4):258–263
Cui Z, Wang R, Huang J, Wang H, Zhao J, Luo N, Li J, Zhang Z, Zhang L (2014) Cryptosporidiosis caused by Cryptosporidium parvum subtype IIdA15G1 at a dairy farm in Northwestern China. Parasit Vectors 7(1):1–4
Dixon B, Parrington L, Cook A, Pintar K, Pollari F, Kelton D, Farber J (2011) The potential for zoonotic transmission of Giardia duodenalis and Cryptosporidium spp. from beef and dairy cattle in Ontario, Canada. Vet Parasitol 175(1–2):20–26
Dong H, Zhao Z, Zhao J, Fu Y, Lang J, Zhang J, Liang G, Zhang L, Li J, Zhao G (2022) Molecular characterization and zoonotic potential of Enterocytozoon bieneusi in ruminants in northwest China. Acta Trop 234:106622
Fan Y, Wang X, Yang R, Zhao W, Li N, Guo Y, Xiao L, Feng Y (2021) Molecular characterization of the waterborne pathogens Cryptosporidium spp., Giardia duodenalis, Enterocytozoon bieneusi, Cyclospora cayetanensis and Eimeria spp. in wastewater and sewage in Guangzhou, China. Parasit Vectors 14:1–10
Fantinatti M, Bello AR, Fernandes O, Da-Cruz AM (2016) Identification of Giardia lamblia assemblage E in humans points to a new anthropozoonotic cycle. J Infect Dis 214(8):1256–1259
Feng Y, Xiao L (2011) Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev 24(1):110–140
Feng Y, Xiao L (2017) Molecular epidemiology of cryptosporidiosis in China. Front Microbiol 8:1701
Feng Y, Ortega Y, He G, Das P, Xu M, Zhang X, Fayer R, Gatei W, Cama V, Xiao L (2007) Wide geographic distribution of Cryptosporidium bovis and the deer-like genotype in bovines. Vet Parasitol 144(1–2):1–9
Feng Y, Ryan UM, Xiao L (2018) Genetic diversity and population structure of Cryptosporidium. Trends Parasitol 34(11):997–1011
Geurden T, Geldhof P, Levecke B, Martens C, Berkvens D, Casaert S, Vercruysse J, Claerebout E (2008) Mixed Giardia duodenalis assemblage A and E infections in calves. Int J Parasitol 38(2):259–264
Geurden T, Vanderstichel R, Pohle H, Ehsan A, von Samson-Himmelstjerna G, Morgan E, Camuset P, Capelli G, Vercruysse J, Claerebout E (2012) A multicentre prevalence study in Europe on Giardia duodenalis in calves, with molecular identification and risk factor analysis. Vet Parasitol 190(3–4):383–390
Gillhuber J, Rügamer D, Pfister K, Scheuerle MC (2014) Giardiosis and other enteropathogenic infections: a study on diarrhoeic calves in Southern Germany. BMC Res Notes 7:1–9
Gong C, Cao X, Deng L, Li W, Huang X, Lan J, Xiao Q, Zhong Z, Feng F, Zhang Y (2017) Epidemiology of Cryptosporidium infection in cattle in China: a review. Parasite 24:1
Guo Y, Ryan U, Feng Y, Xiao L (2022) Emergence of zoonotic Cryptosporidium parvum in China. Trends Parasitol 38(4):335–343
Hamnes IS, Gjerde B, Robertson L (2006) Prevalence of Giardia and Cryptosporidium in dairy calves in three areas of Norway. Vet Parasitol 140(3–4):204–216
Hatam-Nahavandi K, Ahmadpour E, Carmena D, Spotin A, Bangoura B, Xiao L (2019) Cryptosporidium infections in terrestrial ungulates with focus on livestock: a systematic review and meta-analysis. Parasit Vectors 12:1–23
Helmy YA, Krücken J, Nöckler K, von Samson-Himmelstjerna G, Zessin K-H (2013) Molecular epidemiology of Cryptosporidium in livestock animals and humans in the Ismailia province of Egypt. Vet Parasitol 193(1–3):15–24
Heyworth MF (2016) Giardia duodenalis genetic assemblages and hosts. Parasite 23:13
Huang J, Yue D, Qi M, Wang R, Zhao J, Li J, Shi K, Wang M, Zhang L (2014) Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalisin dairy cattle in Ningxia, northwestern China. BMC Vet Res 10(1):1–5
Huey CS, Mahdy MA, Al-Mekhlafi HM, Nasr NA, Lim YA, Mahmud R, Surin J (2013) Multilocus genotyping of Giardia duodenalis in Malaysia. Infect Genet Evol 17:269–276
Ichikawa-Seki M, Aita J, Masatani T, Suzuki M, Nitta Y, Tamayose G, Iso T, Suganuma K, Fujiwara T, Matsuyama K (2015) Molecular characterization of Cryptosporidium parvum from two different Japanese prefectures, Okinawa and Hokkaido. Parasitol Int 64(2):161–166
Jia R, Huang W, Huang N, Yu Z, Li N, Xiao L, Feng Y, Guo Y (2022) High infectivity and unique genomic sequence characteristics of Cryptosporidium parvum in China. PLoS Negl Trop Dis 16(8):e0010714
Jiang Y, Liu L, Yuan Z, Liu A, Cao J, Shen Y (2023) Molecular identification and genetic characteristics of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in human immunodeficiency virus/acquired immunodeficiency syndrome patients in Shanghai, China. Parasit Vectors 16(1):53
Kiani-Salmi N, Fattahi-Bafghi A, Astani A, Sazmand A, Zahedi A, Firoozi Z, Ebrahimi B, Dehghani-Tafti A, Ryan U, Akrami-Mohajeri F (2019) Molecular typing of Giardia duodenalis in cattle, sheep and goats in an arid area of central Iran. Infect Genet Evol 75:104021
Lalle M, Pozio E, Capelli G, Bruschi F, Crotti D, Cacciò SM (2005) Genetic heterogeneity at the β-giardin locus among human and animal isolates of Giardia duodenalis and identification of potentially zoonotic subgenotypes. Int J Parasitol 35(2):207–213
Li J, Luo N, Wang C, Qi M, Cao J, Cui Z, Huang J, Wang R, Zhang L (2016) Occurrence, molecular characterization and predominant genotypes of Enterocytozoon bieneusi in dairy cattle in Henan and Ningxia, China. Parasit Vectors 9:1–5
Li N, Wang R, Cai M, Jiang W, Feng Y, Xiao L (2019a) Outbreak of cryptosporidiosis due to Cryptosporidium parvum subtype IIdA19G1 in neonatal calves on a dairy farm in China. Int J Parasitol 49(7):569–577
Li W, Feng Y, Santin M (2019b) Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol 35(6):436–451
Li N, Zhao W, Song S, Ye H, Chu W, Guo Y, Feng Y, Xiao L (2022a) Diarrhoea outbreak caused by coinfections of Cryptosporidium parvum subtype IIdA20G1 and rotavirus in pre-weaned dairy calves. Transbound Emerg Dis 69(5):e1606–e1617
Li W, Feng Y, Xiao L (2022b) Enterocytozoon bieneusi. Trends Parasitol 38(1):95–96
Li J, Karim MR, Siddiki SFMF, Chen Y, Qin Z, Rume FI, Zhang L (2023) Potential Zoonotic Transmission of Giardia duodenalis between Children and Calves in Bangladesh. Transbound Emerg Dis 2023:8224587
Liang N, Wu Y, Sun M, Chang Y, Lin X, Yu L, Hu S, Zhang X, Zheng S, Cui Z (2019) Molecular epidemiology of Cryptosporidium spp. in dairy cattle in Guangdong Province, South China. Parasitology 146(1):28–32
Lichtmannsperger K, Harl J, Roehl SR, Schoiswohl J, Eibl C, Wittek T, Hinney B, Wiedermann S, Joachim A (2023) Enterocytozoon bieneusi in fecal samples from calves and cows in Austria. Parasitol Res 122(1):333–340
Liu A, Wang R, Li Y, Zhang L, Shu J, Zhang W, Feng Y, Xiao L, Ling H (2009) Prevalence and distribution of Cryptosporidium spp. in dairy cattle in Heilongjiang Province, China. Parasitol Res 105(3):797–802
Ma J, Li P, Zhao X, Xu H, Wu W, Wang Y, Guo Y, Wang L, Feng Y, Xiao L (2015) Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Vet Parasitol 207(3–4):220–227
Ma N, Wang H-X, Tao W-F, Xue N-Y, Bai J-Y, Zhao Q, Jiang J, Lyu C (2021) Detection of Point Prevalence and Assemblages of Giardia spp. in Dairy Calves and Sika Deer, Northeast China. Vector Borne Zoonotic Dis 21(9):685–691
Minetti C, Taweenan W, Hogg R, Featherstone C, Randle N, Latham S, Wastling J (2014) Occurrence and diversity of Giardia duodenalis assemblages in livestock in the UK. Transbound Emerg Dis 61(6):e60–e67
Mravcová K, Štrkolcová G, Mucha R, Barbušinová E, Goldová M, Kačírová J, Maďar M (2020) Cryptosporidium parvum–zoonotic subtype IIdA15G1 in a Slovakian patient. Ann Agric Environ Med 27(3):485–488
Ng J, Yang R, McCarthy S, Gordon C, Hijjawi N, Ryan U (2011) Molecular characterization of Cryptosporidium and Giardia in pre-weaned calves in Western Australia and New South Wales. Vet Parasitol 176(2–3):145–150
Peng J, Zou Y, Li Z, Liang Q, Song H, Li T, Ma Y, Zhu X, Zhou D (2019) Occurrence of Enterocytozoon bieneusi in Chinese Tan sheep in the Ningxia Hui autonomous region, China. Parasitol Res 118:2729–2734
Peng J, Zou Y, Li Z, Liang Q, Song H, Li T, Ma Y, Zhu X, Zhou D-H (2020) Prevalence and multilocus genotyping of Giardia duodenalis in Tan sheep (Ovis aries) in northwestern China. Parasitol Int 77:102126
Qi M, Wang H, Jing B, Wang D, Wang R, Zhang L (2015) Occurrence and molecular identification of Cryptosporidium spp. in dairy calves in Xinjiang, Northwestern China. Vet Parasitol 212(3–4):404–407
Qi M, Wang H, Jing B, Wang R, Jian FC, Ning CS, Zhang LX (2016) Prevalence and multilocus genotyping of Giardia duodenalis in dairy calves in Xinjiang, Northwestern China. Parasit Vectors 9(1):546
Qi M, Zhang K, Huang M, Wang S, Xu C, Wang T, Jing B, Li J (2020) Longitudinal detection of Cryptosporidium spp. in 1–10-week-old dairy calves on a farm in Xinjiang, China. Parasitol Res 119(11):3839–3844
Qin Y, Chen C, Qin Y, Yang X, Li M, Meng X, Zhao Z, Ma N, Cai Y, Zhang Y (2022) Prevalence and related factors of Enterocytozoon bieneusi in cattle: a global systematic review and meta-analysis. Prev Vet Med 208:105775
Qiu L, Xia W, Li W, Ping J, Ding S, Liu H (2019) The prevalence of microsporidia in China: a systematic review and meta-analysis. Sci Rep 9(1):3174
Ruan Y, Xu X, He Q, Li L, Guo J, Bao J, Pan G, Li T, Zhou Z (2021) The largest meta-analysis on the global prevalence of microsporidia in mammals, avian and water provides insights into the epidemic features of these ubiquitous pathogens. Parasit Vectors 14(1):1–14
Ryan U, Cacciò SM (2013) Zoonotic potential of Giardia. Int J Parasitol 43(12–13):943–956
Ryan U, Hijjawi N, Feng Y, Xiao L (2019) Giardia: an under-reported foodborne parasite. Int J Parasitol 49(1):1–11
Ryan U, Zahedi A, Feng Y, Xiao L (2021) An update on zoonotic Cryptosporidium species and genotypes in humans. Animals (Basel) 11(11):3307
Santin M (2020) Cryptosporidium and Giardia in ruminants. Vet Clin North Am Food Anim Pract 36(1):223–238
Santín M, Fayer R (2011) Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res Vet Sci 90(3):363–371
Scheuerle MC, Pfister K, Thompson RA, Ash A, Pallant L, Gillhuber J (2013) Molecular identification of zoonotic and livestock-specific Giardia-species in faecal samples of calves in Southern Germany. Parasit Vectors 6(1):346
Shrivastava AK, Kumar S, Smith WA, Sahu PS (2017) Revisiting the global problem of cryptosporidiosis and recommendations. Trop Parasitol 7(1):8
Sulaiman IM, Fayer R, Bern C, Gilman RH, Trout JM, Schantz PM, Das P, Lal AA, Xiao L (2003) Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg Infect Dis 9(11):1444–1452
Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, Iqbal J, Khalid N, Xiao L (2005) Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol 43(6):2805–2809
Taghipour A, Bahadory S, Abdoli A (2022a) A systematic review and meta-analysis on the global prevalence of cattle microsporidiosis with focus on Enterocytozoon bieneusi: an emerging zoonotic pathogen. Prev Vet Med 200:105581
Taghipour A, Sharbatkhori M, Tohidi F, Ghanbari MR, Karanis P, Olfatifar M, Majidiani H, Khazaei S, Bahadory S, Javanmard E (2022b) Global prevalence of Giardia duodenalis in cattle: a systematic review and meta-analysis. Prev Vet Med 203:105632
Tao W, Li Y, Yang H, Song M, Lu Y, Li W (2018) Widespread occurrence of zoonotic Cryptosporidium species and subtypes in dairy cattle from northeast China: public health concerns. J Parasitol 104(1):10–17
Trout JM, Santín M, Greiner E, Fayer R (2004) Prevalence of Giardia duodenalis genotypes in pre-weaned dairy calves. Vet Parasitol 124(3–4):179–186
Trout JM, Santín M, Greiner E, Fayer R (2005) Prevalence and genotypes of Giardia duodenalis in post-weaned dairy calves. Vet Parasitol 130(3–4):177–183
Wang R, Wang H, Sun Y, Zhang L, Xiao L (2010) Characteristics of cryptosporidium transmission in preweaned dairy cattle in Henan, China. J Clin Microbiol 49(3):1077–1082
Wang R, Zhao G, Gong Y, Zhang L (2017a) Advances and perspectives on the epidemiology of Bovine Cryptosporidium in China in the past 30 years. Front Microbiol 8:1823
Wang X, Cai M, Jiang W, Wang Y, Jin Y, Li N, Guo Y, Feng Y, Xiao L (2017b) High genetic diversity of Giardia duodenalis assemblage E in pre-weaned dairy calves in Shanghai, China, revealed by multilocus genotyping. Parasitol Res 116:2101–2110
Wang S, Wang R, Fan X, Liu T, Zhang L, Zhao G (2018) Prevalence and genotypes of Enterocytozoon bieneusi in China. Acta Trop 183:142–152
Wang R, Li N, Jiang W, Guo Y, Wang X, Jin Y, Feng Y, Xiao L (2019) Infection patterns, clinical significance, and genetic characteristics of Enterocytozoon bieneusi and Giardia duodenalis in dairy cattle in Jiangsu, China. Parasitol Res 118(10):3053–3060
Wang N, Wang K, Liu Y, Zhang X, Zhao J, Zhang S, Zhang L (2022a) Molecular characterization of Cryptosporidium spp., Enterocytozoon bieneusi and Giardia duodenalis in laboratory rodents in China. Parasite 29:46
Wang P, Zheng L, Liu L, Yu F, Jian Y, Wang R, Zhang S, Zhang L, Ning C, Jian F (2022b) Genotyping of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi from sheep and goats in China. BMC Vet Res 18(1):361
Wang D, Gao H, Zhao L, Lv C, Dou W, Zhang X, Liu Y, Kang X, Guo K (2023) Detection of the dominant pathogens in diarrheal calves of Ningxia, China in 2021–2022. Front Vet Sci 10:1155061
Wu Y, Zhang K, Zhang Y, Jing B, Chen Y, Xu C, Wang T, Qi M, Zhang L (2020) Genetic diversity of Cryptosporidium parvum in neonatal dairy calves in Xinjiang, China. Pathogens 9(9):692
Xiao L, Feng Y (2017) Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol 8–9:14–32
Yang H, Mi R, Cheng L, Huang Y, An R, Zhang Y, Jia H, Zhang X, Wang X, Han X (2018) Prevalence and genetic diversity of Enterocytozoon bieneusi in sheep in China. Parasit Vectors 11(1):1–10
Yang F, Ma L, Gou J-M, Yao H-Z, Ren M, Yang B-K, Lin Q (2022) Seasonal distribution of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in Tibetan sheep in Qinghai, China. Parasit Vectors 15(1):394
Yasouri SR, Ghane M, Doudi M, Rezaei A, Naghavi NS (2020) A study of leptospirosis epidemiology in Iran and diagnostic techniques for human, livestock and environment samples. Med Lab Technol 14(6):1–9
Zahedi A, Ryan U (2020) Cryptosporidium–an update with an emphasis on foodborne and waterborne transmission. Res Vet Sci 132:500–512
Zahedi A, Field D, Ryan U (2017) Molecular typing of Giardia duodenalis in humans in Queensland–first report of Assemblage E. Parasitology 144(9):1154–1161
Zhang X, Tan Q, Zhou D, Ni X, Liu G, Yang Y, Zhu X (2015) Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle, northwest China. Parasitol Res 114(7):2781–2787
Zhang X, Tan Q, Zhao G, Ma J, Zheng W, Ni X, Zhao Q, Zhou D, Zhu X (2016) Prevalence, risk factors and multilocus genotyping of Giardia intestinalis in dairy cattle, Northwest China. J Eukaryot Microbiol 63(4):498–504
Zhang Y, Mi R, Yang J, Wang J, Gong H, Huang Y, Wang X, Han X, Zhou H, Chen Z (2020) Enterocytozoon bieneusi genotypes in farmed goats and sheep in Ningxia, China. Infect Genet Evol 85:104559
Zhang X, Dan J, Wang L, Liu H, Zhou Z, Ma X, Ren Z, Fu H, Geng Y, Luo Y (2021) High genetic diversity of Giardia duodenalis assemblage E in Chinese dairy cattle. Infect Genet Evol 92:104912
Zhang Z, Su D, Meng X, Liang R, Wang W, Li N, Guo Y, Guo A, Li S, Zhao Z (2022) Cryptosporidiosis outbreak caused by Cryptosporidium parvum subtype IIdA20G1 in neonatal calves. Transbound Emerg Dis 69(2):278–285
Zhao G, Ren W, Gao M, Bian Q, Hu B, Cong M, Lin Q, Wang R, Qi M, Qi MZ (2013) Genotyping Cryptosporidium andersoni in cattle in Shaanxi province, Northwestern China. Plos One 8(4):e60112
Zhao G, Du S, Wang H, Hu X, Deng M, Yu S, Zhang L, Zhu X (2015) First report of zoonotic Cryptosporidium spp., Giardia intestinalis and Enterocytozoon bieneusi in golden takins (Budorcas taxicolor bedfordi). Infect Genet Evol 34:394–401
Zhong Z, Dan J, Yan G, Tu R, Tian Y, Cao S, Shen L, Deng J, Yu S, Geng Y (2018) Occurrence and genotyping of Giardia duodenalis and Cryptosporidium in pre-weaned dairy calves in central Sichuan province, China. Parasite 25:45
Funding
This study was funded by the National Natural Science Foundation of China (32260887) and Supported by National Natural Science Foundation of Inner Mongolia (2022MS03023).
Author information
Authors and Affiliations
Contributions
MYW, SZ, ZSZ, LZ, and YHL conceived and designed the study and critically revised the manuscript. HLC, ZSZ, WXH, LZ, and YHL performed sample collection. MYW, SZ, ZSZ, XYQ, HLC, YW, WJF, CY, YLD and LZ conducted the laboratory experiments. All the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Declaration of generative AI in scientific writing
Not applicable.
Ethics approval
Our study was performed in strict accordance with the international standards published in the Guide to the Feeding, Management and Use of Experimental Animals (8th Edition) and followed the Regulations on the Management of Experimental Animals and other relevant laws and regulations. The study was approved by the Biomedical Research Ethics Committee of Inner Mongolia Agricultural University (approval no. 2020 [081]). Additionally, permission was obtained from the farm owners prior to specimen collection, and all efforts were made to minimize animal suffering.
Competing interests
The authors declare no competing interests.
Additional headings
All authors consent to participate and consent to publish.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, MY., Zhang, S., Zhang, ZS. et al. Prevalence and molecular characterization of Cryptosporidium spp., Enterocytozoon bieneusi, and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. Vet Res Commun 48, 2629–2643 (2024). https://doi.org/10.1007/s11259-024-10364-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11259-024-10364-6