Abstract
Two new copper(II) complexes, [Cu(Hhida)(BenzImH)]·H2O (1) and [Cu(Hhida)(EtImH)]·2H2O (2) have been synthesized by the interaction of basic copper carbonate with N-(2-hydroxyethyl)-2-aminodiacetic acid (Hhida2−) and benzimidazole (BenzImH) or (2-ethylimidazole (EtImH)). The structures of both complexes were studied by single crystal X-ray diffraction. The geometry around the copper(II) atom can be best described as distorted square pyramidal as indicated by the value of the trigonal index τ = 0.32 for (1) and 0.21 for (2). Spectroscopic and electrochemical studies were performed in order to correlate structural features of the complexes with their superoxide scavenger activity. The ability of the complexes to scavenge superoxide anions was also evaluated.
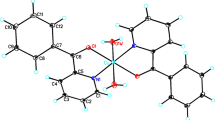
Graphical Abstract
Two new copper(II) complexes [Cu(Hhida)(BenzImH)]·H2O (1) and [Cu(Hhida)(EtImH)]·2H2O (2) have been synthesized by the interaction of basic copper carbonate with N-(2-hydroxyethyl)-2-aminodiacetic acid (Hhida2−) and benzimidazole(BenzImH) (2-ethylimidazole(EtImH)). Spectroscopic and electrochemical studies were performed in order to correlate structural feature of the complexes (Figs. 1 and 2) with their superoxide scavenger activity.


Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Synthetic superoxide dismutase (SOD) mimetics have emerged as a potential novel class of drugs for the treatment of oxidative stress–related diseases. Among these agents, metal complexes with flexible ligands constitute an important group. The synthesis of low-molecular weight copper(II) complexes [1] showing SOD activity has been challenging for bio-inorganic chemists and for many years efforts have been made to obtain compounds with high catalytic activity [2]. Important requirements for SOD-like activity are medium strength donor power and flexibility of the ligand, in order to facilitate the reduction and the accommodation of copper(I), which is known to prefer tetrahedral or linear environments. McCard and Fridovich [3] found that the SOD enzyme might protect cells as a scavenger of superoxide anion free radicals by transferring the O2 − into O2 and H2O2 through a redox reaction.
Several copper complexes with N-heterocyclic ligands such as benzimidazole, imidazole and thiazole have been reported. Some of them have shown significant cytotoxic activity [4]. Imidazole is of considerable interest as a ligand due to its presence in many biological systems. Imidazole also forms a number of inorganic materials with interesting structural and magnetic properties [5]. It is generally a monodentate ligand that forms complexes through its tertiary N atom [6].
The N-(2-hydroxyethyl)-2-aminodiacetic acid (Hhida2−) ligand is generally tetradentate and chelates the metal atom through 3O + N atoms (Scheme 1). Complexes of Hhida2− have some significantly different properties compared to those of the related ligands nta (nitrilotriacetate), pmida (N-(2-pyridylmethyl)iminodiacetate) and S-peida (N-(S)-[1-(2-pyridyl)ethyl]iminodiacetate). Structurally and magnetically, the solid state properties of complexes of Hhida2− are similar to those of nta complexes [7]. In solution, the Hhida2− deviates from the analogous complexes with respect to UV–Vis, EPR and NMR properties.
To our knowledge, there are no examples of structurally characterized copper(II) complexes containing these ligands. In this paper, we describe the syntheses and solid state and solution properties of copper complexes with the ligand H3hida and imidazole derivatives. We report also the preparation, crystal structure and superoxide dismutase activity of two new copper(II) complexes with Hhida2− and benzimidazole or 2-ethylimidazole.
Experimental
Copper carbonate was purchased from Acros Chemicals. All other chemicals were of synthetic grade and used without further purification. Elemental analyses were performed on an Elementar Vario EL III Carlo Erba 1108 analyser. FAB mass spectra were recorded on a JEOL SX 102/DA 6000 Mass Spectrometer using argon/xenon (6 kV, 10 mA) as the FAB gas. The accelerating voltage was 10 kV and the spectra were recorded at room temperature. UV–Vis spectra were recorded at 25 °C on a Shimadzu UV–visible recording spectrophotometer UV-160 in quartz cells. IR spectra were recorded in KBr medium on a Perkin–Elmer 783 spectrophotometer in the 4,000–400 cm−1 region. Cyclic voltammetry was carried out with a BAS-100 Epsilon electrochemical analyzer having an electrochemical cell with a three-electrode system. Ag/AgCl was used as a reference electrode, glassy carbon as working electrode and platinum wire as an auxiliary. 0.1 M NaClO4 was used as supporting electrolyte and DMSO as solvent. All measurements were taken at 298 K under a nitrogen atmosphere. Molar conductivities of freshly prepared 2 × 10−3 of DMSO solutions were measured on a Systronics conductivity TDS meter 308. Room temperature magnetic susceptibilities were measured with a Gouy balance using mercury(II) tetrathiocynato cobaltate(II) as calibrating agent (χg = 16.44 × 10−6 c.g.s. units). X-band epr spectra were recorded on a Varian E-line Century Series Spectrometer equipped with a dual cavity and operating at the X-band (~9.4 GHz) with 100 kHz modulation frequency. TCNE was used as field marker.
Synthesis of [Cu(Hhida)(BenzImH)].H2O 1 and [Cu(Hhida)(EtImH)].2H2O 2
A solid mixture of copper carbonate (CuCO3·Cu(OH)2) (0.1 g, 0.5 mmol) and H3hida (N-2-hydroxyethyl)-2-iminodiacetic acid (0.2 g, 1.0 mmol) was added to water (50 ml). The reaction mixture was stirred for 2 h. An aqueous solution (10 ml) of benzimidazole (0.1 g, 1.0 mmol) was then added dropwise and stirring was continued for 2 h. The resulting blue solution was filtered to remove undissolved solids. After 2–3 months, blue crystals of 1 were collected by filtration and washed with MeOH. These were dried in air at room temperature and stored in a CaCl2 desiccator. Yield: 0.3 g (80%) Anal. Found for C26H28CuN3O7.: C, 41.8; H, 3.7; N, 11.2%. Calcd.: C, 41.9; H,3.8; N,11.3%. Complex 2 was synthesized in a similar manner to that of the benzimidazole complex by employing EtImH in place of BenzImH. In the case of complex 2, blue crystals were also collected. Anal. Found for: C11H15CuN3O7: C,36.2; H,4.2; N,11.5%. Calcd.: C, 36.1; H, 4.1; N, 11.4%.
Crystal structure determination
Crystals suitable for single crystal X-ray analysis for both complexes were grown by slow evaporation of the reaction mixtures at room temperature. Single crystals suitable for single crystal X-ray analysis were mounted on glass fibers and used for data collection. Crystal data were collected on an Enraf–Nonius MACH3 diffractometer using graphite monochromatized MoKα radiation (λ = 0.71073 Å). The crystal orientation, cell refinement and intensity measurements were made using the program CAD-4PC performing ψ-scan measurements. The structures were solved by direct methods using the program SHELXS-97 [8] and refined by full-matrix least-square techniques against F 2 using SHELXL-97 [9]. All non-hydrogen atoms were refined anisotropically. All the hydrogen atoms were geometrically fixed and allowed to refine using a riding model.
SOD activity
The in vitro SOD activity was measured using alkaline DMSO as a source of superoxide radical (O2 −) and nitro blue tetrazolium chloride (NBT) as O2 − scavenger [10]. In general, 400 μl of the sample to be assayed was added to a solution containing 2.1 ml of 0.2 M potassium phosphate buffer (pH 8.6) and 1.0 ml of 56 μM of alkaline DMSO solution was added while stirring. The absorbance was then monitored at 540 nm against a sample prepared under similar conditions except that NaOH was absent from the DMSO. A unit of superoxide dismutase (SOD) activity is taken as the concentration of complex that causes 50% inhibition of alkaline DMSO mediated reduction of nitrobluetetrazolium chloride (NBT).
Results and discussion
Both complexes were prepared according to Scheme 1.
Both complexes 1 and 2 are blue, stable in air and soluble in water and most organic solvents. They were initially characterized by microanalysis and FAB+ mass spectrometry and then by X-ray crystallography.
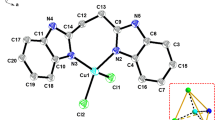
The molecule of complex 1 crystallizes with two monomers per asymmetric unit in the monoclinic crystal system. As seen from Table 1 complex 1 crystallizes as very low quality crystals. The ORTEP [11] view of the complex is shown in Fig. 1. The main bond angles and distances are presented in Table 2. The H3eida ligand is coordinated to the copper(II) by three oxygen atoms and one nitrogen atom and the five-coordinated sphere of Cu(II) is completed by the tertiary N atom of benzimidazole. The imidazolyl Cu–N bond distances in both complexes 1 and 2 are in the range found for similar benzimidazole and imidazolated complexes [12].
The structure of complex 1 consists of two crystallographically independent but chemically equivalent [Cu(Hhida)(BenzImH)]·H2O molecules. The copper(II) center in complex 1 has a distorted square pyramidal geometry and O(1), N(2), O(3) and N(3) occupy one basal plane [O(3)–Cu(2)–O(1) = 158.4(5)°, N(2)–Cu(2)–N(3) = 178.0(5)°]. The Cu–N distances are almost equal and are consistent with the distances usually found for square pyramidal Cu(II) compounds with nitrogen donor ligands. The angles around the copper atom defining the square basal plane N(2)–Cu(2)–O(5) = 99.1(5), O(5)–Cu(2)–N(3) = 81.3(5), O(5)–Cu(2)–O(3) = 95.4(5), O(3)–Cu(2)–N(3) = 84.8(5), N(3)–Cu(2)–O(1) = 83.9(5) and O(1)–Cu(2)–N2) = 94.1(5)° are in the expected range and similar in pairs. All angles around the copper deviate from 90° revealing the distortion of the basal plane. The trans bond angles deviate from the expected 180°: O(3)–Cu(2)–O(1) = 158.4(5)° and N(2)–Cu(2)–N(3) = 178.0(5)°, further confirming the distortion. The imidazole nitrogen can be viewed as strongly coordinated to the metal (Cu–N = 1.948(13) Å), while the hydroxyl oxygen forms a weak bond to Cu(II) (Cu(2)–O(5) = 2.2746 Å(2)) in the axial direction.
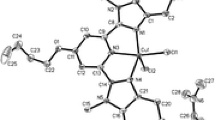
The copper(II) geometry in compound 2 is similar to that of 1 (Fig. 2). The axial position in 2 is occupied by Cu(1)–O(5) = 2.465(7) Å and this bond length is much longer than the equivalent distance in 1. The basal plane of the Cu(II) center in 2 is more planar than that in 1, as, indicated by the O(1)–Cu(1)–O(3) = 166.0(2) and N(2)–Cu(1)–N(1) = 178.7(4)° which can be attributed to the weaker axial Cu(1)–O(5) bond. The important bond angles and distances are presented in Table 3.
The equatorial Cu–O and Cu–N bond lengths are consistent with a square pyramidal CuNO4 coordination sphere [13]. The axial Cu(1)–O(5) bond lengths are consistent with the trends observed in other square pyramidal Cu(II) complexes [14]. In complex 2, the heterocyclic ring of the EtImH ligand and 5-membered chelate ring of the H3hida ligand are both planar, with a dihedral angle between them of 39.50°(N2–C7–C8–C9/Cu1–O1–C1–C2–N1). The other two 5-membered chelate rings, Cu1–N1–C3–C4–O3 and Cu1–N1–C5–C6–O5, both exist in envelope form, where the dihedral angle formed between the mean planes Cu–N1–C3–C4–O3 and Cu–N1–C5–C6–O5 is 67.58°. The rms deviations of the N2–C7–C8–N3–C9, Cu1–O1–C1–C2–N1, Cu–N1–C3–C4–O3 and Cu1–N1–C5–C6–O5 rings are 0.0049, 0.1171, 0.1742 and 0.2114, respectively, for complex 2.
The copper geometry of complexes 1 and 2 is between a square pyramid and a trigonal pyramid, using the distortion index established by Addison et al. [13] (τ = β−α/60), where β and α are the two largest angles. The O(5) atoms of 1 and 2 are both apically coordinated (Fig. 3) with weaker axial bonding at distances of 2.274(12) and 2.465(7) Å, respectively. The copper centers in both 1 and 2 exhibit a slightly distorted square pyramidal geometry (τ = 0.32 for 1 and 0.21 for 2) in which the carboxylate oxygens (O(1) and O(3)) and amine nitrogen (N(3) for 1/N(1) for 2) and imidazolyl nitrogen N2 each occupy a corner of the equatorial plane and the hydroxyl oxygen O(5) of one hydroxyl group exists at the axial position. The crystals of 1 and 2 consist of layers of [Cu(Hhida)(BenzImH)]·H2O and [Cu(Hhida)(EtImH)]·2H2O stacked over each other, held together by HOH…O(carboxyl) interactions. Neighboring mononuclear units are held together through hydrogen bonds involving water molecules in the crystals.
Physico-chemical characterization
The magnetic susceptibilities of complexes 1 and 2 were determined in the solid state at room temperature as 1.81 and 1.83 BM, respectively. These values are characteristic of d9 electronic configuration of planar Cu(II) [15] and are in fair agreement with the spin only value S = 1/2.
The room temperature electronic spectra of the complexes were recorded in DMSO solution. The spectra of the two complexes are very similar and show a low energy ligand field (LF) band at 705 ± 5 nm. A shoulder due to a LMCT (imidazole π1 → Cu2+) band in the region 320–340 nm is also observed [12].
The IR spectra of the complexes are as expected. The ν(COO−) band appears at 1,616 cm−1 [16] for both complex. The bands in the regions 508–5,345 and 280–332 cm−1 are attributed to (Cu–N) [17], while vibrations at 485 cm−1 (weak) can be attributed to M–O bands [18]. A broad band in the range 3,300–3,400 cm−1 assigned to υ(O–H stretch) indicates the presence of water molecules as solvent of crystallization. Molar conductivities (∧m) of DMSO solutions of the complexes indicate that they are non-electrolytes; the values were 8.2 ohm−1cm2mol−1 for complex 1 and 8.2 ohm−1cm2mol−1 for complex 2 [19].
X-band epr spectra of the complexes were recorded in the polycrystalline state and also in pure DMSO. A representative epr spectrum is shown in Fig 4. Derived epr parameters are given in Table 4. The values of the hyperfine coupling constant A∥ are quite low in comparison with the normal range found for other Cu(II) complexes [20], employing a marked tetrahedral distortion of the tetragonal site [21]. No nitrogen superhyperfine splitting could be observed. These one-electron paramagnetic mononuclear Cu(II) complexes display X-band epr spectra in DMSO at 77 K giving g ∥ > g ⊥ > 2.0023, indicating a d x2−y2 ground state [22] in square pyramidal geometry.
The ratio g ∥/A ∥ is a sensitive index showing distortion from planarity for the copper site. The values of g ∥/A ∥ (Table 4) for both complexes are very close to that of Cu-ZnSOD which has a g ∥/A ∥ value of 162 cm [23]. Pierre et al. [24] have synthesized one of the best Cu–Zn SOD models so far, with a g ∥/A ∥ value of 145 cm.
The polycrystalline and frozen solution spectra of both the present complexes are clearly axial (g ∥ > g ⊥ > 2.0) and suggest a d x2−y2 ground state [25] for Cu(II) in square based geometry. The frozen solution epr spectra showed well-resolved epr signals out of one M I =+3/2 component of copper, giving A ∥ values of 155 for 1 and 150 for 2, thereby suggesting a minor distortion in the square pyramidal geometry [26]. Since the values of g ∥ and A ∥ are known to increase and decrease, respectively, with increase [27] in either tetrahedral distortion or axial interaction, the higher g ∥ values observed for the present complexes, suggest the presence of strong axial interactions, in line with the electronic spectral results. The geometric parameter G, which is a measure of the exchange interaction between the copper centers in a polycrystalline solid has been calculated. According to Hathaway [28], if G > 4 the exchange interaction is negligible. The values of G for the present complexes are 5.1 for 1 and 4.2 for 2, indicating negligible exchange interactions in the solid state.
The epr parameters and d–d transition energies were used to evaluate the bonding parameters α2, β2 and γ2, which may be regarded as a measure of the covalency of the in-plane σ bonding and the in-plane π-and out-of-plane π bonding, respectively. The in-plane σ bonding parameter α2 was calculated by using the expression [29]:
The orbital reduction factors K ∥ and K ⊥ were estimated from the expressions [30].
where K ∥ = α2 β2, K ⊥ = α2 γ2 and λ0 represents the one-electron spin–orbit coupling constant for the free ion, equal to −828 cm−1. For both of these complexes, K ∥ > K ⊥ which indicates the presence of out-of-plane bonding. The values of α2, β2 and γ2 are consistent with both strong in-plane σ and in-plane π bonding. The empirical factor f = gII/AII cm−1 is an index of tetragonal distortions and its value may vary from 105-135 for small to extreme distortions in square planar complexes depending on the nature of the coordinated atoms [31]. The f values of these complexes are 152 for 1 and 155 for 2, indicating significant distortion from planarity.
According to ligand field theory [32], the g ∥ value increases and the A ∥ value decreases as the planar ligand field becomes weaker or as the axial ligand field becomes stronger and this occurs with the simultaneous red shift of the d–d absorption bands in the electronic spectra. This sequence, in principle, parallels the degree of distortion from square planar to square pyramidal, C 4v and then to octahedral (O h) or tetragonal (D 4h) geometries. The g ∥ and A ∥ value parameters are in accord with two nitrogen and two oxygen donor atoms coordinated to the metal center in the equatorial plane. These parameters and also the value of the maximum of the visible band suggest a square pyramidal structure where the equatorial plane is determined by the two nitrogen and two oxygen atoms and one oxygen atom is in the apical position.
Electrochemical studies
The electrochemical properties of the two complexes have been studied by cyclic voltammetry (CV) under a nitrogen atmosphere in DMSO solution in the potential range +8.00 to −1.40 V versus Ag/AgCl reference electrode. The electrochemical properties of metal complexes particularly have been studied in order to monitor spectral and structural changes accompanying electron transfer [33]. Both the Cu(II) complexes showed similar electrochemical behavior. The oxidation peak in the 0.4–0.48 V region associated with reduction peak in the 0.120–0.18 V region are observed for both complexes and is assigned to the Cu(II)–Cu(I) redox couple. In the cathodic region of both complexes, an irreversible peak at 0.050–0.680 V and oxidation peak in the 0.116–0.133 V associated region are observed corresponding to Cu(II)/Cu(I) and Cu(I)/Cu(0) [34]. During the reverse scan, the oxidation Cu(I)/Cu(II) skips over and passes to the Cu(II) state. This was confirmed by the ratio of anodic to cathodic current value of the Cu(II)/Cu(III) couple (I pa/I pc = 2). Voltammetric studies gave evidence to a quasi-reversible process that could be attributed to a dynamic rearrangement of the coordination environment during the redox process. The separation between potential peak (ΔE p = E pa−E pc) varies from 100 to 300 mV. This behavior is characteristic of quasi-reversible processes (1). On the basis of the width of the peaks it is possible that two redox steps are present, but only one broad peak is detected. Repetitive scans did not show a different behavior, thus a disproportionation process can be ruled out.
Superoxide dismutase activity
The SOD mimetic activities of the present complexes were examined by the NBT assay [35] following kinetically the reduction of NBT to MF+ at 560 nm. Superoxide was enzymatically supplied from alkaline DMSO. The count fraction causing 50% inhibition of NBT reduction is called IC50. The IC50 are 43 and 47 μmol for complexes 1 and 2 respectively. The observed IC50 values of the present complexes are comparable with the various reported values for the copper(II) complexes but are less active than the native SOD. In our case, the reduction to Cu(I) is assumed to result in the lengthening of the Cu–O(hydroxyl) bonds to better fit the copper ion. It is, however, difficult to deduce the coordination number of Cu(I). One can recognize that these bonds are broken during the reduction of the complex giving rise to four-coordinated copper. This assumption can be related to the SOD activity of the two complexes. The mechanism of dismutation of superoxide anion by native SOD has been found to involve binding of O2 − to copper in place of weakly bound water at the fifth coordination site.
Antibacterial activity
The present complexes were evaluated against the bacterium E. coli, as a function of concentration of these complexes. The susceptibility of the bacteria was determined by measuring the size of inhibition diameter. Both complexes are effective against E. coli, with the zone of inhibition being less for a concentration of 1 mM than for 3 mM concentration. For complex 1, the diameter of inhibition zone was 18 mm at concentration 3 mM while for complex 2 it was 10 mm at concentration 3 mM. Similar antimicrobial results were reported by Tarafder et al. [36] and also by our school [37] for simple copper(II) binary and ternary complexes.
Conclusion
Mononuclear copper(II) complexes of the ligand N-(2-hydroxyethyl)-2-iminodiacetic acid (H3hida) have been characterized. The Hheida2− ligand is monoprotonated in these complexes and their structural and magnetic characteristics are as expected for five-coordinated copper(II). Both complexes are active SOD mimics but at physiological pH the complexation with copper(II) is incomplete. This precludes the applicability as SOD mimics. An interesting structural feature in these complexes is the fact that the two carboxylate moieties are coordinated trans to one another. The tetradentate role of H3hida in both complexes reveals its noticeable conformational flexibility. Voltammetric studies gave evidence for a quasi-reversible process that could be attributed to a dynamic rearrangement of the coordination environment during the redox process.
Supplementary material
CCDC 764419 and 764420 contain the supplementary crystallographic data for [Cu(Hhida)(BenzImH)].H2O 1 and [Cu(Hhida)(EtImH)].2H2O 2. These data can be obtained free of charge via http://www.ccdc.cam.ac.uk/conts/retrieving.html, or from the Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK; fax (+44) 1223-336-033; or e-mail: deposit@ccdc.cam.ac.uk.
References
Palniko I (2008) Functional and structural mimics of superoxide dismutase enzymes. In: JG Hughes, AJ Robinson (eds) Research Progress, Chap. 10. Nova Science Publishers, Inc., New York, 281
Murphy BP (1993) Coord Chem Rev 124:63
Mc Cord JM, Fridovich J (1969) J Biol Chem 244:6049
Budzisz E, Lorenz P, Mayer P, Panety P, Szalkowski L, Krajewska U, Rozalski M, Miernicka M (2009) New J Chem 32:2238
Atria AM, Cortes PC, Garland TM, Baggio R (2003) Acta Cryst 59:m396
Ucar I, Bulut A, Yesilel OY, Olmez H, Buyukgungor O (2004) Acta Cryst C60:m5663
Polyakova IN, Sergienko VS, Poznyak AL (2002) Crystallogr Rep 47(2):246
Shedrick GM (1990) SHELXS-97 Program for the solution for crystal structures. University of Gottingen, Gottingen, Germany, 1997. Acta Crystallogr Sec A 46: 467
Sheldrick GM (1997) SHELXL-97. Program for the refinement of crystal structures. University of Goettingen, Germany
Patel RN, Gundla VLN, Patel DK (2008) Polyhedron 27:1054
Johnson CK (1976) ORTEP, III Report ORNL-5138. Oak Ridge National laboratory, Oak ridge
Monzani E, Quinti L, Perotti A, Faleschini P, Tabbi G (1998) Inorg Chem 37:553
Addison AW, Rao TN, Reedijk J, Rijn JV, Verschoor GC (1984) J Chem Soc Dalton Trans 1349
Hathaway BJ, Billing DE (1970) Coord Chem 5:143
Patel RN, Singh N, Gundla VLN (2007) Polyhedron 26:757
Tahir MM, Keramidas AD, Goldferb RB, Anderson OP, Miller MM, Crans DC (1997) Inorg Chem 36:1667
Nakamoto K (1986) Infrared and Raman spectra of inorganic and coordination compounds, 4th edn. Wiley, New York
Garcia-Raso A, Friol JJ, Adrover B, Tauler P, Pons A, Mota I, Espinosa E, Molins E (2003) Polyhedron 22:3255
Phaniband A, Dhamwad SD (2009) J Coord Chem 62(14):2399
Malmstrom BC, Vamngard T (1960) J Mol Biol 2:118
Batra G, Mathur P (1992) Inorg Chem 31:1575
Klinman JP (1996) Chem Rev 96:2541
Weser U, Schubotz LM, Lengfetder E (1981) J Mol Cat 13:249
Pierre JL, Chautemps P, Refaif S, Beguin C, Marzouki AE, Serratrice G, Saint-Aman E, Ray P (1995) J Am Chem Soc 117:1965
Murali M, Palaniandavar M, Pandiyan T (1994) Inorg Chim Acta 224:19
Faster CL, Liu X, Kilner CA, Pett MT, Halcrow MA (2000) J Chem Soc Dalton Trans 4563
Vilapana R, Garcia-Basallote M, Gonzalez-Vilchez F (1988) Am Chem Soc Natl INOR 322
Hathaway BJ, Billing DE (1970) Coord Chem Rev 5:143
Bryce GF (1966) J Phys Chem 70:3549
West DX (1984) Inorg Nucl Chem 43:3169
Pogni R, Bartoo MC, Diaz A, Basosi R (2000) J Inorg Biochem 79:333
Fernandes AS, Gaspar J, Cabral MF, Caneiras C, Guedes R, Rueff J, Castro M, Costa J, Oliveria NG (2007) J Inorg Biochem 101:849
Kulkarni AD, Patil SA, Badami PS (2009) J Sulf Chem 30(2):145
Srinivasan S, Athappan P, Rajagopal G (2001) Transition Met Chem 26:588
Bhirud RG, Shrivastava TS (1991) Inorg Chim Acta 179:125
Tarafder MTH, Chew KB, Crouse KA, Ali AM, Yamin BM, Fun HK (2002) Polyhedron 21:2683
Patel RN, Singh N, Shukla KK, Chouhan UK, Niclos-Gutierrez J, Castineiras A (2004) Inorg Chim Acta 357(9):2476
Acknowledgments
Our grateful thanks are due to the National Diffraction Facility, X-ray Division, and RSIC (SAIF), IIT Mumbai for single crystal data collection and epr measurements, respectively. The Head RSIC (SAIF), Central Drug Research Institute, Lucknow is also thankfully acknowledged for providing analytical and spectral facilities. Financial assistance from UGC [Scheme no. 36-28/2008 (SR)] and CSIR [Scheme no. 01 (2094)/07/EMR-II], New Delhi are also thankfully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Patel, R.N., Singh, A., Shukla, K.K. et al. Copper(II) complexes with N-(2-hydroxyethyl)-2-iminodiacetic acid and imidazoles: crystal structure, spectroscopic studies and superoxide dismutase activity. Transition Met Chem 35, 577–584 (2010). https://doi.org/10.1007/s11243-010-9366-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11243-010-9366-x