Abstract
In Platanus acerifolia, the SEP3 homolog PlacSEP3 may possess multiple functions, including roles in floral organ identity, development, and vegetative development. This work aimed to characterize the function of PlacSEP3 promoter and analyzed whether PlacSEP3 promoter could be used for genetic engineering flowerless London plane cultivars. The PlacSEP3 promoter was isolated by TAIL-PCR and cis-acting regulatory elements were predicted using the PLACE database and PlantCARE database. Three 5′-deletions of PlacSEP3 promoter were fused to the GUS gene and transformed into tobacco to analyze the functional by Histochemical staining and fluorometric determination. The pPlacSEP3-3::Barnase and pPlacSEP3-3::Barnase-mic35S-Barstar vectors were constructed and transformed into tobacco to test whether pPlacSEP3 could be used for genetic engineering flowerless London plane cultivars. A 1491-bp sequence was isolated, this sequence contained multiple putative cis-regulatory elements related to flower, pollen, and embryo/endosperm development, light responsive. Histochemical staining and fluorometric determination showed that GUS was strongly expressed in reproductive organs and apical buds, and was slightly expressed in vegetative tissues, except the roots of pPlacSEP3-1::GUS and pPlacSEP3-3::GUS transgenic tobacco. However, GUS was not expressed in floral organs and was just slightly expressed in fruits of pPlacSEP3-2::GUS transgenic tobacco. Interestingly, transgenic tobacco of all three constructs showed GUS staining at the bud side of semi-lignified and old stems. The pPlacSEP3-3::Barnase-mic35S-Barstar transgenic tobacco showed various degrees of sterility and vegetative developmental defects. The pPlacSEP3 involved in flower initiation, development and some aspects of vegetative development, such as lateral bud initiation and development. The pPlacSEP3 had the potential application for genetic engineering flowerless cultivars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
London plane (Platanus acerifolia Willd.) is a famous landscape plant that is commonly planted along streets and as shade trees in temperate and subtropical regions of the world owing to its rapid growth, strong stress resistance, and broad, dense crown. However, London plane also has negative properties: its seed hairs and pollen disperse widely in the spring and early summer, which not only pollutes the environment, but also affects human health, e.g., by causing allergic reactions (Subiza et al. 1994; Varela et al. 1997). This disadvantage has badly tarnished public perception of the plant and has limited its application in urban greening. Transgenic technologies provide a powerful tool to solve these problems. As vegetative propagation is preferred over sexual reproduction for most commercial cultivars of London plane (Grolli et al. 2005; Vlachov 1987), engineering flowerless London plane could provide an effective means by which to address the pollen- and seed hair-mediated environmental pollution and allergy issues.
Driving the flower- or anther/pollen-specific expression of a cytotoxic gene, such as barnase or DTA (Diphtheria toxin A), is an effective way to create flowerless or male- and female-sterile plants. This strategy has been successfully used in many species, e.g., for the production of flowerless transgenic birch (Lännenpää et al. 2005; Lemmetyinen et al. 2004) and poplar (Skinner et al. 2003; Wei et al. 2007), the creation of male-sterile cabbage (Lee et al. 2003), petunia (Yue et al. 2016), rice (Luo et al. 2006), wheat (Block et al. 1997), Kalanchoe blossfeldiana (Garcia-Sogo et al. 2010), Pelargonium (Garcia-Sogo et al. 2012), Cryptomeria japonica (Manabu et al. 2011), pine and Eucalyptus (Zhang et al. 2012), and the generation of both male- and female-sterile transgenic tobacco and Arabidopsis (Gardner et al. 2009; Huang et al. 2016). A flower-specific expression promoter is necessary to generate flowerless London plane. However, flower-specific expression promoter has not been identified in London plane, other than the PlacLFY and B-class MADS-box genes promoters (Li et al. 2017; Lu et al. 2012), but PlacLFY promoter is not only active in reproductive tissues, but also in vegetative tissues, and is not suitable for engineering flowerless London plane (Lu et al. 2012). In B-class MADS-box genes, pPaAP3::BARNASE had serious defects in transgenic tobacco, and pPaPI::BARNASE showed the potential application in London plane because of it depauperate flower buds formed in transgenic tobacco (Li et al. 2017).
The SEP3 gene belongs to the SEP subfamily of MADS-box genes, whose members have nearly redundant functions for the specification of floral meristem and organ identities. Accumulating evidence shows that SEP3 plays a more critical role than other SEP genes, since the SEP3 protein binds to the regulatory regions of thousands of potential target genes (Kaufmann et al. 2009) and is sufficient to loop DNA in ‘floral quartet’-like complexes in vitro (Melzer et al. 2009). In Arabidopsis, SEP3 is expressed throughout flower development and is restricted to the inner three whorls (Mandel and Yanofsky 1998). Despite variation in its expression pattern, SEP3 expression shows highly reproductive organs specific in many species, such as petunia (Ferrario et al. 2003), tobacco (Jang et al. 1999), tomato (Pnueli et al. 1994), Alpinia hainanensis (Song et al. 2010; Li et al. 2014), Taihangia rupestris (Wang et al. 2007), Trilliaceae (Kubota and Kanno 2015), and peach (Xu et al. 2008), in which expression is restricted to the inner three whorls, similar to in Arabidopsis. It is expressed in four whorls in some species, such as Antirrhinum (Davies et al. 1996), Gerbera (Kotilainen et al. 2000), citrus (Endo et al. 2006), Prunus mume (Zhou et al. 2017), Dendrobium orchids (Yu and Goh 2000), saffron crocus (Tsaftaris et al. 2011), and lilies (Tzeng et al. 2003). These previous studies have shown that the promoter of SEP3 is a good candidate for engineering flowerless plants.
In a previous study, the London plane SEP3-like gene (PlacSEP3) and its allele (PlacSEP3.1) were characterized, and an expression analysis showed that PlacSEP3 and its allele are expressed obviously in the male and female inflorescence during initiation stages and throughout the flower and fruit development process, and are slightly expressed in vegetative tissues, such as shoot apical buds and vegetative subpetiolar buds of adult trees (Li et al. 2012; Zhang et al. 2017). Here, the PlacSEP3 promoter was isolated and its function was investigated by examining the stable expression of the β-glucuronidase (GUS) reporter gene in transgenic tobacco, and its possible use for sterility breeding in Platanus acerifolia was examined by controlling the Barnase-Barstar and its modified system Barnase-mic35S-Barstar in transgenic tobacco. Our results provide a candidate promoter and a useful Barnase-mic35S-Barstar system for plant sterility genetic engineering.
Materials and methods
Plant materials
Young leaves were collected for DNA extraction from adult field-grown London plane trees at Huazhong Agricultural University, Wuhan, China. The tobacco (Nicotiana tabacum) cultivar ‘Xanthi’ used for transformation was cultured in Murashige and Skoog solid medium (Murashige and Skoog 1962) at 25 °C under a 14-h photoperiod.
Isolation and sequence analysis of the PlacSEP3 promoter
London plane genomic DNA was extracted from young leaves, following the cetyltrimethyl ammonium bromide (CTAB) isolation procedure (Li et al. 2007). The PlacSEP3 promoter was isolated by thermal asymmetric interlaced PCR (TAIL PCR) (Liu and Huang 1998; Liu et al. 1995). According to the cDNA sequence of PlacSEP3, three specific primers, SP1, SP2 and SP3, and eight arbitrary degenerate (AD) primers, AD1–8 (Liu et al. 1995; Lu et al. 2012), were designed (Table 1). An initial DNA segment from the 5′-terminus of PlacSEP3 was isolated by TAIL PCR with the specific primers SP1, SP2, and SP3 and eight AD primers. To obtain a longer promoter sequence of PlacSEP3, another TAIL PCR was performed using the specific primers SP4, SP5, and SP6 (Table 1), which were designed according to the sequence of the first TAIL PCR product. Two TAIL PCR products were assembled and termed pPlacSEP3, and confirmed and sequencing by primers pPlacSEP3F and pPlacSEP3R which designed according to the assembled sequence (Table 1).
The cis-acting regulatory elements in pPlacSEP3 were predicted using the PLACE database (Higo et al. 1999) and PlantCARE database (Lescot et al. 2002).
Vector construction
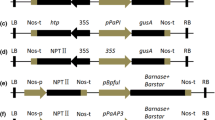
Based on the motif location, different 5′-deletion fragments were selected and amplified for expression vector construction. Regulatory regions 683, 1160, and 1441 bp upstream of the translation initiation site of PlacSEP3 sequences were amplified by PCR using the forward primers pPlacSEP3-1, pPlacSEP3-2, and pPlacSEP3-3 (Table 1), combined with common reverse primer pPlacSEP3R (Table 1), respectively. The 5′ end of the primers was extended 8 bp to generate a PstI in the forward primers and NcoI in the reverse primer (shown in bold and italic), respectively. The amplified products were digested with PstI and NcoI, and then ligated to the likewise digested plasmid pCAMBIA1303. The constructed vectors were verified by digestion with PstI and NcoI, and by PCR amplification using the primers PlacSEP3-1, PlacSEP3-2, PlacSEP3-3, and 1303R (Table 1).
To test whether pPlacSEP3 could be used for genetic engineering flowerless London plane cultivars with the cytotoxic gene Barnase, the pPlacSEP3::Barnase and pPlacSEP3::Barnase-mic35S-Barstar vectors were constructed. Barnase/Barstar was amplified from the BpFUL::Barnase plasmid (Lännenpää et al. 2005) using the primers BarnaseVF and BarstarVR (Table 1), which contained NcoI and BstPI digestion sites. Additionally, the Barnase-mic35S-Barstar construct was built based on the pMD18-Barnase-Barstar as follows. The 110-bp mic35S and Barstar were amplified using 35SVF and 35SVR (Table 1) and BarstarVF and BarstarVR (Table 1), and were then built into pMD18-T, respectively. The mic35S fragment was digested with KpnI and XbaI and then cloned into the pMD18-Barstar plasmid in front of the Barstar gene. Based on the constructed pMD18-Barnse-Barstar, the Barstar was removed (PstI/SphI) and replaced by mic35S-Barstar to obtain the Barnase-mic35S-Barstar construct. Based on the constructed pPlacSEP3-3::GUS plasmid, the GUS gene was removed (NcoI/BstPI) and replaced with Barnase-Barstar and Barnase-mic35S-Barstar, respectively, yielding the pPlacSEP3-3::Barnase and pPlacSEP3-3::Barnase-mic35S-Barstar constructs. Subsequently, the constructed vectors were introduced into the Agrobacterium tumefaciens strain EHA105 by the electroporation method (Mahmood et al. 2008).
Plant transformation
Agrobacterium tumefaciens strain EHA105 containing the constructed plasmids were grown at 28 °C in LB liquid medium supplemented with 100 mg/L kanamycin. Tobacco leaf disc transformation was performed according to the methods of Horsch et al. (1985). The transformants were selected in 25 mg/L hygromycin and were then transplanted into soil and grown in greenhouse in normal conditions. The transgenic plants were confirmed by PCR using the primers pPlacSEP3ZF and GUSVR, BarnaseVR (Table 1).
Histochemical GUS assays
Histochemical localization of GUS activity in pPlacSEP3-1::GUS, pPlacSEP3-2::GUS, and pPlacSEP3-3::GUS transgenic plants was performed essentially as described previously (Jefferson et al. 1987). Various tissues of the transgenic and control plants, including apical buds, young leaves, old leaves, young stem, semi-lignified stem, old stem, root, young flower, fully expanded flower (divided into sepal, petal, stamen, and stigma), young fruit, and semi-mature fruit, were incubated in X-gluc buffer (50 mM sodium phosphate, pH 7.0, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, 10 mM Na2-EDTA, 0.1% w/v Triton X-100 and 1 mg/mL X-Gluc) at 37 °C for 4–7 h. After staining, the samples were immersed in 70% ethanol, incubated at room temperature to remove the chlorophyll, and photographed.
Quantification of GUS activity
For the quantitative measurement of GUS activity in transgenic tobacco, plant tissues were homogenized in GUS extraction buffer (50 mM NaH2PO4, pH 7.0, 10 mM EDTA, 0.1% Triton X-100, 0.1% sodium lauryl sarcosine, and 10 mM β-mercaptoethanol). The homogenate was centrifuged for 10 min at 12,000 rpm at 4 °C, and the GUS activity of the supernatant was assessed according to the methods described by Jefferson et al. (1987). Protein content was assessed by the Bradford method (Bradford 1976) using bovine serum albumin as a standard. GUS activity was normalized to the protein concentration in each of the crude extracts and was calculated as the 4-methylumbelliferone (4-MU) (pmol) produced per minute, per microgram of soluble protein.
Results
Sequence analysis of the PlacSEP3 promoter
By two TAIL-PCRs and validation of successful amplification, a 1491-bp PlacSEP3 putative promoter was cloned, termed pPlacSEP3. The sequence was searched against PlantCARE and PLACE to detect putative cis-elements involved in the regulation of gene expression. Several types of motifs were detected in the promoter (Table 2), including three putative TATA boxes (− 13, − 1222, − 1407), nine CAAT-boxes (CCAAT and CAAT, − 397, − 882, − 1155, − 1167, − 1208, − 1213, − 1235, − 1306, − 1348), one MADS-box protein-binding motif CArG-box (CAAAAATAAG, − 343), several endosperm- and embryo-specific elements (Skn-1_motif, GTCAT, − 334, − 1401; DOFCOREZM, AAAG, − 35, − 247, − 286, − 500, − 677, − 804, − 839, − 863, − 917, − 923, − 952; SEF3MOTIFGM, AACCCA, − 1191; CACGTG motif, CACGTG, − 457; − 300CORE, TGTAAAG, − 839; ACGTCBOX, GACGTC, − 228), transcriptional control and tissue-specific expression elements (GATA-box, GATA, − 400, − 734, − 824, − 865), cis-acting regulatory elements related to meristem-specific activation (CCGTCC-box, CCGTCC, − 591), several pollen development regulatory elements (GTGANTG10, GTGA, − 327, − 455, − 762, − 869, − 919, − 935, − 1391; POLLEN1LELAT52, AGAAA, − 87, − 340; QELEMENTZMZM13, AGGTCA, − 1402), several light response elements (Table 2), and hormone response elements (Table 2). Moreover, some negative regulatory elements were also detected in pPlacSEP3 (WBOXATNPR1, TTGAC, − 1162, − 1296; WBOXNTERF3, TGACY, − 963, − 1140, − 1295; WRKY71OS, TGAC, − 327, − 585, − 651, − 963, − 1140, − 1162, − 1296), an axillary bud outgrowth element (UP1ATMSD, CCCAAT/GCCCATT/CCCATT/GGCCCA/GCCCAA, − 249, − 262, − 362, − 553, − 1306), as well as a MYB binding site involved in drought-inducibility (MBS, TAACTG, − 357), and a cis-acting regulatory element involved in circadian control (Circadian clock, CAANNNNATC, − 1114, − 1443).
Histological expression analysis
After selection on hygromycin and confirmation by PCR analysis, 21, 17, and 24 independent tobacco transformants of pPlacSEP3-1::GUS, pPlacSEP3-2::GUS, and pPlacSEP3-3::GUS were obtained, respectively. All of the positive transgenic plants were assayed by histochemical staining. In pPlacSEP3-1::GUS transgenic tobacco (Fig. 1A), GUS was strongly expressed in four whorls of the young flower (Fig. 1Aa) and mature flower (Fig. 1Ab–d), young fruit (Fig. 1Ae), semi-mature fruit (Fig. 1Af), and apical bud (Fig. 1Ag) and was slightly expressed in the young stem (Fig. 1Ah), semi-lignified stem (Fig. 1Ai), old stem (Fig. 1Aj), young leaf (Fig. 1Ak), and old leaf (Fig. 1Al). No GUS staining was found in root (Fig. 1Am). Moreover, in pPlacSEP3-1::GUS transgenic tobacco, GUS was detected in the young stem (Fig. 1Ah), but was just slightly expressed on the side of lateral bud initiation in the semi-lignified stem (Fig. 1Ai) and old stem (Fig. 1Aj).
Histochemical GUS staining of pPlacSEP3-1::GUS (A), pPlacSEP3-2::GUS (B) and pPlacSEP3-3::GUS (C) transgenic tobacco plants. a-m represent young flower, sepal, petal and stamen, stigma, young fruit, semi-mature fruit, apical stem, young stem, semi-lignification stem, old stem, young leaf, old leaf and root, respectively
The pPlacSEP3-2::GUS transgenic tobacco showed a different GUS staining pattern from that of pPlacSEP3-1::GUS (Fig. 1B). In reproductive tissues, GUS was slightly expressed in the young fruit and semi-mature fruit (Fig. 1Be, f), and absent in the young flower (Fig. 1Ba) and the sepal, petal, stamen, and sigma of mature flowers (Fig. 1Bb–d). In vegetative tissues, GUS staining was observed in the apical bud (Fig. 1Bg), the lateral bud side of the young stem and semi-lignified stem (Fig. 1Bh, i), and young leaf (Fig. 1Bk), and was absent in the old stem (Fig. 1Bj), old leaf (Fig. 1Bl), and root (Fig. 1Bm). Interestingly, the GUS expression pattern in the stem was similar to that of pPlacSEP3-1::GUS.
In pPlacSEP3-3::GUS transgenic tobacco, the GUS expression pattern was similar to that of pPlacSEP3-1::GUS transgenic tobacco. In reproductive tissues, the GUS expression pattern was similar to that of pPlacSEP3-1::GUS, i.e., it was highly expressed in young flowers and the sepal, petal, stamen, and stigma of mature flowers, young fruits, and semi-mature fruits (Fig. 1Ca–f). In vegetative tissues, GUS was strongly expressed in the apical bud (Fig. 1Cg) and young stem (Fig. 1Ch) and was slightly expressed on the lateral bud side of the semi-lignified stem (Fig. 1 Ci), old stem (Fig. 1Cj), young leaf, and old leaf (Fig. 1Ck, l). The GUS expression pattern in the stem was similar to that of pPlacSEP3-1::GUS and pPlacSEP3-2::GUS. No GUS staining was found in the root (Fig. 1Cm).
Quantification of GUS activity
Quantitative GUS assays were performed to determine the expression level of different pPlacSEP3 fragments in different tissues of transgenic tobacco by measuring the specific GUS activity in tobacco. Five lines for each construct of transgenic tobacco were used for the quantitative GUS activity.
In pPlacSEP3-1::GUS transgenic tobacco (Fig. 2), GUS exhibited high activity in reproductive organs, and had the highest activity in the petal, followed by the stamen, young flower, sepal, fruit, and carpel, with GUS activity levels of 3.09 ± 0.17, 2.81 ± 0.15, 2.72 ± 0.09, 2.29 ± 0.12, 2.01 ± 0.11, and 1.75 ± 0.10 pmol 4-MU/min/µg protein, respectively. In vegetative organs, the young stem showed the highest GUS activity, followed by the apical bud, young leaf, semi-lignified stem, and old leaf, with GUS activity levels of 1.51 ± 0.08, 1.26 ± 0.07, 1.22 ± 0.07, 1.06 ± 0.08, and 1.01 ± 0.06 pmol 4-MU/min/µg protein, respectively. The root and old stem showed similar GUS activity levels to those of wild-type control plants.
Fluorometric analysis of GUS activity in different tissues of wild-type tobacco and transgenic tobacco harboring pPlacSEP3-1::GUS, pPlacSEP3-2::GUS and pPlacSEP3-3::GUS constructs. YF, S, P, ST, C, F, AB, YS, SLS, OS, YL, OL and R represent Young flower, Sepal, Petal, Stamen, Carpel, Fruit, Apical bud, Young stem, Semi-lignification stem, Old stem, Young leaf, Old leaf and Root, respectively
In pPlacSEP3-2::GUS transgenic tobacco (Fig. 2), the reproductive organs showed similar GUS activity levels to those of the wild-type control, except in the fruit (1.44 ± 0.08 pmol 4-MU/min/µg protein). In vegetative tissues, the young stem showed the highest GUS activity, followed by the semi-lignified stem, young leaf, apical bud, and old leaf, with GUS activity levels of 1.59 ± 0.09, 1.46 ± 0.05, 1.24 ± 0.07, 1.16 ± 0.06, and 1.10 ± 0.06 pmol 4-MU/min/µg protein, respectively. Similar to pPlacSEP3-1::GUS transgenic tobacco, the root and old stem showed similar GUS activity levels to those of wild-type control plants.
Similar to pPlacSEP3-1::GUS transgenic tobacco (Fig. 2), GUS exhibited high activity in the reproductive organs in pPlacSEP3-3::GUS transgenic tobacco. Maximum GUS activity was observed in the sepal, followed by the stamen, petal, fruit, young flower, and carpel, with GUS activity levels of 3.89 ± 0.09, 3.65 ± 0.20, 3.24 ± 0.18, 3.03 ± 0.17, 3.01 ± 0.19, and 2.51 ± 0.14 pmol 4-MU/min/µg protein, respectively. In vegetative tissues, the young stem showed the highest activity, followed by the apical bud, young leaf, semi-lignified stem, old leaf, and old stem, with GUS activity levels of 2.27 ± 0.13, 1.62 ± 0.09, 1.57 ± 0.09, 1.40 ± 0.10, 1.31 ± 0.07, and 1.06 ± 0.06 pmol 4-MU/min/µg protein, respectively. The root showed a similar GUS activity level to that of the wild-type control roots.
In general, pPlacSEP3-3::GUS showed higher GUS activity than that of other pPlacSEP3 deletion lines. GUS activity in reproductive tissues was higher than that in vegetative tissues in pPlacSEP3-1::GUS and pPlacSEP3-3::GUS, and pPlacSEP3-1::GUS and pPlacSEP3-2::GUS showed high GUS activity in vegetative tissues, except in the semi-lignified stem.
Prevention of flower formation in tobacco via pPlacSEP3::Barnase-mic35S-Barstar
To further analyze the function of pPlacSEP3 and to test whether pPlacSEP3 could be used for genetic engineering flowerless London plane cultivars, plants were transformed with the cytotoxic gene Barnase and Barnase-mic35S-Barstar driven by pPlacSEP3-3. After selection on hygromycin and confirmation by PCR, ten pPlacSEP3-3::Barnase-mic35S-Barstar transgenic tobacco lines were obtained, but no pPlacSEP3-3::Barnase transgenic tobacco was obtained.
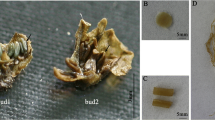
Of these ten pPlacSEP3-3::Barnase-mic35S-Barstar transgenic tobacco lines, three lines showed serious defects with respect to vegetative development in vitro, including degenerated, wrinkled, narrow, and browned leaves and shortened stem internodes (Fig. 3a). These three lines died within a couple of weeks after they were transplanted.
The prevention of flower development in pPlacSEP3-3::Barnase-mic35S-Barstar transgenic tobacco. In some non-flowering lines there were serious vegetative defects in vitro culture stage: degenerated, wrinkled, narrow and browned leaves, shorten stem internodes (a). Non-flowering line showed dramatic defects in vegetative growth after three weeks of transplant: narrow, pale and wrinkled leaves (b). Two non-flowering lines showed defects in leaf and apical bud growth, continuously produced lateral buds, and resulted in dwarfish cluster phenotype (c–e). Wild-type tobacco showed normal phenotype in young stage (f). Some transgenic tobacco lines showed flowering prevention and just minor defects in vegetative development stage: deflexed and slightly wrinkled leaves (g), these transgenic tobacco lines showed dramatic disorders at the flower formation stage: browning young leaves and apical bud (h) and then the apical bud and young leaves turning withered (i)
The other seven lines were developmentally normal within 3 weeks after transplantation. However, one line showed dramatic defects in vegetative growth after 3 weeks; it showed a dwarfish phenotype with very narrow, pale, and wrinkled leaves, and it died within 5 weeks after it was transplanted (Fig. 3b). Two lines showed a non-flowering phenotype, but also exhibited defects in leaf and apical bud growth, continuously produced lateral buds, and had a dwarfish cluster phenotype (Fig. 3c–e). Four lines showed non-flowering and no serious phenotypic defects in vegetative growth, only minor defects, i.e., the leaves were deflexed and wrinkled (Fig. 3g). At the time when the wild-type control plants formed flowers, the transgenic tobacco showed defects, with browning young leaves and apical buds (Fig. 3h); the apical bud and young leaves then withered (Fig. 3i), while the other tissues remained normal.
Discussion
In theory, the pattern and level of gene expression are primarily regulated by cis-acting elements in gene promoters. In this study, we isolated the promoter of PlacSEP3, and a putative cis-element analysis showed that it contains a core promoter element (TATA-box and CAAT-box) and several elements related to flower-, pollen-, and embryo/endosperm-specific development. For example, the CArG-box is the binding site of MADS-box transcription factors (Riechmann et al. 1996; Riechmann and Meyerowitz 1997) and could be the site for SOC1-like protein binding, as observed in Arabidopsis (Lee et al. 2008), the pollen-specific expression motifs GTGANTG10, POLLEN1LELAT52 and QELEMENTZMZM13 are involved in pollen-specific development in tobacco, tomato, and maize (Bate and Twell 1998; Hamilton et al. 1998; Rogers et al. 2001), and the motifs Skn-1-motif, DOFCOREZM, -300CORE, SEF3MOTIFGM, and CACGTGMOTIF are related to endosperm/embryo development in rice, maize, wheat, soybean, and Arabidopsis (Allen et al. 1989; Chandrasekharan et al. 2003; Forde 1994; Washida et al. 1999). Some light responsive elements, hormone regulation motifs, and negative regulatory motifs were also observed.
Although London plane is transformable (Li et al. 2007), the time to flowering is long; accordingly, it is necessary to use tobacco as a model system. A histological expression analysis and quantification of GUS activity of pPlacSEP3-3::GUS in transgenic tobacco showed that GUS had high expression in the sepal, stamen, petal, and carpel of fully expanded flowers, unlike SEP3 homologs of most plants, for which expression is restricted to the inner three floral whorls. These findings indicated that PlacSEP3 may not only be involved in the regulation of the development of the inner three floral whorls, but also in the outer floral whorl in London plane. This expression pattern may be regulated by the CArG motif within the promoter, and is similar to that of citrus CiMADS3 (Endo et al. 2006), peach PPERSEP3 (Tani et al. 2009), lily LMADS3 (Tzeng et al. 2003), and Dendrobium DOMADS1 (Yu and Goh 2000), all of which are expressed in the sepal, in addition to the inner three whorls. Moreover, the young and semi-mature fruits showed strong GUS expression, which may be regulated by the embryo/endosperm-specific motifs in the PlacSEP3 promoter. The expression of GUS in young fruits of transgenic tobacco was consistent with the high expression of PlacSEP3 in mature embryos and in the female inflorescence in April in London plane, at which time the female flowers have completed pollination and embryo development has begun (Li et al. 2012; Zhang et al. 2017). In addition to its expression in reproductive organs, GUS was also detected vegetative tissues, except the root, in pPlacSEP3-3::GUS transgenic tobacco, similar to the PlacSEP3 and PlacSEP3.1 expression pattern in London plane, which are also slightly expressed in vegetative organs (Li et al. 2012; Zhang et al. 2017). SEP3-like homologs from many other plants are indeed expressed at a considerably low level in vegetative tissues. For example, PPERSEP3 transcripts were detected in the leaf in addition to the sepal, inner three whorls, and fruit (Tani et al. 2009), LbSEP3 transcripts accumulated in vegetative tissues in addition to the sepal and inner three whorls (Zeng et al. 2011). Furthermore, transgenic tobacco showed an interesting GUS expression pattern in the stem; staining was detected wholly and deeply in the young stem, and on the side of the lateral bud of the semi-lignified stem, and slight staining was detected at the location of the initiation of the lateral bud and the lateral bud in the old stem. This pattern was coincident with PlacSEP3 expression in London plane, which is expressed in the subpetiolar bud of juvenile and adult plant (Li et al. 2012; Zhang et al. 2017). A putative element analysis of pPlacSEP3 indicated that it includes the UP1ATMSD motif, which up-regulates gene expression during the initiation of axillary bud outgrowth in Arabidopsis (Tatematsu et al. 2005). Therefore, it was reasonable to speculate that pPlacSEP3 may regulate the roles of PlacSEP3 in subpetiolar bud initiation and development in London plane.
A promoter deletion analysis showed that pPlacSEP3-1::GUS and pPlacSEP3-3::GUS had similar GUS expression patterns, but pPlacSEP3-2::GUS showed repressed GUS expression in floral organs and reduced GUS expression levels in fruits. These results indicate that some productive organ-specific expression repressors are located between − 683 and − 1160 bp of pPlacSEP3 (− 744 to − 1224 bp from ATG). A putative elements analysis showed that there were some negative regulatory elements between − 744 and − 1224 bp (WBOXNTERF3, − 1025, − 1202; WRKY71OS, − 1026, − 1203). A previous study showed that the WBOXNTERF3 motif and WRKY71OS motif are transcriptional repressors of the gibberellin and ABA signaling pathway (Xie et al. 2005; Zhang et al. 2004). However, it is not clear whether these motifs function as repressors in the regulation of gene-specific expression in flowers, and further studies are needed to confirm this speculation.
One of the original aims of this research was to determine whether pPlacSEP3 could be used to synthesize a cytotoxic transgene for the genetic engineering of sterility. Lemmetyinen et al. (2001, 2004) showed that BpMADS1::BARNASE (BpMADS1 is an SEP3 homolog in silver birch) could prevent flowering in transgenic Arabidopsis, tobacco, and silver birch. However, for BpMADS1::GUS, which exhibited virtually inflorescence-specific expression in transgenic Arabidopsis and tobacco (Lemmetyinen et al. 2001), a GUS histological expression analysis showed that pPlacSEP3 was not regulated specifically in reproductive organs, but also in vegetative tissues. As a result, no pPlacSEP3-3::Barnase transgenic tobacco was obtained. Therefore, based on the observed pattern of GUS expression in pPlacSEP3-3::GUS transgenic tobacco, the Barnase-mic35S-Barstar construct was adopted in this research to minimize cytotoxicity in vegetative tissues. The phenotype of pPlacSEP3-3::Barnase-mic35S-Barstar transgenic tobacco showed that the cytotoxicity in vegetative tissues could be eliminated, and flower formation could be prevented, despite some vegetative side effects in most of the non-flowering lines. Four lines of pPlacSEP3-3::Barnase-mic35S-Barstar transgenic tobacco showed serious defects and died within 5 weeks after transplant, similar to most BpFULL1::BARNASE transgenic Arabidopsis (Lännenpää et al. 2005). This may be explained by the high Barnase expression and Barstar, which when promoted by mic35S, could not completely eliminate the cytotoxicity in vegetative tissues. The phenotype was characterized by continuous production of lateral buds and a dwarfish cluster, which may result from the high activation of pPlacSEP3 in the apical bud, and the death of the apex leading to lateral bud initiation and development; when the lateral branches also start to development in the same way and produce lateral branches, the total number of lateral branches increased dramatically. The same phenomenon was also observed in BpFULL1::BARNASE transgenic birch (Lännenpää et al. 2005). The increase in the number of lateral branches and the dwarf cluster phenotype might be useful for some practical applications, e.g., when more lateral branches is the breeding goal.
The prevention of flower formation and maintenance of vegetative growth was the aim of this research. Four transgenic tobacco lines were non-flowering and showed no serious phenotypic defects in vegetative growth at the vegetative development stage. However, in the flowering transition stage, the transgenic tobacco showed defects in vegetative tissues. These defects might be caused by the low activation of pPlacSEP3 at the vegetative development stage, while at the flowering transition stage, pPlacSEP3 activation increased and Barstar was unable to completely eliminate the cytotoxicity in vegetative tissues. However, whether pPlacSEP3-3::Barnase-mic35S-Barstar transgenic woody plants generate this phenotype is still unknown, and additional studies are needed to clarify this.
Taken together, pPlacSEP3 exhibited high activation in reproductive organs and moderate activation in vegetative tissues. It may be involved in vegetative tissue development, in addition to reproductive organ initiation and development. Moreover, it may be related to lateral bud initiation and development. The pPlacSEP3-3::Barnase-mic35S-Barstar transgenic tobacco analysis showed that pPlacSEP3-3::Barnase-mic35S-Barstar could prevent tobacco flower formation, but more studies are needed to confirm whether it could be used in genetic engineering sterility breeding programs for woody plants.
References
Allen RD, Bernier F, Lessard PA, Beachy RN (1989) Nuclear factors interact with a soybean beta-conglycinin enhancer. Plant Cell 1(6):623–631. https://doi.org/10.1105/tpc.1.6.623
Bate N, Twell D (1998) Functional architecture of a late pollen promoter: pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol Biol 37(5):859–869. https://doi.org/10.1023/A:1006095023050
Block MD, Debrouwer D, Moens T (1997) The development of a nuclear male sterility system in wheat. Expression of the barnase gene under the control of tapetum specific promoters. Theor Appl Genet 95(1–2):125–131. https://doi.org/10.1007/s001220050
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Chandrasekharan MB, Bishop KJ, Hall TC (2003) Module-specific regulation of the beta-phaseolin promoter during embryogenesis. Plant J 33(5):853–866. https://doi.org/10.1046/j.1365-313X.2003.01678.x
Davies B, Di Rosa A, Eneva T, Saedler H, Sommer H (1996) Alteration of tobacco floral organ identity by expression of combinations of Antirrhinum MADS-box genes. Plant J 10(4):663–677. https://doi.org/10.1046/j.1365-313X.1996.10040663.x
Endo T, Shimada T, Fujii H, Omura M (2006) Cloning and characterization of 5 MADS-box cDNAs isolated from citrus fruit tissue. Sci Hortic-Amsterdam 109:315–321. https://doi.org/10.1016/j.scienta.2006.06.008
Ferrario S, Immink RG, Shchennikova A, Busscher-Lange J, Angenent GC (2003) The MADS box gene FBP2 is required for SEPALLATA function in petunia. Plant Cell 15(4):914–925. https://doi.org/10.1105/tpc.010280
Forde BG (1994) AT-rich elements (ATREs) in the promoter regions of nodulin and other higher plant genes: a novel class of cis-acting regulatory element? Results Probl Cell Differ 20:87–103. https://doi.org/10.1007/978-3-540-48037-2_4
Garcia-Sogo B, Pineda B, Castelblanque L, Anton T, Medina M, Roque E, Torresi C, Beltran JP, Moreno V, Canas LA (2010) Efficient transformation of Kalanchoe blossfeldiana and production of male-sterile plants by engineered anther ablation. Plant Cell Rep 29(1):61–77. https://doi.org/10.1007/s00299-009-0798-8
Garcia-Sogo B, Pineda B, Roque E, Anton T, Atares A, Borja M, Beltran JP, Moreno V, Canas LA (2012) Production of engineered long-life and male sterile Pelargonium plants. BMC Plant Biol 12:156. https://doi.org/10.1186/1471-2229-12-156
Gardner N, Felsheim R, Smith AG (2009) Production of male- and female-sterile plants through reproductive tissue ablation. J Plant Physiol 166(8):871–881. https://doi.org/10.1016/j.jplph.2008.10.002
Grolli P, Morini S, Loreti F (2005) Propagation of Platanus acerifolia Willd. by cutting. J Hortic Sci Biotech 80(6):705–710. https://doi.org/10.1080/14620316.2005.11512002
Hamilton DA, Schwarz YH, Mascarenhas JP (1998) A monocot pollen-specific promoter contains separable pollen-specific and quantitative elements. Plant Mol Biol 38(4):663–669. https://doi.org/10.1023/A:1006083725102
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27(1):297–300. https://doi.org/10.1093/nar/27.1.297
Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231. https://doi.org/10.1126/science.227.4691.1229
Huang J, Smith AR, Zhang T, Zhao D (2016) Creating completely both male and female sterile plants by specifically ablating microspore and megaspore mother cells. Front Plant Sci 7:30. https://doi.org/10.3389/fpls.2016.00030
Jang S, Hong MY, Chung YY, An G (1999) Ectopic expression of tobacco MADS genes modulates flowering time and plant architecture. Mol Cells 9(6):576–586
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6(13):3901–3907. https://doi.org/10.1002/j.1460-2075.1987.tb02730.x
Kaufmann K, Muino JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC (2009) Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol 7(4):e1000090. https://doi.org/10.1371/journal.pbio.1000090
Kotilainen M, Elomaa P, Uimari A, Albert VA, Yu D, Teeri TH (2000) GRCD1, an AGL2-like MADS box gene, participates in the C function during stamen development in Gerbera hybrida. Plant Cell 12(10):1893–1902. https://doi.org/10.1105/tpc.12.10.1893
Kubota S, Kanno A (2015) Analysis of the floral MADS-box genes from monocotyledonous Trilliaceae species indicates the involvement of SEPALLATA3-like genes in sepal-petal differentiation. Plant Sci 241:266–276. https://doi.org/10.1016/j.plantsci.2015.10.013
Lännenpää M, Hassinen M, Ranki A, Hölttä-Vuori M, Lemmetyinen J, Keinonen K, Sopanen T (2005) Prevention of flower development in birch and other plants using a BpFULL1::BARNASE construct. Plant Cell Rep 24(2):69–78. https://doi.org/10.1007/s00299-004-0903-y
Lee YH, Chung KH, Kim HU, Jin YM, Kim HI, Park BS (2003) Induction of male sterile cabbage using a tapetum-specific promoter from Brassica campestris L. ssp. pekinensis. Plant Cell Rep 22(4):268–273. https://doi.org/10.1007/s00299-003-0688-4
Lee J, Oh M, Park H, Lee I (2008) SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. Plant J 55(5):832–843. https://doi.org/10.1111/j.1365-313X.2008.03552.x
Lemmetyinen J, Pennanen T, Lännenpää M, Sopanen T (2001) Prevention of flower formation in dicotyledons. Mol Breed 7:341–350. https://doi.org/10.1023/A:1011602504959
Lemmetyinen J, Keinonen K, Sopanen T (2004) Prevention of the flowering of a tree, silver birch. Mol Breed 13(3):243–249. https://doi.org/10.1023/B:MOLB.0000022525.96200.53
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327. https://doi.org/10.1093/nar/30.1.325
Li ZN, Fang F, Liu GF, Bao MZ (2007) Stable Agrobacterium-mediated genetic transformation of London plane tree (Platanus acerifolia Willd.). Plant Cell Rep 26(5):641–650. https://doi.org/10.1007/s00299-006-0271-x
Li Z, Zhang J, Liu G, Li X, Lu C, Zhang J, Bao M (2012) Phylogenetic and evolutionary analysis of A-, B-, C- and E-class MADS-box genes in the basal eudicot Platanus acerifolia. J Plant Res 125(3):381–393. https://doi.org/10.1007/s10265-011-0456-4
Li X, Fan T, Song J, Sun W, Xia K, Liao J, Zhang M (2014) Functional conservation and divergence of four ginger AP1/AGL9 MADS-box genes revealed by analysis of their expression and protein-protein interaction, and ectopic expression of AhFUL gene in Arabidopsis. PLoS ONE 9(12):e114134. https://doi.org/10.1371/journal.pone.0114134
Li Z, Liu G, Zhang J, Zhang S, Bao M (2017) Functional analysis of the promoters of B-class MADS-box genes in London plane tree and their application in genetic engineering of sterility. Plant Cell Tiss Org 130(2):279–288. https://doi.org/10.1007/s11240-017-1222-7
Liu YG, Huang N (1998) Efficient amplification of insert end sequences from bacterial artificial chromosome clones by thermal asymmetric interlaced PCR. Plant Mol Biol Rep 16:175–181. https://doi.org/10.1023/A:1007420918645
Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8(3):457–463. https://doi.org/10.1046/j.1365-313X.1995.08030457.x
Lu S, Li Z, Zhang J, Yi S, Liu L, Bao M, Liu G (2012) Isolation and expression analysis of a LEAFY/FLORICAULA homolog and its promoter from London plane (Platanus acerifolia Willd.). Plant Cell Rep 31(10):1851–1865. https://doi.org/10.1007/s00299-012-1299-8
Luo H, Lee JY, Hu Q, Nelson-Vasilchik K, Eitas TK, Lickwar C, Kausch AP, Chandlee JM, Hodges TK (2006) RTS, a rice anther-specific gene is required for male fertility and its promoter sequence directs tissue-specific gene expression in different plant species. Plant Mol Biol 62(3):397–408. https://doi.org/10.1007/s11103-006-9031-0
Mahmood T, Zar T, Naqvi SMS (2008) Multiple pulses improve electroporation efficiency in Agrobacterium tumefaciens. Electron J Biotechnol 11(1):1–4. https://doi.org/10.2225/vol11-issue1-fulltext-1
Manabu K, Atsushi W, Ken-ichi K, Miyoko T, Tomonori H, Katsuaki I, Toru T (2011) Towards male sterility in Cryptomeria japonica using the male strobilus-specific genes of C. japonica. BMC Proc 5(Suppl 7):P180. https://doi.org/10.1186/1753-6561-5-s7-p180
Mandel MA, Yanofsky MF (1998) The Arabidopsis AGL9 MADS box gene is expressed in young flower primordia. Sex Plant Reprod 11:22–28. https://doi.org/10.1007/s004970050116
Melzer R, Verelst W, Theissen G (2009) The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in ‘floral quartet’-like complexes in vitro. Nucleic Acids Res 37(1):144–157. https://doi.org/10.1093/nar/gkn900
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15(3):473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Pnueli L, Hareven D, Broday L, Hurwitz C, Lifschitz E (1994) The TM5 MADS box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell 6(2):175–186. https://doi.org/10.1105/tpc.6.2.175
Riechmann JL, Meyerowitz EM (1997) Determination of floral organ identity by Arabidopsis MADS domain homeotic proteins AP1, AP3, PI, and AG is independent of their DNA-binding specificity. Mol Biol Cell 8(7):1243–1259. https://doi.org/10.1091/mbc.8.7.1243
Riechmann JL, Krizek BA, Meyerowitz EM (1996) Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA 93(10):4793–4798. https://doi.org/10.1073/pnas.93.10.4793
Rogers HJ, Bate N, Combe J, Sullivan J, Sweetman J, Swan C, Lonsdale DM, Twell D (2001) Functional analysis of cis-regulatory elements within the promoter of the tobacco late pollen gene g10. Plant Mol Biol 45(5):577–585. https://doi.org/10.1023/A:1010695226241
Skinner JS, Meilan R, Ma C, Strauss SH (2003) The Populus PTD promoter imparts floral-predominant expression and enables high levels of floral-organ ablation in Populus,Nicotiana and Arabidopsis. Mol Breed 12:119–132. https://doi.org/10.1023/A:1026044927910
Song J, Chen Z, Liao J (2010) Isolation and characterization of a SEP-like gene from Alpinia hainanensis (Zingiberaceae). Mol Biol Rep 37(2):917–922. https://doi.org/10.1007/s11033-009-9719-8
Subiza J, Cabrera M, Valdivieso R, Subiza JL, Jerez M, Jimenez JA, Narganes MJ, Subiza E (1994) Seasonal asthma caused by airborne Platanus pollen. Clin Exp Allergy 24(12):1123–1129. https://doi.org/10.1111/j.1365-2222.1994.tb03317.x
Tani E, Polidoros AN, Flemetakis E, Stedel C, Kalloniati C, Demetriou K, Katinakis P, Tsaftaris AS (2009) Characterization and expression analysis of AGAMOUS-like, SEEDSTICK-like, and SEPALLATA-like MADS-box genes in peach (Prunus persica) fruit. Plant Physiol Biochem 47(8):690–700. https://doi.org/10.1016/j.plaphy.2009.03.013
Tatematsu K, Ward S, Leyser O, Kamiya Y, Nambara E (2005) Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiol 138(2):757–766. https://doi.org/10.1104/pp.104.057984
Tsaftaris A, Pasentsis K, Makris A, Darzentas N, Polidoros A, Kalivas A, Argiriou A (2011) The study of the E-class SEPALLATA3-like MADS-box genes in wild-type and mutant flowers of cultivated saffron crocus (Crocus sativus L.) and its putative progenitors. J Plant Physiol 168(14):1675–1684. https://doi.org/10.1016/j.jplph.2011.03.015
Tzeng TY, Hsiao CC, Chi PJ, Yang CH (2003) Two lily SEPALLATA-like genes cause different effects on floral formation and floral transition in Arabidopsis. Plant Physiol 133(3):1091–1101. https://doi.org/10.1104/pp.103.026997
Varela S, Subiza J, Subiza JL, Rodriguez R, Garcia B, Jerez M, Jimenez JA, Panzani R (1997) Platanus pollen as an important cause of pollinosis. J Allergy Clin Immunol 100(6 Pt 1):748–754. https://doi.org/10.1016/S0091-6749(97)70268-9
Vlachov D (1987) Vegetative propagation of sp. Platanus L. through rooting of cuttings. Int Symp Propag Ornam Plants 226:375–378
Wang Y, Tian H, Du X, Lü S, Lu W, Chong K, Meng Z (2007) Isolation and characterization of a putative class E gene from Taihangia rupestris. J Integr Plant Biol 49(3):343–350. https://doi.org/10.1111/j.1672-9072.2007.00431.x
Washida H, Wu CY, Suzuki A, Yamanouchi U, Akihama T, Harada K, Takaiwa F (1999) Identification of cis-regulatory elements required for endosperm expression of the rice storage protein glutelin gene GluB-1. Plant Mol Biol 40(1):1–12. https://doi.org/10.1023/A:1026459229671
Wei H, Meilan R, Brunner AM, Skinner JS, Ma C, Gandhi HT, Strauss SH (2007) Field trial detects incomplete barstar attenuation of vegetative cytotoxicity in Populus trees containing a poplar LEAFY promoter::barnase sterility transgene. Mol Breed 19:69–85. https://doi.org/10.1007/s11032-006-9045-y
Xie Z, Zhang ZL, Zou X, Huang J, Ruas P, Thompson D, Shen QJ (2005) Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol 137(1):176–189. https://doi.org/10.1104/pp.104.054312
Xu Y, Zhang L, Xie H, Zhang Y, Oliveira MM, Ma R (2008) Expression analysis and genetic mapping of three SEPALLATA-like genes from peach (Prunus persica (L.) Batsch). Tree Genet Genomes 4(4):693–703. https://doi.org/10.1007/s11295-008-0143-3
Yu H, Goh CJ (2000) Differential gene expression during floral transition in an orchid hybrid Dendrobium Madame Thong-In. Plant Cell Rep 19:926–931. https://doi.org/10.1007/s002990000227
Yue Y, Yang D, Sun J, Peng H, Yin C, Guo R, Ning G, Hu H (2016) A novel PhLRR gene promoter is sufficient for engineering male sterility in Petunia. Plant Mol Biol Rep 34(5):970–977. https://doi.org/10.1007/s11105-016-0977-z
Zeng SH, Xu YQ, Wang Y (2011) Isolation and characterization of two MADS-box genes from Lycium barbarum. Biol Plant 55(3):567–571. https://doi.org/10.1007/s10535-011-0127-2
Zhang ZL, Xie Z, Zou X, Casaretto J, Ho TH, Shen QJ (2004) A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol 134(4):1500–1513. https://doi.org/10.1104/pp.103.034967
Zhang C, Norris-Caneda KH, Rottmann WH, Gulledge JE, Chang S, Kwan BY, Thomas AM, Mandel LC, Kothera RT, Victor AD, Pearson L, Hinchee MA (2012) Control of pollen-mediated gene flow in transgenic trees. Plant Physiol 159(4):1319–1334. https://doi.org/10.1104/pp.112.197228
Zhang S, Lu S, Yi S, Han H, Liu L, Zhang J, Bao M, Liu G (2017) Functional conservation and divergence of five SEPALLATA-like genes from a basal eudicot tree, Platanus acerifolia. Planta 245(2):439–457. https://doi.org/10.1007/s00425-016-2617-0
Zhou Y, Xu Z, Yong X, Ahmad S, Yang W, Cheng T, Wang J, Zhang Q (2017) SEP-class genes in Prunus mume and their likely role in floral organ development. BMC Plant Biol 17(1):10. https://doi.org/10.1186/s12870-016-0954-6
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (Grant Nos. 31200526, 31272206), the Ministry of education ‘new century excellent talents support program’ (NCET-12-0867), the Ministry of education ‘innovation team development plan’ (IRT13065) and the Forestry Industry Research Special Funds for Public Welfare Projects (Grant No. 201304103).
Author information
Authors and Affiliations
Contributions
GL, MB and SL designed the research. SL performed most experiments. SY performed tobacco transformation. JZ performed vector construction. LL performed histological expression analysis. SL and GL analyzed the data and wrote the manuscript. All of the authors approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Maria Margarida Oliveira.
Rights and permissions
About this article
Cite this article
Lu, S., Yi, S., Zhang, J. et al. Isolation and functional characterization of the promoter of SEPALLATA3 gene in London plane and its application in genetic engineering of sterility. Plant Cell Tiss Organ Cult 136, 109–121 (2019). https://doi.org/10.1007/s11240-018-1497-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-018-1497-3