Abstract
Salt and drought stress are important abiotic factors that negatively affect plant growth and productivity. Defense mechanisms, which plants have developed to cope with stress, are followed by alterations in a genome expression profile that in turn result in qualitative and quantitative change of the proteome. Although proteomic-based approach for studies of plant responses to salinity and drought has already been successfully employed in several plants, for cactus species such analyses have not been done so far. Therefore, in this study we have performed proteomic analysis of Mammillaria gracilis Pfeiff. in vitro-grown cultures, callus and tumor, exposed to iso-osmotic NaCl and mannitol. Obtained results differed among analyzed tissues. The higher number of differentially expressed proteins after either salt or mannitol treatment was revealed in tumor compared to callus. According to classification to different functional categories, majority of the identified callus responsive proteins belongs to protein synthesis and processing category, while the highest number of identified tumor proteins belongs to category of metabolism, which suggest that the mechanisms that mediate responses to salt- and mannitol-induced stress in cactus callus and tumor are dependent on tissue type. Down-regulation of proteins involved in cell protection suggests the inability of tumor to activate protective processes against salinity and osmotic stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Throughout the centuries of evolution, plants have developed finely regulated mechanisms to sense and respond to changes in their environment. Namely, due to their sessile nature, plants lack the ability to migrate and avoid unfavorable environmental conditions. Therefore, they have to cope with a variety of abiotic and biotic stresses, some of which can seriously limit plant growth and development. Among most important environmental factors that reduce plant productivity worldwide are drought and salinity in combination with heat waves (Mittler et al. 2012), hitting hard especially in arid and semiarid regions where fresh water is scarce and the use of saline water in agriculture has increased. High salt depositions in the soil generate a low water potential zone, making it difficult for plants to acquire both water and nutrients. Therefore, salt stress essentially results in a water deficit and takes the form of a physiological drought (Mahajan and Tuteja 2005). Drought and salt stress impose an osmotic stress on plants, resulting in dehydration of cells and osmotic imbalance. Apart from this osmotic effect, salinity also provokes ionic stress due to the specific toxicity of ions; therefore, the physiological mechanisms that plants use to respond to salinity or drought may differ between different plants species (Erdei et al. 1990; Lefèvre et al. 2001).
Biochemical and physiological changes in plants growing under saline and drought conditions have been broadly investigated in many crop (Castillo et al. 2007; Hu et al. 2007; Teixeira and Pereira 2007; Wang et al. 2012), as well as in noncrop plants (Tonon et al. 2004; Lokhande et al. 2011; Balen et al. 2013). The plant response to abiotic stress, salinity and drought included, often occurs as a consequence of change in protein expression rates. Since the stress defense is usually accompanied by an alteration in the pattern of gene expression, this inevitably leads to qualitative and quantitative changes in proteins. However, the genomic studies monitor the response to salinity and drought only at transcriptional level (i.e. mRNA level) (Kempa et al. 2008; Teige et al. 2010), which does not always correlate with the changes at the translational levels (i.e. protein level). It has been shown that mRNA levels in a particular biological sample cannot always be used to predict the changes in protein expression (Mazzucotelli et al. 2008; Rogers et al. 2008). In addition, protein expression is also regulated not only at the translational, but also at posttranslational levels via glycosylation, phosphorylation, sumoylation or ubiquitination (Hirano et al. 2004). Therefore, only the study of proteins themselves can provide information on their real amount and activity (Zivy and de Vienne 2000). Current proteomics-based techniques offer powerful approaches for the identification of proteins associated with a particular environmental and/or developmental signal (Gygi and Aebersold 2000).
Proteomic-based approach for studying the plant responses to salinity and drought has been successfully employed for several plant species (Hajheidari et al. 2005; Wang et al. 2008; Lagana et al. 2009); however, for cactus species, to our knowledge, such analyses have not been done so far. As plants with a crassulacean acid metabolism (CAM), cacti are characterized by high water use efficiency and tolerance to drought. They have been cultivated in the arid and semiarid regions of many countries for food, feed, and medicinal and therapeutic use (Murillo-Amador et al. 2001). On the other hand, cacti are sensitive to salinity; the growth of the majority of cactus species is drastically inhibited by saline water and soil (Murillo-Amador et al. 2001; Silva-Ortega et al. 2008). Therefore, there is an increasing interest in investigating the physiological and biochemical mechanisms that could increase cactus tolerance to salinity. Various in vitro techniques, including micropropagation as well as callus and cell suspension cultures, have been established for numerous cactus species (Elias-Rocha et al. 1998; Llamoca-Zarate et al. 1999; Krsnik-Rasol and Balen 2001) and can be used for the characterization of cell behavior under stress conditions, independently of the regulatory systems that take place at the whole plant level (Errabii et al. 2007).
The objects of this study were two in vitro-grown cultures of cactus Mammillaria gracilis, callus and tumor, subjected to salt- and mannitol-induced stress. Callus tissue is hormone independent (habituated) and regenerable, while tumor also grows on the medium without the addition of growth regulators, but does not express any organogenic potential (Krsnik-Rasol and Balen 2001). Both tissues are considered hyperhydric because of their high water content (Balen et al. 2009). In our previous paper we reported that both callus and tumor were found to be sensitive to osmotic stress caused by either NaCl or mannitol. In response to salt stress, both tissues accumulated Na+, which contributed to osmotic adjustment to a certain extent, as revealed by a less-negative impact on growth compared to mannitol treatment. However, at the cellular level, more prominent oxidative damage was induced by NaCl compared to mannitol in tumor, which could be related to ion toxicity. Results indicated that mechanisms that mediate responses to salt- and mannitol-induced stress were different and dependent on tissue type (Balen et al. 2013). In the present study we have analyzed the effect of 250 mM NaCl and iso-osmotic 500 mM mannitol on proteomes of M. gracilis callus and tumor tissues and identified stress-related proteins with the aim to clarify the mechanisms involved in response of different cactus tissues to imposed stresses.
Materials and methods
Plant material and stress treatment
The objects of this study were callus and tumor tissues of the cactus M. gracilis. Callus spontaneously formed at the base of the cactus plants propagated in vitro on hormone-free Murashige and Skoog (MS) nutrient medium (Murashige and Skoog 1962), containing 3 % sucrose and solidified with 0.9 % agar (agar plant cell culture tested, Sigma-Aldrich, Germany). Callus tissue was detached from the plants and subcultured on the same nutrient medium as a hormone independent habituated tissue. It had a snowy surface with a rather compact yellowish or light green inner portion and expressed high morphogenic capacity. Tumor tissue was obtained after transformation of cactus plants with Agrobacterium tumefaciens, strain B6S3 (Vervliet et al. 1974), harboring the wild-type tumor-inducing (Ti) plasmid. For tumor formation, cross-sections were removed from in vitro-grown M. gracilis plants and inoculated with a bacterial suspension. After the primary tumor was induced, it was maintained as a stable line of tumor tissue (Krsnik-Rasol and Balen 2001). Tumors were yellowish to orange brown, and never expressed any organogenic potential. Both tissues were subcultured for 12 years prior to this research.
Three weeks after subculture of callus and tumor tissues to fresh nutrient medium, stress was initiated by transferring the tissues to solid MS medium supplemented with either NaCl or mannitol. To discriminate between the effects of ionic and osmotic components of salinity on callus and tumors, the effects of iso-osmotic concentrations of NaCl (75, 250, and 350 mM) and mannitol (150, 500, and 700 mM) were evaluated after 1 week by means of gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Control tissues were grown on solid MS medium with no addition of either salt or mannitol for the entire treatment period. All tissues were maintained under a 16/8-h light/dark photoperiod (light intensity, 90 μE m−2 s−1) at 24 °C.
Protein extraction and quantification
For protein extraction, fresh callus and tumor tissues were ground to a fine powder in liquid nitrogen using precooled mortar and pestle. For the gradient SDS-PAGE, approximately 0.5 g of tissue was used and homogenized at 4 °C in 1.2 mL of 0.1 M Tris/HCl extraction buffer, pH = 8.0 (Staples and Stahman 1964), which was supplemented with protease inhibitors (Complete Mini EDTA-free protease inhibitor cocktail, Roche, USA) to protect proteins against a broad range of proteases. The insoluble PVP (10 mg) was added to tissue samples prior to grinding. The homogenates were centrifuged at 20,800×g and 4 °C for 15 min. Obtained supernatants were centrifuged again at 20,800×g and 4 °C for 60 min. The supernatant was collected and protein content was determined according to Bradford (1976) using bovine serum albumin (BSA) as a standard.
Based on the results obtained by gradient SDS-PAGE, callus and tumor tissues exposed to 250 mM NaCl and iso-osmotic 500 mM mannitol were chosen for further analysis with two-dimensional electrophoresis (2-DE). For 2-DE, approximately 1.5 g of tissue was assayed by phenol extraction protocol as initially described by Faurobert et al. (2007) and modified by Pavoković et al. (2011). For protein quantification, dried protein pellets were dissolved in 500 µL of isoelectric focusing (IEF) buffer consisting of 9 M urea, 2 % (w/v) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 0.5 % (v/v) pH 3–10 carrier ampholytes and 0.2 % (w/v) dithiothreitol (DTT). Samples were incubated in IEF buffer for 10 min at room temperature, and sonicated for 30–45 s in an ice bath. Incubation and sonication were repeated until the pellets dissolved. After incubation, samples were centrifuged at 20,800×g for 5 min at room temperature and collected supernatants were used for determination of protein content according to modified Bradford assay (Faurobert et al. 2007) using BSA as a standard.
Electrophoretic separation of proteins
Tissue extracts were analyzed using vertical gradient 8–18 % T (2.67 % C) polyacrylamide gels (gradient PAGE; Protean II xi cell, BioRad, USA) with addition of SDS and using the buffer system of Laemmli (1970). Equal amount of protein, 5 µg per well, was loaded for each sample. Protein bands were visualized using silver staining procedure (Blum et al. 1987).
For 2-DE, dry immobilized pH gradient (IPG) strips (17 cm, pH 3–10 NL, BioRad, USA) were rehydrated overnight (approx. 14 h) at room temperature in 300 µL of IEF buffer containing 300 µg of proteins. IEF was performed in the Ettan IPGphor 3 system (GE Healthcare, UK) using the following stepped gradient: 0–500 V over 1 h, 500–1000 V over 1 h, 1000–8000 V over 1 h, and then 8000 V step until a total run of 45,000 V h. IPG strips were stored at −80 °C until use. Prior to second dimension, the strips were equilibrated for 15 min in equilibration buffer (6 M urea, 50 mM Tris–HCl (pH = 6.8), 2 % (w/v) SDS, 30 % (v/v) glycerol) containing 130 mM dithiothreitol (DTT) and then again for 15 min in the same buffer but with 135 mM iodoacetamide instead of DTT. Second dimension was performed using vertical polyacrylamide slab gels (12 % T, 2.67 % C), with addition of SDS and using the buffer system of Laemmli (1970). Protein spots were visualized by Commassie Brilliant Blue (CBB) R-250 staining solution (0.1 % (w/v) CBB, 45 % (v/v) methanol and 10 % (v/v) glacial acetic acid).
Image acquisition
Gradient SDS gels were scanned at 600 dpi on a flatbed scanner (Epson Perfection, V700 Photo, USA), and protein pattern was analyzed by image processing software Quantity One (BioRad, USA).
For proteomes analysis, the whole experiment (from initiation of stress to 2-DE) was repeated three times for each sample using three technical replicates. 2-D gels were scanned with VersaDoc Imaging System device (model 4000, BioRad, USA) with exposition time of 30 s. Images acquired this way were quantitatively and qualitatively analyzed using Discovery Series PDQuest program (version 7.4.0., BioRad, USA).
The abundance of each spot on 2-D gels was estimated based on the volume percentage. Only those with significant and biological reproducible changes (abundance variation at least twofold, Student’s t test, P < 0.05) were considered as differentially expressed protein spots.
Mass spectrometry analysis
Protein spots, which showed significant differences after analysis of 2-D gels, were excised with a pipette tip from the gels and washed thoroughly using destaining buffer (10 % (v/v) glacial acetic acid and 40 % (v/v) methanol). Proteins were digested using sequencing-grade trypsin (Roche, USA). After overnight in-gel digestion with trypsin (γ = 20 µg mL−1 in 25 mM ammonium bicarbonate) and in-gel protein extraction (in solution containing 50 % (v/v) trifluoroacetic acid (TFA, φ = 5 %) in acetonitrile), peptides were purified using ZipTip C18 columns (Agilent Technologies, USA) and dried.
Obtained tryptic peptides were resuspended in 4 µL of 5 mg mL−1 α-cyano-4-hydroxycinnamic acid (CHCA) matrix prepared in solution consisting of 50 % TFA (φ = 0.1 %) in acetonitrile, and spotted onto the MALDI plate. Mass spectra were obtained using matrix-assisted laser desorption/ionization–time-of-flight mass spectrometer (4800 Plus MALDI TOF/TOF analyzer, Applied Biosystems, USA) in positive reflector mode. For each spot, 1600 shots per spectrum were taken in MS analysis and 2000 shots in MS/MS analysis, covering the mass range of 800–4000 Da, focus mass 2000 Da and delay time 450 ns. Trypsin autolysis peaks were used as internal standards. Automated spectrum interpretation was performed, choosing ten most intense peaks of each MS spectrum (excluding peaks generating from trypsin autolysis, matrix or acrylamide) for subsequent MS/MS analysis. MS/MS was achieved by 1 kV collision-induced dissociation (CID).
Proteins were identified by applying global protein server (GPS) explorer software (version 3.6, Applied Biosystems, USA) for Mascot (Matrix Science version 2.1, UK) search against National Center for Biotechnology Information database (NCBInr) database. Monoisotopic peptide masses were used for combined MS and MS/MS database searches with the following search parameters: maximum allowed peptide mass error, 50 ppm; fragment mass tolerance, ±0.3 Da; minimum 5 S/N; and a maximum of two incomplete cleavages per peptide. All searches were evaluated based on the significant scores obtained from Mascot. The number of trypsin digested peptides matched for each protein was between 9 and 13. All identified proteins were included in the Tables 2 and 3, but only proteins whose score exceeded threshold score were taken for further consideration in discussion of their possible involvement in response of cacti tissues to salinity and osmotic stress. The protein score confidence interval percentage and total ion score confidence interval percentage were both set above 95 %, and the significance threshold was P = 0.05 for the MS/MS. Statistical analysis of changes in protein abundance between stressor and control was performed using Mann–Whitney U test, with a significance level of P < 0.01, using the STATISTICA12 (Stat Soft Inc., USA) software. Gene ontology (GO, http://www.geneontology.org) analysis was derived through Universal Protein Resource (UniProt) hit accessions for all protein identifications according to three categories which describe Biological process, Cellular component and Molecular function.
Results
Protein separation
Protein separation in gradient SDS polyacrylamide gels showed distinct protein patterns for each cactus tissue (Fig. 1). Among each tissue type only a few differences in protein expression were recorded between treatments and control. In callus tissue, the most prominent differences were noticed after the treatment with 250 mM NaCl (Fig. 1, lane 3), which was characterized with the expression of new bands (arrows 1–5 and 7–9 in lane 3), while one protein (Fig. 1, arrow 6, lane 1) was missing when compared to control. In tumor tissue, the greatest differences in protein pattern were also observed in sample obtained after the exposure to 250 mM NaCl (Fig. 1, arrows 10–12, lane 10) as well as to 500 mM mannitol (Fig. 1, arrows 13–16, lane 13). Moreover, protein samples obtained from the tissues exposed to the highest investigated concentrations of both stressors showed signs of protein degradation, which was most pronounced in tumor treated with 700 mM mannitol (Fig. 1, lane 14). Therefore, for further analyses protein extracts obtained from treatments with 250 mM NaCl and 500 mM mannitol were chosen for both tissues.
SDS-PAGE analysis of callus and tumor proteins after exposure to either NaCl or mannitol. Mr molecular weight markers, 1 callus control, 2 callus 75 mM NaCl, 3 callus 250 mM NaCl, 4 callus 350 mM NaCl, 5 callus 150 mM mannitol, 6 callus 500 mM mannitol, 7 callus 700 mM mannitol, 8 tumor control, 9 tumor 75 mM NaCl, 10 tumor 250 mM NaCl, 11 tumor 350 mM NaCl, 12 tumor 150 mM mannitol, 13 tumor 500 mM mannitol, and 14 tumor 700 mM mannitol. Arrows 1–16 indicate differentially expressed proteins
Changes in proteome profiles under salt- and mannitol-induced stress
For each tissue and treatment, triplicate 2-D gels were obtained from three independent experiments. Gels from the different experiments had high reproducibility. Representative gels from the controls and treated callus and tumor tissues are shown in Figs. 2 and 3, respectively. More than 500 protein spots were reproducibly detected in the pH range of 3–10 through the CBB staining from nine independent gels out of three different experiments (three technical replicates per experiment). In terms of the protein distribution patterns in 2-D image, they were very similar among different applied stresses and tissues.
2-DE analysis of callus proteins. Proteome profiles of callus control tissue (a) and tissue treated with either 250 mM NaCl (b) or 500 mM mannitol (c) were compared. Differentially expressed proteins (at least twofold than the control) are indicated by the arrows. Down-regulated proteins (spots market with D letter) are indicated on the gel of control tissue. Up-regulated proteins (spots marked with U letter) are indicated on the gels representing callus proteome after exposure to either 250 mM NaCl or 500 mM mannitol. The numbers correspond to the numbers listed in Tables 1 and 2. Mr molecular weight markers
2-DE analysis of tumor proteins. Proteome profiles of tumor control tissue (a) and tissue treated with either 250 mM NaCl (b) or 500 mM mannitol (c) were compared. Differentially expressed proteins (at least twofold than the control) are indicated by the arrows. Down-regulated proteins (spots market with D letter) are indicated on the gel of control tissue. Up-regulated proteins (spots marked with U letter) are indicated on the gels representing tumor proteome after exposure to either 250 mM NaCl or 500 mM mannitol. The numbers correspond to the numbers listed in Tables 1 and 3. Mr molecular weight markers
To select proteins differentially expressed in response to salinity and mannitol-induced stress, protein profiles of callus and tumor were compared using Discovery Series PDQuest program. Analysis of differential protein expression in both tissues is presented in Table 1 and Fig. 2.
Out of more than 500 reproducibly detected protein spots, 37 spots showed significant and at least twofold change in abundance between control and callus tissue exposed to either salt or mannitol. Enhanced expression was revealed for 8 proteins induced by NaCl (spots 30–37 in Table 1; Fig. 2), although the same treatment down-regulated synthesis of 7 proteins (spots 9–15 in Table 1; Fig. 2) in comparison to control. Exposure to mannitol down-regulated the expression of 11 proteins (spots 1–8 and 16–18 in Table 1; Fig. 2) when compared to control tissue, while the same treatment induced up-regulation of 11 proteins (spots 19–29 in Table 1; Fig. 2). Analysis of responsiveness of callus proteins to either salt or mannitol revealed that mannitol treatment induced changes in expression of 60 % of the proteins, among which the equal number was up- and down-regulated (Fig. 4a). Exposure to 250 mM NaCl also resulted with both enhanced and suppressed expression of callus proteins, although a slightly higher number of up-regulated proteins was recorded (21 %) (Fig. 4a).
Table 1 and Fig. 3 display the variation in protein expression in cactus tumor exposed to 250 mM NaCl and 500 mM mannitol compared to control tissue. Out of more than 500 reproducibly detected protein spots, 57 spots showed significant and at least twofold change in abundance between control and tumor tissue exposed to either salt or mannitol. Salt treatment up-regulated 9 proteins (spots 86–89 and 90–94 in Table 1; Fig. 3) in tumor tissue compared to control, while at the same time it down-regulated 20 proteins (spots 38 and 44–62 in Table 1; Fig. 3). Osmotic stress provoked by mannitol induced the synthesis of 12 proteins (spots 74–85 in Table 1; Fig. 3), while the same treatment decreased the synthesis of 16 proteins (spots 39–43 and 63–73 in Table 1; Fig. 3) in comparison to control tissue. 2-D gel analysis revealed that between two stressors salinity induced a slightly higher response in tumor proteins (51 %), among which majority was down-regulated (35 %), while after exposure to mannitol similar amount of responsive proteins was up- and down-regulated (Fig. 4b).
Identification of proteins responsive to salt- and mannitol-induced stress
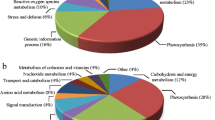
Out of 37 differentially expressed proteins revealed in callus tissue after either salt or mannitol treatment, 18 were successfully identified by searching against the Viridiplantae subset of the NCBInr. In tumor tissue, 57 differentially expressed proteins were revealed after either salt or mannitol treatment, out of which 35 were identified. For a broader classification, the entire set of identified proteins from both tissues was subjected to gene ontology (GO) analysis. Ten callus and 48 tumor proteins were associated with the GO category Biological process (Fig. 5a). Common categories of callus and tumor proteins within Biological process were “metabolic process”, “oxidation–reduction process”, “cell wall organization” and “cell wall modification”. The majority of the proteins from both tissues belonged to the category of “metabolic process” (30 and 29 % in callus and tumor, respectively), although category “oxidation–reduction process” was also highly represented (20 and 17 % in callus and tumor, respectively). Tumor proteins were associated with 15 different categories of Biological process, while callus proteins belonged to only seven. Callus specific categories were “carbon fixation”, “reductive pentose-phosphate cycle” and “photosynthesis”. Callus and tumor proteins were associated with ten different Cellular compartment categories (Fig. 5b). Majority of the callus proteins were assigned to “plastid” (4 out of 5), while one protein was from the “ribosome” category, which was specific for callus tissue. Tumor proteins were associated with nine different Cellular compartments categories, among which the most abundant was “cytoplasm” (40 %). As for the molecular function categories, tumor proteins were associated with 16 different, while callus protein belonged to only 11 categories (Fig. 5c). The most abundant categories for proteins from both tissues were “catalytic activity” (17 and 14 % in callus and tumor, respectively) and “oxidoreductase activity” (13 and 14 % in callus and tumor, respectively). Categories “ATP-binding”, “nucleotide binding” and “metal ion binding” were also common for both tissues. Callus specific categories were “cation binding” (highly abundant, 17 %) and much less abundant “ribulose-bisphosphate carboxylase activity” (7 %) and “RNA binding”, “alpha-amylase activity”, “antioxidant activity” and “chaperone” (4 %, one protein in each category). Eleven categories were specific for tumor proteins, which were mostly associated with “hydrolase activity” (10 %) as well as with “isomerase activity” and “NAD binding” (6 %).
GO analysis of identified proteins from callus and tumor. Differentially expressed proteins were identified by MALDI-TOF/TOF MS according to the NCBInr database. GO analysis was derived through Uniprot hit accessions for all protein identifications according to three categories which describe a biological process, b cellular component and c molecular function
In further analysis identified proteins from both tissues were classified into categories according to their putative physiological functions. Callus proteins were classified into six categories (Table 2), including energy, metabolism, oxidation and detoxification, protein synthesis and processing, disease and defense and category of unknown proteins. By calculating the relative proportions, it was found that the most abundand category was protein synthesis and processing (28 %, Fig. 6a), in which three out of five proteins showed changes in expression due to NaCl treatment (heat shock protein 70, chaperone protein DnaJ and C2 domain-containing protein) and were mostly up-regulated (Table 2). Energy category was represented with 22 % of all identified proteins (Fig. 6a), among which two out of four proteins were up-regulated (both identified as partial RUBISCO large subunits) in response to salinity as well as mannitol. Metabolism as well as oxidation and detoxification classes were represented with 11 % of identified proteins out of which three were mannitol responsive (alpha amylase, cytosolic glyceraldehyde-3 phosphate dehydrogenase and catalase) and mostly up-regulated. Only one detected protein, thaumatin, belonged to the disease and defense category (5 %, Fig. 6a) and exhibited enhanced expression after exposure to salinity (Table 2). Four of the identified proteins belonged to the group of unknown proteins. When all categories are taken together it is obvious that majority of the identified proteins were up-regulated (78 %) and mannitol-responsive (61 %). Only proteins identified as partial RUBISCO large subunits responded to both stressors.
Proteins from tumor tissue, were also classified into different functional categories (Table 3), including categories previously mentioned for callus and one additional, cell structure. The most abundant category was found to be metabolism (54 %, Fig. 6b), in which approximately two thirds of the proteins were down-regulated as a result of either NaCl or mannitol treatment (Table 3). Protein synthesis and processing group was represented with 11 % of all identified proteins (Fig. 6b), which were all down-regulated regardless of the stress imposed (Table 3). The same response was observed in two proteins belonging to the disease and defense category (both identified as subtilisin-like serine proteinases) as well as in those belonging to oxidation and detoxification (cytosolic monodehydroascorbate reductase and lactoylglutathione lyase), which in both cases were represented by 6 % of all identified proteins (Fig. 6b). Category of cell structure was represented by only one identified protein, actin, which was down-regulated by salt treatment, while protein belonging to the energy group (adenosine kinase 2) was up-regulated by mannitol. Six of the identified proteins belonged to the group of unknown proteins. From the analysis of all categories taken together it can be deduced that majority of the proteins were down-regulated (68 %) but similarly responsive to either NaCl (49 %) or mannitol (51 %). Only several proteins, phosphoglycerat mutase, glutamine synthetase and glyceraldehyde-3-phosphate dehydrogenase, belonging to the metabolism category, and subtilisin-like serine proteinase, responded to both stressors.
Discussion
Proteomic tools using 2-DE and MS are useful for the separation, visualization and identification of stress responsive proteins in plants (Zivy and de Vienne 2000). Positive identification of these proteins leads to the discovery of expressed genes that play a role in stress tolerance (Salekdeh et al. 2002). Therefore, plant stress proteomics has the potential of identifying possible candidate genes that can be used for the genetic improvement of plants against stresses (Cushman and Bohnert 2000). Much of the knowledge obtained on plant developmental processes and stress response mechanisms has been gained from work using Arabidopsis and rice (Jorrín et al. 2007; Jorrín-Novo et al. 2009), mainly because of their completed genome sequences. Namely, in proteomics, genome sequences are important resource tools for the identification of proteins. Where fully annotated sequences are not yet available, protein identification can be obtained through similarity searches of homologous proteins in closely related species (Carpentier et al. 2008). Alternatively, expressed sequence tags (ESTs), which represent partial gene sequences, can also be used (Aebersold and Goodlett 2001). However, for the other plant species without significant amounts of published genomic DNA or ESTs sequences, protein identification success rates are lowered resulting in limited proteomic data being available (Jorrín et al. 2007).
In the present study, we tried to elucidate differences in proteomes in two M. gracilis in vitro-grown tissues after salt- and mannitol-induced stress. Despite the fact that in some economies cactus species are industrially important, the proteomic information regarding response of these plants to abiotic stress can not be found. Moreover, it is a plant species whose genome is not sequenced. However, in the present study we have successfully identified numerous cellular proteins involved in response to salt- and mannitol-induced stress. The higher number of responsive genes was detected in tumor tissue compared to callus after exposure to either salt- or mannitol-induced stress. Moreover, only two identified proteins, cytosolic glyceraldehyde-3-phosphate dehydrogenase and heat shock protein 70, were detected in both tissues. These results suggest that the mechanisms that mediate responses to salt- and mannitol-induced stress in cactus callus and tumor are dependent on tissue type. Higher number of changes in proteins in tumor cells than in callus cells could be ascribed to the higher degree of uniformity of the cells in the completely unorganized tumor culture compared to callus tissue which expresses high morphogenic capacity. Difference in responsiveness to applied stresses was particularly pronounced in proteins belonging to the metabolism functional category. This result can be correlated with our previous findings in which more biochemical deviations, as well as changes in photosynthetic features, were reported for cactus tumor compared to callus (Poljuha et al. 2003; Balen et al. 2009, 2012).
Responses to both, salinity and drought, are known to involve the alterations of expression of a multitude of genes, the products of stress-inducible genes functioning both in the initial stress responses and in establishing plant stress tolerance (Chaves et al. 2009). Early responses to water-deficit and salt stress have been considered mostly identical (Munns 2002) because both stresses impose osmotic effects on plants. In addition to alterations in photosynthesis and cell growth, both stresses often induce osmotic adjustment which is considered an important mechanism to allow the maintenance of water uptake and cell turgor under stress conditions. Additionally, accumulation of compatible solutes plays a role in membrane and protein protection, and scavenging of reactive oxygen species (ROS). Under salinity, in addition to water deficits, plants endure salt-specific effects. Salt response follows a biphasic model, with current metabolic data indicating an early similarity with drought, whereas in the long-term plants are responding to ion toxicity (Chaves et al. 2009). In cactus callus, mannitol-induced stress changed the abundance of slightly more proteins than salinity indicating that callus is more sensitive to water deficit than to salinity as already suggested by our previous report (Balen et al. 2013). Under salinity, ions can act as osmolytes and less energy is needed to produce solutes for osmotic adjustment as compared with an organic compatible solute such as proline. Present proteome analysis revealed that both stresses increased the abundance of proteins involved with metabolic and oxidation–reduction processes. In the study of Cramer et al. (2007) proteins involved in energy metabolism and reactive oxygen scavenging exhibited increased abundance with water deficit at an earlier date than in salinized plants, suggesting that water deficit-treated plants appear to have a higher demand than salinized plants to adjust osmotically, detoxify ROS, and cope with photoinhibition. In cactus tumor tissue, both stresses changed abundance of similar amount of proteins, causing down-regulation of many proteins involved with metabolism, protein fate and cellular defense. These indicate severe metabolic and biochemical changes in tumor, an unorganized tissue with different metabolic demands compared to organized plant tissue.
We describe below some of the most important proteins identified, whose identification score exceeded threshold score, as well as their putative biological functions.
Energy
Salinity is known to inhibit photosynthesis; due to osmotic stress stomata become closed and CO2 availability reduced, thus leading to decreased CO2 assimilation. A reduced CO2 assimilation rate is reflected by a decreased abundance of Rubisco (Kosová et al. 2013). In this study, down-regulation of chloroplast Rubisco activase 1 (RbcA) was reported in mannitol-treated callus, while at the same time Rubisco large subunit (RbcL) seemed to be up-regulated in the same tissue exposed to both salt and mannitol. However, identified RbcL proteins appeared to be the products of degradation because their observed molecular masses of approximately 38 kDa on both 2-D gels (spots U22 and U35, Fig. 2) were smaller than the theoretical mass of the native RbcL (53 kDa). It is known that breakdown product of RbcL has a lower molecular mass than the native protein (Taylor et al. 2009). An enhanced degradation of Rubisco subunits was observed in several glycophytic plants and crops exposed to salt (Kosová et al. 2013). Similar observations were also reported by Yan et al. (2006) during rice chilling stress as well as in sugar beet (Hajheidari et al. 2005), rice (Salekdeh et al. 2002) and maritime pine seedlings (Costa et al. 1998) under water stress. Oxidative-stress conditions, which have been proven to exist in callus tissue during exposure to osmotic stress (Balen et al. 2013), accelerate degradation of RbcL thus indicating involvement of active oxygen as a trigger in the degradation of Rubisco (Ishida et al. 1997).
Adenosine kinase 2 (ADK) was found to be up-regulated in tumor exposed to mannitol. ADK catalyzes the transfer of gamma-phosphate from adenosine triphosphate (ATP) to adenosine, leading to formation of adenosine monophosphate (AMP). ADK was found to be up-regulated by salt-induced stress in several glycine betaine accumulators (Weretilnyk et al. 2001; Aghaei et al. 2008; Sobhanian et al. 2010). It maintains methylation activities during salt stress and is responsible for adenosine removal if plants require higher levels of methylated products. Weretilnyk et al. (2001) proposed that ADK is not a stress-responsive enzyme per se, but plays a pivotal role in sustaining transmethylation reactions in general by serving as a coarse metabolic control to reduce the cellular concentration of free adenosine.
Metabolism
Glycolysis and carbohydrate metabolism
Plant responses to salinity pose enhanced demands on energy production. Therefore, an increased relative abundance of several proteins involved in metabolic processes leading to energy release such as glycolysis, tricarboxylic (TCA) acid cycle, photorespiratory pathway and pentose phosphate pathway (PPP) has been found in response to salt stress (Kosová et al. 2013). In accordance, up-regulation of the glycolysis proteins, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and triosephosphate isomerase (TPI) was observed in cactus tissues exposed to NaCl and mannitol. However, in tumor exposed to salinity and mannitol we have also observed the significant down-regulation of two glycolysis enzymes, phosphoglycerat mutase (iPGAM) and enolase. In correlation with lower expression of fructokinase, alcohol dehydrogenase and pyruvate dehydrogenase, glycolysis-related enzymes, these results suggest that the primary metabolism in cactus tissues was significantly affected by salinity and mannitol-induced stress. It is possible that in response to salinity and osmotic stress, energy metabolism is decreased in order to reduce the excess production of ROS, which could induce oxidative stress (Gao et al. 2008). Down-regulation of pyruvate dehydrogenase, which links the glycolysis metabolic pathway to the citric acid cycle and contributes to releasing energy via NADH, was found in cucumber seedling roots subjected to salt stress (Du et al. 2010). iPGAM was down-regulated during the salt stress in Thellungiella halophila (Gao et al. 2008), while Yan et al. (2005) reported lower expression of enolase in rice roots exposed to salt stress. Moreover, lower expression of these enzymes and of fructokinase was also related to osmotic stress (Cramer et al. 2013; Yang et al. 2013). Down-regulation of fructokinase may lead to fructose accumulation, and in this sense may be regarded as a part of osmotic adjustment. Indeed, in soybean roots, Toorchi et al. (2009) also found that PEG-induced osmotic stress reduced the formation of a fructokinase 2 protein. It appears that a decrease in fructokinase may result in a reduction in cell wall polysaccharide content in tissue, impeding tissue growth under osmotic stress, since fructokinase catalyses the transfer of fructose into fructose-6-phosphate. Odanaka et al. (2002) reported that suppression of the fructokinase encoding gene Frk2 inhibited root growth of tomato. In our previous study, inhibited growth was recorded for both cactus tissues exposed to either salt- or mannitol-induced stress (Balen et al. 2013).
It should also be emphasized that several glycolysis proteins, for example GAPDH and TPI, which were up-regulated in cactus tissues exposed to NaCl and mannitol, are multifunctional proteins which display important functional diversity in mammals and plants (Rius et al. 2008; Zhang et al. 2011). Therefore, they are considered as moonlighting proteins, which exhibit activities distinct from their classically identified functions; one of the roles most frequently assigned to these enzymes is their involvement in cell response to oxidative stress (Sirover 2011). In Arabidopsis it was found that cytosolic GAPDH reacted directly with hydrogen peroxide (H2O2), which suggests that this enzyme may mediate ROS signaling in plants (Hancock et al. 2005). It has been reported that cytosolic GAPDH plays an important role in plant salt stress tolerance, including Zea mays (Zörb et al. 2004), potato (Jeong et al. 2001) and rice (Zhang et al. 2011), probably through the regulation of H2O2 levels. As for TPI, its transcript level was regulated in response to various abiotic stresses including salinity and drought (Salekdeh et al. 2002; Yan et al. 2005) as well as methylglyoxal (MG), a cytotoxic byproduct of glycolysis, which can modify proteins and DNA through the formation of advanced glycation end products and contribute to formation of ROS. Increased TPI could bring down the toxic levels of methylglyoxal formed under stress conditions, thus defending the plants against abiotic stress (Sharma et al. 2012). Up-regulation of GAPDH and TPI, found in callus and tumor tissue exposed to NaCl and mannitol, could be therefore involved in defense against oxidative stress, which was previously found to be induced by both stresses in investigated cactus tissues (Balen et al. 2013).
Amino acid metabolism
The regulation of glutamate metabolism appears to be of considerable importance in the nitrogen economy of plants. Glutamine synthetase (GS) catalyzes the combination of ammonia and glutamate into glutamine (Glu) (Nam et al. 2012), while glutamate dehydrogenase (GDH) catalyzes the reversible amination of 2-oxoglutarate to form Glu and therefore it is able to either assimilate or liberate ammonium. It was found that GDH is a stress-responsive protein that may reflect an additional/alternative route to the GS pathway for ammonia assimilation under intracellular hyperammonia conditions induced by salinity (Skopelitis et al. 2006). Numerous reports indicate that GDH and GS are salt stress responsive proteins in rice and Arabidopsis roots (Nam et al. 2012; Jiang et al. 2007; Yan et al. 2005). Moreover, Wang et al. (2012) reported that salt stress induced a strong up-regulation on the expression of GS and GDH genes in rice leaves. Skopelitis et al. (2006) found that salt stress-generated ROS induce GDH expression, and increase assimilation of ammonia by GDH isoenzymes, which could therefore act as anti-stress enzymes in ammonia detoxification and production of Glu for proline synthesis. Proline is well-known osmo-protectant involved in stress resistant mechanisms in plants. Up-regulation of both GS and GDH found in this study in tumor tissue exposed to NaCl could be correlated with significantly increased proline content previously found in M. gracilis tumor exposed to salinity (Balen et al. 2013).
γ-Aminobutyric acid (GABA), is also derived from glutamate via the activity of glutamate decarboxylase (GAD). This molecule is known to accumulate in response to a wide range of environmental stimuli including NaCl exposure, (Kinnersley and Turano 2000). γ-Aminobutyrate transaminase (GABA-transaminase), the first enzyme of the GABA catabolism, was up-regulated in response to NaCl in Arabidopsis (Renault et al. 2010). Moreover, it was also reported that GABA-transaminase deficient mutant was oversensitive to ionic stress in spite of higher GABA levels (Renault et al. 2010) and that GABA-transaminase deficiency impairs central carbon metabolism (Renault et al. 2013). In our study, this enzyme was down-regulated in tumor treated with NaCl, which can be correlated with observed sensitivity of cacti tissues to NaCl.
In plants, serine is synthesized through a couple of pathways. 3-Phosphoglycerate dehydrogenase (PGDH), the first enzyme that is involved in the phosphorylated pathway of serine biosynthesis, is responsible for the oxidation of 3-phosphoglycerate to phosphohydroxypyruvate. In our study, PGDH was found to be down-regulated in tumor exposed to NaCl which indicates that the serine biosynthesis pathway from 3-phosphoglycerate was down-regulated. Cramer et al. (2013) also reported down-regulation of PGDH in grapevine in response to water deficit.
Cell wall metabolism
The plant cell wall is modified in coordination with almost all plant developmental processes. Modifications in the cell wall are thought to be mediated by cell wall hydrolases, including those encoded by a large family of genes specifying endo-1,4-beta-d-glucanases (EGases), which participate in the breakdown of beta-1,4 glucosidic linkages (Sasidharan et al. 2011). Since their potential substrates include xyloglucan and cellulose, EGases could cause wall loosening via action on the xyloglucan-cellulose network, thereby regulating the cellular expansion process (Rose et al. 2002). In our study, cellulase/β-1,4-glucanase was found to be up-regulated in tumor tissues exposed to mannitol-induced osmotic stress. A proteomic analysis on water-stressed maize roots identified a number of drought-induced cell wall modifying proteins including endo-1,3;1,4-β-d-glucanases (Zhu et al. 2007), while Spollen et al. (2008) reported the up-regulation of an endo-1,3;1,4-beta-d-glucanase in the actively growing apical tip region of the roots experiencing water deficit.
Oxidation and detoxification
It is well-known that abiotic stresses induce accumulation of ROS, which can damage DNA, proteins and carbohydrates, resulting in cell death (Mittler 2002). To cope with this, plants have developed several antioxidant enzymes to detoxify ROS and protect their cells from oxidative injury (Mittler 2002; Lokhande et al. 2011). In this study, antioxidative enzymes catalase (CAT) and dehydroascorbate reductase (DHAR) were found to be enhanced in callus treated with salt and mannitol, respectively. The increase in activities of CAT, PPX, APX and DHAR, as major H2O2-detoxifying enzymes, has been related to better resistance to salt and water deficits in many plant species (Rout and Shaw 2001; Cramer et al. 2013). In our previous work we found prominent oxidative damage in correlation to increased activity of several antioxidative enzymes, including CAT, in mannitol-treated callus (Balen et al. 2013). Expression of DHAR, responsible for regenerating ascorbic acid from an oxidized state, regulates the cellular ascorbate redox state, which in turn affects cell responsiveness and tolerance to environmental ROS (Chen and Gallie 2006). Omar et al. (2012) reported that DHAR plays an important role in scavenging H2O2 during dehydration of Jatropha curcas seeds, while Wang et al. (2014) found increases in DHAR abundance in leaves of mangrove Kandelia candel during exposure to salinity. Moreover, up-regulation of both CAT and DHAR was found in two barley cultivars as the effect of salinity stress (Pérez-López et al. 2009). Also, expression of cytosolic monodehydroascorbate reductase (MDHAR), one of the key enzymes in the ascorbate–glutathione pathway that plays a major role in the detoxification of ROS, was found to be increased several times under salt stress and osmotic stress (Lunde et al. 2006). However, in our study tumor tissue exposed to salinity was characterized by the down-regulation of MDHAR which together with lower activity of antioxidant enzymes, particularly APX and CAT, observed previously in tumor after exposure to high salinity resulted in more prominent oxidative stress, probably due to ion toxicity (Balen et al. 2013).
Lactoylglutathione lyase, also known as glyoxalase I, is an enzyme involved in detoxification of MG. Yadav et al. (2005) reported that MG level in plants increases twofold to sixfold in response to salinity, drought, and cold stress conditions. In our study, lactoylglutathione lyase was found to be down-regulated in tumor exposed to mannitol, which suggests that detoxification of MG via glyoxalase system in cells under mannitol-induced osmotic stress was not sufficient. This might resulted with increased level of MG and subsequent inactivation of antioxidative enzymes, which is in accordance with decreased activities of PPX and APX found in our previous study (Balen et al. 2013). Consequently, it probably led to the up-regulation of TPI as discussed earlier.
Protein synthesis and processing
In order to neutralize stress influence, plants can change gene expression and protein accumulation, transcription, protein biosynthesis as well as protein degradation (Yan et al. 2005). One of the protein families whose expression is regulated by stress are heat shock proteins (Hsps). They can play a crucial role in protecting plants against stress by reestablishing normal protein conformation and thus cellular homeostasis (Wang et al. 2004). The Hsp70 and Hsp60 families are known to prevent the aggregation of stress-denatured or nascent proteins in the cell and thus “chaperone” the correct native folding and/or assembly of other proteins (Sharma et al. 2010; Finka et al. 2011; Priya et al. 2013a, b). Although in majority of the studies conducted on plants exposed to either salinity or drought-induced stress these proteins were found to be enhanced, in our study, Hsp70 was down-regulated in callus and tumor tissues treated with either NaCl or mannitol. Moreover, chaperonin 60 kDa (Hsp60), that is crucial to achieve native forms by newly synthesized proteins, was also down-regulated in tumor exposed to NaCl. Bogeat-Triboulot et al. (2007) reported that the abundances of Hsp70 and Hsp60 were significantly decreased by water deficit in leaves of Populus euphratica, arguing that the down-regulation of Hsps is acclimatization to the prolonged stress. Hsp70 proteins were found to be down-regulated in plants exposed to drought (Hajheidari et al. 2005; Ashoub et al. 2013; Kausar et al. 2013). Decreased expression of Hsp70 was also found in salt sensitive genotypes of barley exposed to salt and osmotic stress (Ueda et al. 2004) and Brassica napus in response to salinity (Bandehagh et al. 2013).
Translation elongation in eukaryotes requires a set of soluble non-ribosomal proteins known as eukaryotic elongation factors or eEFs (Sasikumar et al. 2012). In our study, putative eukaryotic elongation factor 1B (eEF1B) was found to be down-regulated in tumor tissues treated with NaCl. Similarly, in salt-treated Arabidopsis cell culture, a decreased relative abundance of several proteins involved in protein biosynthesis such as eukaryotic translation initiation factor eIF-4E2, putative elongation factor EF2 or tRNA synthase class II was observed, indicating a suppression of protein biosynthesis upon salt stress. However, this result could be also correlated with one of the moonlighting functions of this protein. Namely, outside of the translational apparatus, it was found that the eEF1B plays a role in the oxidative stress response pathway in yeast; deletion of the two genes encoding eEF1Bγ in S. cerevisiae gave an additive resistance to oxidative stress (Olarewaju et al. 2004). Moreover, total protein analysis by 2-DE indicated a constitutively active stress response pathway in the absence of eEF1Bγ (Esposito and Kinzy 2010). Namely, change in protein levels suggests alterations in protein turnover in eEF1Bγ-deficient strains that may result in prolonged presence of specific stress response proteins in these mutants. Also, the yeast eEF1Bγ-deficient strain accumulated a greater amount of protein carbonylation when compared with the wild type strain exposed to the same sublethal level of oxidative stress (Esposito and Kinzy 2010). This is in good correlation with results of our previous study in which tumor tissue exposed to salt stress exhibited significantly elevated carbonyl content (Balen et al. 2013). Thus, the stress is affecting the cellular components; however, the lack of turnover pathways leads to their accumulation. In accordance, 26S proteasome subunit α, a component of the large proteasome complex that selectively degrades various cellular proteins (Kurepa and Smalle 2008) was found to be down-regulated in tumor tissue treated with NaCl. Given the fact that during the stress the number of misfolded and damaged proteins increases, one would expect an up-regulation of 26S proteasome subunit (Kosová et al. 2013). However, in Brachypodium distachyon leaves during salt stress the abundance of 26S proteasome subunit α decreased (Lv et al. 2014). As protein degradation is an energy-consuming process, the same authors speculated that plants under stress treatment handle misfolded proteins mainly through refolding.
Disease and defense proteins
Subtilisin-like serine proteinase (subtilase) was found to be down-regulated in tumor tissue after exposure to either salt or mannitol. An involvement in general protein turnover has been predicted for these proteins (Rautengarten et al. 2005), although plant subtilases may also fulfill highly specific functions in plant development and signaling cascades (Rautengarten et al. 2005). It was reported that during the salt stress response in Arabidopsis, subtilase AtS1P targets a membrane-associated bZIP factor, AtbZIP17, which functions as a stress sensor/transducer (Liu et al. 2007).
Conclusion
In conclusion, in this study it was confirmed that in vitro-grown cactus tissues were sensitive to stress caused by either mannitol or NaCl. The difference in the number of responsive genes belonging to different functional categories between callus and tumor exposed to either salt- or mannitol-induced stress suggest that the mechanisms that mediate responses to both stresses are dependent on tissue type. Down-regulation of proteins involved in cell protection suggests the inability of tumor to activate protective processes against salinity and osmotic stress. However, unique flexibility of plant carbohydrate and energy metabolism may help in coping with unavoidable environmental stresses.
References
Aebersold R, Goodlett DR (2001) Mass spectrometry in proteomics. Chem Rev 101:269–295
Aghaei K, Ehsanpour AA, Komatsu S (2008) Proteome analysis of potato under salt tress. J Proteome Res 7:4858–4868
Ashoub A, Beckhaus T, Berberich T, Karas M, Brüggemann W (2013) Comparative analysis of barley leaf proteome as affected by drought stress. Planta 237:771–781
Balen B, Tkalec M, Pavoković D, Pevalek-Kozlina B, Krsnik-Rasol M (2009) Growth conditions in in vitro culture can induce oxidative stress in Mammillaria gracilis tissues. J Plant Growth Regul 28:36–45
Balen B, Tkalec M, Rogić T, Šimac M, Peharec Štefanić P, Vidaković-Cifrek Ž, Krsnik-Rasol M (2012) In vitro conditions affect photosynthetic performance and crassulacean acid metabolism in Mammillaria gracilis Pfeiff. tissues. Acta Physiol Plant 34:1883–1893
Balen B, Tkalec M, Rogić T, Šimac M, Peharec Štefanić P, Rončević S, Pitarević Svedružić L (2013) Effects of iso-osmotic NaCl and mannitol on growth, proline content, and antioxidant defense in Mammillaria gracilis Pfeiff. in vitro-grown cultures. In Vitro Cell Dev Biol Plant 49:421–432
Bandehagh A, Uliaie ED, Salekdeh GH (2013) Proteomic analysis of rapeseed (Brassica napus L.) seedling roots under salt stress. Ann Biol Res 4:212–221
Blum H, Beier H, Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93–99
Bogeat-Triboulot M-B, Brosché M, Renaut J, Jouve L, Le Thiec D, Fayyaz P, Vinocur B, Witters E, Laukens K, Teichmann T, Altman A, Hausman J-F, Polle A, Kangasjarvi J, Dreyer E (2007) Plant gradual soil water depletion results in reversible changes of gene expression, protein profiles, ecophysiology, and growth performance in Populus euphratica, a poplar growing in arid regions. Plant Physiol 143:876–892
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–524
Carpentier SC, Panis B, Vertommen A, Swennen R, Sergeant K, Renaut J, Laukens K, Witters E, Samyn B, Devreese B (2008) Proteome analysis of non-model plants: a challenging but powerful approach. Mass Spectrom Rev 27:354–377
Castillo E, Tuong TP, Ismail AM, Inubushi K (2007) Response to salinity in rice: comparative effects of osmotic and ionic stresses. Plant Prod Sci 10:159–170
Chaves MM, Flexas J, Pinheiro C (2009) Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot 103:551–560
Chen Z, Gallie DR (2006) Dehydroascorbate reductase affects leaf growth, development, and function. Plant Physiol 142:775–787
Costa P, Bahrman N, Frigerio J-M, Kremer A, Plomion C (1998) Water-deficit-responsive proteins in maritime pine. Plant Mol Biol 38:587–596
Cramer GR, Ergül A, Grimplet J, Tillett RL, Tattersall EA, Bohlman MC, Vincent D, Sonderegger J, Evans J, Osborne C, Quilici D, Schlauch KA, Schooley DA, Cushman JC (2007) Water and salinity stress in grapevines: early and late changes in transcript and metabolite profiles. Funct Integr Genomics 7:111–134
Cramer GR, Van SluyterSC, Hopper DW, Pascovici D, Keighley T, Haynes PA (2013) Proteomic analysis indicates massive changes in metabolism prior to the inhibition of growth and photosynthesis of grapevine (Vitis vinifera L.) in response to water deficit. BMC Plant Biol 13:49
Cushman JC, Bohnert HJ (2000) Genomic approaches to plant stress tolerance. Curr Opin Plant Biol 3:117–124
Du CX, Fan HF, Guo SR, Tezuka T, Li J (2010) Proteomic analysis of cucumber seedling roots subjected to salt stress. Phytochemistry 71:1450–1459
Elias-Rocha MA, Santos-Diaz MD, Arredondo-Gomez A (1998) Propagation of Mammillaria candida (Cactaceae) by tissue culture techniques. Haseltonia 6:96–101
Erdei L, Trivedi S, Takeda K, Matsumoto H (1990) Effects of osmotic and salt stresses on the accumulation of polyamines in leaf segments from wheat varieties differing in salt and drought tolerance. J Plant Physiol 137:165–168
Errabii T, Gandonou CB, Essalmani H, Abrini J, Idaomar M, Skali-Senhaji N (2007) Effects of NaCl and mannitol induced stress on sugarcane (Saccharum sp.) callus cultures. Acta Physiol Plant 29:95–102
Esposito AM, Kinzy TG (2010) The eukaryotic translation elongation Factor 1Bgamma has a non-guanine nucleotide exchange factor role in protein metabolism. J Biol Chem 285:37995–38004
Faurobert M, Pelpoir E, Chaib J (2007) Phenol extraction of proteins for proteomic studies of recalcitrant plant tissues. Methods Mol Biol 355:9–14
Finka A, Mattoo RUH, Goloubinoff P (2011) Meta-analysis of heat- and chemically upregulated chaperone genes in plant and human cells. Cell Stress Chaperon 16:15–31
Gao F, Zhou Y, Huang L, He D, Zhang G (2008) Proteomic analysis of long-term salinity stress-responsive proteins in Thellungiella halophila leaves. Chin Sci Bull 53:3530–3537
Gygi SP, Aebersold R (2000) Mass spectrometry and proteomics. Curr Opin Chem Biol 4:489–494
Hajheidari M, Abdollahian-Noghabi M, Askari H, Heidari M, Sadeghian SY, Ober ES, Salekdeh GH (2005) Proteome analysis of sugar beet leaves under drought stress. Proteomics 5:950–960
Hancock JT, Henson D, Nyirenda M, Desikan R, Harrison J, Lewis M, Hughes J, Neill SJ (2005) Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol Biochem 43:828–835
Hirano H, Islam N, Kawasaki H (2004) Techical aspects of functional proteomics in plants. Phytochemistry 65:1487–1489
Hu Y, Burucs Z, Tucher SV, Schmidhalter U (2007) Short-term effects of drought and salinity on mineral nutrient distribution along growing leaves of maize seedlings. Environ Exp Bot 60:268–275
Ishida H, Nishimori Y, Sugisawa M, Makino A, Mae T (1997) The large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase is fragmented into 37-kDa and 16-kDa polypeptides by active oxygen species in the lysates of chloroplasts from primary leaves of wheat. Plant Cell Physiol 38:471–479
Jeong MJ, Park SC, Byun MO (2001) Improvement of salt tolerance in transgenic potato plants by glyceraldehyde-3 phosphate dehydrogenase gene transfer. Mol Cell 12:185–189
Jiang Y, Yang B, Harris NS, Deyholos MK (2007) Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J Exp Bot 58:3591–3607
Jorrín JV, Maldonado A, Castillejo MA (2007) Plant proteome analysis: a 2006 update. Proteomics 7:2947–2962
Jorrín-Novo JV, Maldonado AM, Echevarría-Zomeño S, Valledor L, Castillejo MA, Curto M, Valero J, Sghaier B, Donoso G, Redondo I (2009) Plant proteomics update (2007–2008): second-generation proteomic techniques, an appropriate experimental design, and data analysis to fulfill MIAPE standards, increase plant proteome coverage and expand biological knowledge. J Proteomics 72:285–314
Kausar R, Arshad M, Shahzad A, Komatsu S (2013) Proteomics analysis of sensitive and tolerant barley genotypes under drought stress. Amino Acids 44:345–359
Kempa S, Krasensky J, Dal Santo S, Kopka J, Jonak C (2008) A central role of abscisic acid in stress-regulated carbohydrate metabolism. PLoS One. doi:10.1371/journal.pone.0003935
Kinnersley AM, Turano FJ (2000) Gamma aminobutyric acid (GABA) and plant responses to stress. Crit Rev Plant Sci 19:479–509
Kosová K, Prášil IT, Vítámvás P (2013) Protein contribution to plant salinity response and tolerance acquisition. Int J Mol Sci 14:6757–6789
Krsnik-Rasol M, Balen B (2001) Electrophoretic protein patterns and peroxidase activity related to morphogenesis in Mammillaria gracilis tissue culture. Acta Bot Croat 2:219–226
Kurepa J, Smalle JA (2008) Structure, function and regulation of plant proteasomes. Biochimie 90:324–335
Laemmli U (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lagana A, Caruso G, Cavaliere C, Foglia P, Gubbiotti R, Samperi R (2009) Analysis of drought responsive proteins in wheat (Triticum durum) by 2D-PAGE and MALDI-TOF mass spectrometry. Plant Sci 177:570–576
Lefèvre I, Gratia E, Lutts S (2001) Discrimination between the ionic and osmotic components of salt stress in relation to free polyamine level in rice (Oryza sativa). Plant Sci 161:943–952
Liu JX, Srivastava R, Che P, Howell SH (2007) An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane associated transcription factor, AtbZIP28. Plant Cell 19:4111–4119
Llamoca-Zarate RM, Studart-Guimaraes C, Landsmann J, Campos FAP (1999) Establishment of callus and cell suspension cultures of Opuntia ficus-indica. Plant Cell Tissue Organ Cult 58:155–157
Lokhande VH, Nikam TD, Patade VY, Ahire ML, Suprasanna P (2011) Effects of optimal and supra-optimal salinity stress on antioxidative defence, osmolytes and in vitro growth responses in Sesuvium portulacastrum L. Plant Cell Tissue Organ Cult 104:41–49
Lunde C, Baumann U, Shirley NJ, Drew DP, Fincher GB (2006) Gene structure and expression pattern analysis of three monodehydroascorbate reductase (Mdhar) genes in Physcomitrella patens: implications for the evolution of the MDHAR family in plants. Plant Mol Biol J 60:259–275
Lv DW, Subburaj S, Cao M, Yan X, Li X, Appels R, Sun DF, Ma W, Yan YM (2014) Proteome and phosphoproteome characterization reveals new response and defense mechanisms of Brachypodium distachyon leaves under salt stress. Mol Cell Proteomics 13:632–652
Mahajan S, Tuteja N (2005) Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 444:139–158
Mazzucotelli E, Mastrangelo AM, Crosatti C, Guerra D, Stanca AM, Cattivelli L (2008) Abiotic stress response in plants: when post-transcriptional and post-translational regulations control transcription. Plant Sci 174:420–431
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Mittler R, Finka A, Goloubinoff P (2012) How do plants feel the heat? Trends Biochem Sci 37:118–125
Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25:239–250
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol Plant 15:473–479
Murillo-Amador B, Cortés-Avila E, Troyo-Diéguez A, Nieto-Garibay HGJ (2001) Effects of NaCl salinity on growth and production of young cladodes of Opuntia ficus-indica. J Agron Crop Sci 187:269–279
Nam MH, Huh SM, Kim KM, Park WJ, Seo JB, Cho K, Kim DY, Kim BG, Yoon IS (2012) Comparative proteomic analysis of early salt stress-responsive proteins in roots of SnRK2 transgenic rice. Proteome Sci 10:25
Odanaka S, Bennett AB, Kanayama Y (2002) Distinct physiological roles of fructokinase isozymes revealed by gene-specific suppression of Frk1 and Frk2 expression in tomato. Plant Physiol 129:1119–1126
Olarewaju O, Ortiz PA, Chowdhury WQ, Chatterjee I, Kinzy TG (2004) The translation elongation factor eEF1B plays a role in the oxidative stress response pathway. RNA Biol 1:89–94
Omar S, Elsheery N, Kalaji MH, Xu Z-F, Song S-Q, Carpentier R, Lee C-H, Allakhverdiev SI (2012) Dehydroascorbate reductase and glutathione reductase play an important role in scavenging hydrogen peroxide during natural and artificial dehydration of Jatropha curcas seeds. J Plant Biol 55:469–480
Pavoković D, Križnik B, Krsnik-Rasol M (2011) Evaluation of protein extraction methods for proteomic analysis of non-model recalcitrant plant tissues. Croat Chem Acta 85:177–183
Pérez-López U, Robredo A, Lacuesta M, Sgherri C, Muñoz-Rueda A, Navari-Izzo F, Mena-Petite A (2009) The oxidative stress caused by salinity in two barley cultivars is mitigated by elevated CO2. Physiol Plant 135:29–42
Poljuha D, Balen B, Bauer A, Ljubešić N, Krsnik-Rasol M (2003) Morphology and ultrastructure of Mammillaria gracilis (Cactaceae) in in vitro culture. Plant Cell Tissue Organ Cult 75:117–123
Priya S, Sharma SK, Goloubinoff P (2013a) Molecular chaperones as enzymes that catalytically unfold misfolded polypeptides. FEBS Lett 587:1981–1987
Priya S, Sharma SK, Sood V, Mattoo RUH, Finka A, Azem A, De Los RiosP, Goloubinoff P (2013b) GroEL and CCT are catalytic unfoldases mediating out-of-cage polypeptide refolding without ATP. PNAS 110:7199–7204
Rautengarten C, Steinhauser D, Büssis D, Stintzi A, Schaller A, Kopka J, Altmann T (2005) Inferring hypotheses on functional relationships of genes: analysis of the Arabidopsis thaliana subtilase gene family. PLoS Comput Biol. doi:10.1371/journal.pcbi.0010040
Renault H, Roussel V, El Amrani A, Arzel M, Renault D, Bouchereau A, Deleu C (2010) The Arabidopsis pop2-1 mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biol 10:1–16
Renault H, El Amrani A, Berger A, Mouille G, Soubigou-Taconnat L, Bouchereau A, Deleu C (2013) γ-Aminobutyric acid transaminase deficiency impairs central carbon metabolism and leads to cell wall defects during salt stress in Arabidopsis roots. Plant Cell Environ 36:1009–1018
Rius SP, Casati P, Iglesias AA, Gomez-Casati DF (2008) Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiol 148:1655–1667
Rogers S, Girolami M, Kolch W, Waters KM, Liu T, Thrall B, Wiley HS (2008) Investigating the correspondence between transcriptomic and proteomic expression profiles using coupled cluster models. Bioinformatics 24:2894–2900
Rose JKC, Braam J, Fry SC, Nishitani K (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43:1421–1435
Rout NP, Shaw BP (2001) Salt tolerance in aquatic macrophytes: possible involvement of the antioxidative enzymes. Plant Sci 160:415–423
Salekdeh GH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J (2002) Proteomic analysis of rice leaves during drought stress and recovery. Proteomics 2:1131–1145
Sasidharan R, Voesenek LA, Pierik R (2011) Cell wall modifying proteins mediate plant acclimatization to biotic and abiotic stresses. Crit Rev Plant Sci 30:548–562
Sasikumar AN, Perez WB, Kinzy TG (2012) The many roles of the eukaryotic elongation factor 1 complex. Wiley Interdiscip Rev RNA 3:543–555
Sharma SK, De Los RiosP, Christen P, Lustig A, Goloubinoff P (2010) The kinetic parameters and energy cost of the Hsp70 chaperone as a polypeptide unfoldase. Nat Chem Biol 6:914–920
Sharma S, Mustafiza A, Singla-Pareeka SL, Srivastavab PS, Soporya SK (2012) Characterization of stress and methylglyoxal inducible triose phosphate isomerase (OscTPI) from rice. Plant Signal Behav 7:1337–1345
Silva-Ortega CO, Ochoa-Alfaro AE, Reyes-Agüero JA, Aguado-Santacruz GA, Jiménez-Bremont JF (2008) Salt stress increases the expression of P5CS gene and induces proline accumulation in cactus pear. Plant Physiol Biochem 46:82–92
Sirover MA (2011) On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim Biophys Acta 1810:741–751
Skopelitis DS, Paranychianakis NV, Paschalidis KA, Pliakonis ED, Yakoumakis D, Delis ID, Kouvarakis A, Papadakis AK, Stephanou E, Roubelakis-Angelakis KA (2006) Abiotic stress generated ROS signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis. Plant Cell 18:2767–2781
Sobhanian H, Razavizadeh R, Nanjo Y, Ehsanpour AA, Jazii FR, Motamed N (2010) Proteome analysis of soybean leaves, hypocotyls and roots under salt stress. Proteome Sci 8:19–25
Spollen WG, Tao W, Valliyodan B, Chen K, Hejlek L, Kim J-J, LeNoble M, Zhu J, Bohnert H, Henderson D, Schachtman DP, Davis GE, Springer GK, Sharp RE, Nguyen HT (2008) Spatial distribution of transcript changes in the maize primary root elongation zone at low water potential. BMC Plant Biol 8:32
Staples RC, Stahmann MA (1964) Changes in proteins and several enzymes in susceptible bean leaves after infection by the bean rust fungus. Phytopathology 54:1950–1953
Taylor NL, Tan Y-F, Jacoby RP, Millar HA (2009) Abiotic environmental stress induced changes in the Arabidopsis thaliana chloroplast, mitochondria and peroxisome proteomes. J Proteomics 72:367–378
Teige M, Mehlmer N, Wurzinger B, Stael S, Hofmann-Rodrigues D, Csaszar E, Pfister B, Bayer R (2010) The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J 63:484–498
Teixeira J, Pereira S (2007) High salinity and drought act on an organ dependent manner on potato glutamine synthetase expression and accumulation. Environ Exp Bot 60:121–126
Tonon G, Kevers C, Faivre-Rampant O, Grazianil M, Gaspar T (2004) Effect of NaCl and mannitol iso-osmotic stresses on proline and free polyamine levels in embryogenic Fraxinus angustifolia callus. J Plant Physiol 161:701–708
Toorchi M, Yukawa K, Nouri MZ, Komatsu S (2009) Proteomics approach for identifying osmotic-stress-related proteins in soybean roots. Peptides 30:2108–2117
Ueda A, Kathiresan A, Inada M, Narita Y, Nakamura T, Shi W, Takabe T, Bennett J (2004) Osmotic stress in barley regulates expression of a different set of genes than salt stress does. J Exp Bot 55:2213–2218
Vervliet G, Holsters M, Teuchy H, Van Montagu M, Schell J (1974) Characterisation of different plaque-forming and defective temperate phages in Agrobacterium strains. J Gen Virol 26:33–48
Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9:244–252
Wang M-C, Peng Z-Y, Li C-L, Li F, Liu C, Xia G-M (2008) Proteomic analysis on a high salt tolerance introgression strain of Triticum aestivum/Thinopyrum ponticum. Proteomics 8:1470–1489
Wang H, Zhang M, Guo R, Shi D, Liu B, Lin X, Yang C (2012) Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol 12:194
Wang L, Liu X, Liang M, Tan F, Liang W, Chen Y, Lin Y, Huang L, Jianhong X, Chen W (2014) Proteomic analysis of salt-responsive proteins in the leaves of mangrove Kandelia candel during short-term stress. PLoS One. doi:10.1371/journal.pone.0083141
Weretilnyk EA, Alexander KJ, Drebenstedt M, Snider JD, Summers PS, Moffatt BA (2001) Maintaining methylation activities during salt atress. The involvement of adenosine kinase. Plant Physiol 125:856–865
Yadav SK, Singla-Pareek SL, Ray M, Reddy MK, Sopory SK (2005) Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem Biophys Res Commun 337:61–67
Yan S, Tang Z, Su W, Sun W (2005) Proteomic analysis of salt stress responsive proteins in rice root. Proteomics 5:235–244
Yan SP, Zhang QY, Tang ZC, Su WA, Sun WN (2006) Comparative proteomic analysis provides new insights into chilling stress responses in rice. Mol Cell Proteomics 5:484–496
Yang Z-B, Eticha D, Führs H, Heintz D, Ayoub D, Van Dorsselaer A, Schlingmann B, Rao IM, Braun H-P, Horst WJ (2013) Proteomic and phosphoproteomic analysis of polyethylene glycol-induced osmotic stress in root tips of common bean (Phaseolus vulgaris L.). J Exp Bot 64:5569–5586
Zhang XH, Rao XL, Shi HT, Li RJ, Lu YT (2011) Overexpression of a cytosolic glyceraldehydes-3-phosphate dehydrogenase gene OsGAPC3 confers salt tolerance in rice. Plant Cell Tissue Organ Cult 107:1–11
Zhu J, Alvarez S, Marsh EL, Lenoble ME, Cho IJ, Sivaguru M, Chen S, Nguyen HT, Wu Y, Schachtman DP, Sharp RE (2007) Cell wall proteome in the maize primary root elongation zone. II. Region-specific changes in water soluble and lightly ionically bound proteins under water deficit. Plant Physiol 145:1533–1548
Zivy M, de Vienne D (2000) Proteomics: a link between genomics, genetics and physiology. Plant Mol Biol 44:575–580
Zörb C, Schmitt S, Neeb A, Karl S, Linder M, Schubert S (2004) The biochemical reaction of maize (Zea mays L.) to salt stress is characterized by a mitigation of symptoms and not by a specific adaptation. Plant Sci 167:91–100
Acknowledgments
The financial support of this work was provided by The Ministry of Science, Education and Sports of the Republic of Croatia (119-1191196-1200) as well as by the University of Zagreb.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rogić, T., Horvatić, A., Tkalec, M. et al. Proteomic analysis of Mammillaria gracilis Pfeiff. in vitro-grown cultures exposed to iso-osmotic NaCl and mannitol. Plant Cell Tiss Organ Cult 122, 127–146 (2015). https://doi.org/10.1007/s11240-015-0756-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-015-0756-9