Abstract
The photosynthetic membranes of the filamentous anoxygenic phototroph Roseiflexus castenholzii have been studied with electron microscopy, atomic force microscopy, and biochemistry. Electron microscopy of the light-harvesting reaction center complex produced a 3D model that aligns with the solved crystal structure of the RC–LH1 from Thermochromatium tepidum with the H subunit removed. Atomic force microscopy of the whole membranes yielded a picture of the supramolecular organization of the major proteins in the photosynthetic electron transport chain. The results point to a loosely packed membrane without accessory antenna proteins or higher order structure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Optimal function of the photosynthetic apparatus relies on the proteins present and how they are arranged in the membrane. Mega-complexes in cyanobacteria, supercomplexes in purple bacteria, and other kinds of higher order structure or supramolecular organization have been shown to enhance solar energy capture and conversion efficiency (Sturgis and Niederman 2008; Liu et al. 2013). Purple bacteria such as Rhodobacter sphaeroides have tightly packed arrays of light-harvesting antenna LH2 (Bahatyrova et al. 2004; Sturgis et al. 2009; Tucker et al. 2010), that are adjacent to RC–LH1–PufX core complexes, which in turn can form S-shaped dimers in complexes or exist as standalone rings (Sener et al. 2007; Qian et al. 2008, 2013; Cartron et al. 2014). These organized arrays permit the rapid transfer of excitation energy between complexes and to the RC, where this energy is conserved by charge separation (Hu and Ritz 2002; Cogdell and van Grondelle 2003). Here we investigate the structure of individual photosynthetic reaction center–light-harvesting (RC–LH) protein complexes and the supramolecular organization of the photosynthetic membranes of the phototrophic bacterium Roseiflexus castenholzii.
Roseiflexus castenholzii (RFX) is a photosynthetic bacterium belonging to the filamentous anoxygenic phototrophs (FAPs) (Hanada et al. 2002). The FAPs are generally thermophiles, exist in niche environments and have a unique set of photosynthetic proteins that may have been acquired through horizontal gene transfer events (Hohmann-Marriott and Blankenship 2011; van der Meer et al. 2010; Cao et al. 2012). The large light-harvesting antenna structure called the chlorosome is also found in the green sulfur bacteria (GSB) and the type II reaction center and its associated core antenna, RC–LH1, is similar to that found in purple bacteria (Blankenship 2010; Tang et al. 2011; Hohmann-Marriott and Blankenship 2011). Finally, FAPs have a novel replacement for the cytochrome bc 1 complex called the alternative complex III (ACIII) and a membrane-tethered small blue copper protein, auracyanin, as the soluble electron carrier (Tsukatani et al. 2009; Gao et al. 2010).
RFX was isolated from middle layers of hot spring microbial mats, first in Japan and later in Yellowstone National Park (Takaichi et al. 2001; Van ver Meer et al. 2002). It is closely related to the founding organism of the FAPs, Chloroflexus aurantiacus (CFX) (Pierson and Castenholz 1974; Tang et al. 2011). The major difference between CFXs and RFX is that RFX does not have the large accessory light-harvesting antenna, the chlorosome (Hanada et al. 2002). The photosynthetic core complex of RFX has been isolated and biochemically characterized revealing several unique features (Collins et al. 2009). First, this bacterium only has a combined RC–LH complex to absorb light, much like a RC–LH1 system, but does not have an LH2-type antenna or any other accessory antenna (Yamada et al. 2005). Secondly, this RC–LH complex absorbs further into the red than most organisms, maximally at 880 nm, which allows it to survive in the filtered light environments of the microbial mats using this lower energy light to drive photosynthesis (Niedzwiedzki et al. 2010). Third, the RFX RC–LH has been shown to have the ring-like structure of an RC–LH1, but behaves spectroscopically more like an LH2 complex (Collins et al. 2010) in having monomeric bacteriochlorophylls absorbing at 800 nm. Fourth, the RC–LH is lacking the RC–H and PufX or W subunits typically found in the related purple bacteria type II reaction centers and core complexes (Pugh et al. 1998; Qian et al. 2005; Xin et al. 2012). Additionally, the complex contains an integral tetraheme cytochrome c, as first observed in the Blastochloris viridis system, but not ubiquitously found in other purple bacteria or type II RCs (Deisenhofer et al. 1985; Collins et al. 2011).
The other major component of the Roseiflexus membranes is a functional replacement for the cytochrome bc 1 complex called the ACIII (Majumder et al. 2013). ACIII has been shown to be an oxidoreductase oxidizing menaquinone from the RC and returning electrons to the RC special pair via the small blue copper protein auracyanin (Gao et al. 2009, 2013). ACIII has no structural or evolutionary relationship to the cytochrome bc 1 complex but is related to enzymes from the iron–sulfur molybdopterin family, and is therefore a novel protein in photosynthetic membranes (Majumder et al. 2013). Auracyanin, which replaces the typical soluble cytochrome electron carrier, has been previously characterized and crystallized in the CFX system, and the RFX auracyanin has a sequence similar to a mixture of CFX auracyanins A and B, but there is only one copy in the genome and the C-terminal membrane tether tail is predicted to be slightly shorter (Tsukatani et al. 2009).
The basic biochemical characterization of the major protein players of the FAP photosynthetic cyclic electron transport chain (ETC) revealed novel complexes with distinct features. Despite the studies of the RC–LH, ACIII, and auracyanin, the FAP ETC remains poorly understood. No high-resolution crystal structure has been solved of the RC–LH or the ACIII from any FAP. Likewise, the mechanism, cofactor content, and arrangement within each complex remain unknown. A full cyclic electron transport network has not been demonstrated biochemically, specifically the interaction between auracyanin and the membrane protein complexes. It is also not known how these complexes are organized in the membrane or exactly how they function as a unit. However, based on the similarity of the FAP RC–LH to the purple bacterial RC–LH1 systems, we predict that the FAP supramolecular organization mirrors the purple bacterial patterns (Sturgis et al. 2009).
In this work, we made further efforts to determine the structure of the RC–LH and to visualize the components of the photosynthetic system in situ in the membrane. The RFX photosynthetic unit has been studied with both transmission electron microscopy (EM) and atomic force microscopy (AFM), as well as biochemical and in silico techniques. We found the RFX RC–LH to be structurally similar to purple bacterial RC–LH1 complexes, but did not find a highly ordered pattern as seen in the supramolecular organization of the membrane of Blc. viridis (Scheuring and Sturgis 2009). We propose a supramolecular arrangement for RC–LH, ACIII, and auracyanin in the photosynthetic membranes of RFX.
Materials and methods
Purification of RC–LH and LH-only complexes
Isolation and purification of the RC–LH and LH-only complexes of photosynthetically grown R. castenholzii HL08 was carried out by the method described by Collins et al. (2010). Briefly, whole membranes (WM) were isolated from the cells by sonication and ultracentrifugation and then solubilized in 2 % LDAO. The solubilized membranes were centrifuged and filtered to remove debris and loaded onto a QSHP ion exchange column and eluted with a NaCl gradient. For LH-only complexes, the column was washed in 1 % LDAO for at least one column volume before applying the salt gradient. Collected colored fractions were analyzed by UV/Vis spectrophotometry. Those containing the desired complex were concentrated using 100 kDa MWCO spin filters and run over a S200 gel filtration column. Fractions were checked again and the pure complexes were concentrated and verified by SDS-PAGE.
Electron microscopy
Concentration of the purified samples, RC–LH, and LH-only complexes from RFX, were adjusted to 0.1 mg/ml protein. Five μl of the protein solution was loaded on the surface of freshly glow-discharged EM copper grid coated with 16-nm-thick carbon film. After 30 s, the excess protein solution was blotted by the use of filter paper. The grid was washed with distilled water twice, and then stained with 0.75 % (w/v) uranyl formate for 30 s. The stain solution was blotted and the grid was dried in air. The grids were examined using a Philips CM100 electron microscopy under 100 kV accelerating voltage. Micrographs were recorded with a 1K × 1K Gatan Multiscan 794 CCD camera at a normal magnification of 52,000× with variation of under-focus values from 0.5 to 1.5 μm, resulting in a pixel size of 3.93 Å at specimen level.
Particles were picked from selected images. All micrographs that showed drifting, bad staining, or unsuitable under focus were discarded. In total, 18,945 RC–LH and 6086 LH-only particles were collected for data processing initially by the use of EMAN2 (Tang et al. 2007). After CTF correction, two data sets were compiled, and subsets containing 5837 particles for RC–LH and 3242 LH-only complexes were used to produce reference-free 2D class averages. Selected 2D averaged classes were used to establish the initial 3D model, which was used for 3D reconstruction with the corresponding subset data. The full-set data were applied at the second refinement stage. Iteration of the refinement continued until a stable model was obtained. During each iteration, 10 % bad particles were rejected.
The resolution of the final model was estimated by calculating the Fourier shell correlation (FSC) between two 3D reconstruction models that were built up separately from two halves of the 2D classes.
Preparation of intracytoplasmic membranes
Membranes were isolated from whole cells of photosynthetically grown RFX. Whole wet cells were suspended in 20 mM Tris buffer pH 8 with 2 % Triton X-100 and 500 mM EDTA, and 0.1 M NaCl at 2 ml per every gram of cells to remove the glycosylated layer around the cells. The cell suspension was placed in a 37 °C water bath for at least 15 min. The cells were pelleted at 12,000 rpm in 5 min. The supernatant was discarded, and the reddish colored pellets were resuspended in 20 mM Tris buffer pH 8 with 100 mM EDTA and 0.1 M NaCl. Egg white lysozyme was added from a stock solution to a final concentration of 0.8 mg/ml. The suspension was again incubated at 37 °C for 30 min and then pelleted by the same method. If the pellet was colorless/white and the supernatant was red/orange/brown, then patches had been obtained. If the pellet was still colored, the cells were subjected to one round on the French Press at 20,000 psi.
Membrane patches were prepared from the total membrane preparation using 5 optical density units, at 882 nm, of these membranes layered onto a 30/34/37/40/50 % w/w sucrose, 20 mM HEPES pH 7.5, 1 mM EDTA discontinuous gradient and centrifuged in a Beckman SW41 rotor for 2 h at 40,000 rpm, 4 °C and the resultant bands collected. These detergent-free membranes were stored at −20 °C until used for imaging.
The membranes were diluted by between 5- and 10-fold with 20 mM HEPES pH 7.5 and 5 µl of this diluted suspension pipette into 35 µl of absorption buffer, 20 mM HEPES pH 7.5, 150 mM KCl, and 25 mM MgCl2, on a freshly cleaved 6 mm diameter mica disk and mixed by gentle pipetting. The membranes were allowed to adsorb to the mica for a minimum of 1 h at RT in a closed Petri dish with a piece of moist tissue to minimize evaporation. The disk was washed twice with imaging buffer, 20 mM HEPES pH 7.5, 100 mM KCl, immediately prior to imaging in the AFM.
Atomic force microscopy
Peak Force tapping mode imaging was conducted using Bruker SNL probes, 0.24 N/m spring constant, 2 kHz operating frequency mounted in a standard liquid cell in a Nanoscope V Multimode 8 with E scanner. The images were recorded at scan frequencies of 0.5–1.0 Hz and subsequently analyzed as described below.
Topographs were flattened and three-dimensional (3-D) representations generated using Bruker nanoscope analysis software (v1.4). Statistical analysis of high-resolution topographs was performed using the same software to measure peak to peak of height profiles across individual molecules in line with the scan and overall height with respect to the mica substrate or surrounding lipid bilayer surface. Only clearly defined complexes were examined to produce the most accurate analysis.
Heme protein quantitation
WMs were isolated as in section above, but were collected before the detergent addition of the solubilization step. The WMs were diluted 1:1 with 20 mM Tris pH 8 buffer. The WM samples were prepared for SDS-PAGE by the 1:1 mixing of membrane with reductant free SDS-PAGE sample buffer, 30 % glucose, 0.05 % Coomassie Blue 250, 4 % sodium dodecyl sulfate in 150 mM Tris pH 7. The samples were vortexed and allowed to incubate at 65 °C for at least 30 min. Samples were loaded onto an 8 M urea 12 % SDS-PAGE gel with a linearly increasing amount of sample added per well. Two identical gels were run at 30 V for 1 h and 100 V for at least 2 h until the blue dye began to leak from the bottom of the gel.
Gels were stained in the following manner. One was stained with CBR-250 in methanol and water for 4 h and then destained in 10 % acetic acid overnight. The second gel was stained, photographed, and destained with the TMBZ heme staining procedure as described in Goodhew et al. (1986). The quantity of TMBZ staining was measured with the gel analysis function in FIJI-ImageJ.
Results
EM single particle analysis and 3D reconstruction
RFX RC–LH and LH-only complexes were isolated and purified as described above. The complexes were placed on EM grids, negatively stained, and imaged. Under negative stain conditions, the single protein molecules can be seen clearly under the magnification of 60015×. A total of 18945 RCLHC and 6086 LHC-only particles were selected from negative-stained EM micrographs and 28 representative classes identified, most showing face-on orientations (Fig. 1). Different orientations of the RC–LH particles on the EM grid can be distinguished. The molecules facing up or down with respect to the membrane were an elliptical circle with a clear central density attributed to the RC. (Fig. 1a). The molecules lying on their sides showed a protruding density on one side of the LH ring, assigned as the tetraheme cytochrome c subunit (Fig. 1b). In the EM grid of LH-only (Fig. 1c, d), LH complexes were empty elliptical toroids, indicating that the RC was removed successfully from the purified RC–LH. After 2D classification, discrete orientations of the RC–LH and LH-only proteins on the EM grid were revealed as seen in Fig. 1b, d.
Single particles of purified RC–LH and LH-only complexes. a Electron micrograph of the RC–LH complex. Scale bar 50 nm. b Selected 2D averaged classes from single particles of the RC–LH complex. c LH-only electron micrograph and d selected 2D averaged classes of LH-only particles. Box size 28.3 × 28.3 nm
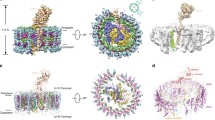
From the 28 classes of EM images of the RC–LH and the LH-only complexes, it was possible to conduct 3D reconstructions to ≤20 Å resolution. The resulting structures are shown in three orientations in Fig. 2. Panels A and B from Fig. 2 are in the plane of the membrane with the periplasmic face up. Panels C and D are rotated 90° to A and B to show the periplasmic face, and panels E and F are rotated a further 180° to show the cytoplasmic face. The 3D reconstructions show the ring structure of the LH-only complex like that of a LH1 complex from purple bacteria, and the central density of the RC in the RC–LH with the tetraheme cytochrome c subunit protruding into the periplasm. On the cytoplasmic face, there is less density than observed in RCs with an H subunit. The reconstructions were generated on the same scale allowing a size comparison. We observe that the LH-only is smaller than the RC–LH by about 10 %. This can be seen more clearly in Fig. 3a, where we have overlaid the RC–LH, mesh, on the LH-only, solid surface. This also shows that the cytoplasmic and periplasmic faces of the LH-only complex protrude less when the RC is removed.
3D models of RC–LH (left) and LH-only (right) complex from RFX. a, b Side view with the periplasmic side uppermost; c, d top view from the periplasmic side; e, f bottom view from the cytoplasmic side. Images were produced using UCFS Chimera (Pettersen et al. 2004)
Comparison of the RC–LH and LH-only with the RC–LH1 T. tepidum crystal structure. a Overlaid 3D EM reconstruction models of the LRHC (mesh) and LH-only (solid) complexes. b RC–LH of the RFX (mesh) and RC–LH1 of the T. tepidum (solid) with a spacefill representation, LH1 is in green, RC in purple, and the cytochrome is in red. The EM model of RC–LH of the RFX was condensed to match the overall shape of the RC–LH1 complex from T. tepidum. The threshold level of the 3D EM model was adjusted such that total volume matches its predicted molecular weight (Collins et al. 2010). This method was applied to EM 3D model of LH as well in a
In addition to the overall shape and conformation, the 3D reconstruction EM models can be used to calculate the resolution limits and dimensions of the complexes. Resolution calculations were carried out based on 0.5 FSC criteria and show that the RC–LH and LH-only models were determined to 14.6 and 19.4 Å resolution, respectively (Fig. S1A, B). Although these resolutions are still not high enough to resolve individual transmembrane helices, the 3D reconstruction models of RC–LH and LH-only complexes reveal interesting structural features. The LH complex embraces the RC to form a complete elliptical ring with a long axis of 15.3 nm and a short axis of 14.1 nm. Removing the RC changes the dimension of the LH complex; it is still a complete elliptical ring, but shrinks to have a long axis of 14.1 nm and a short axis of 12.6 nm, losing over a nanometer in both directions. The C subunit of the RC–LH protrudes from the periplasmic side. The total height measured from the cytoplasmic side of the RC–LH complex to the top of the C subunit is 10.7 nm, while the height of the LH ring in the RC–LH complex is about 7.5 nm. The height of the LH-only complex is 7.1 nm. There is a depressed area on the cytoplasmic side of the RC–LH complex, which in most of the core complexes from purple bacteria is occupied by the RC–H subunit that protrudes from the cytoplasmic side of the LH1 ring (Roszak et al. 2003; Qian et al. 2013; Niwa et al. 2014). The lack of protrusion on the cytoplasmic face confirms the lack of RC–H subunit or any type of replacement density in the core complex from RFX.
Figure 3 compares the 3D reconstruction of the RFX RC–LH with the crystal structure of RC–LH1 from T. tepidum (Niwa et al. 2014). The overall size and shape of the RFX RC–LH model and the T. tepidum crystal structure are a close match with the exception of the bound cytochrome (Fig. 3b). On the basis of the similar diameters, we propose that the LH surrounding the RC in RFX is composed of 16 α1β1bchl3 units in agreement with the number of LH α/β dimer pairs observed in T. tepidum structure and biochemically determined in RFX. The disparity in the cytochrome c subunit is that the RFX model EM density is a couple of nanometers shorter and almost appears folded over compared to the T. tepidum C subunit. This is likely to be due to the negative stain used and the imposition of rotational symmetry on the reconstruction. Despite the size discrepancy in the cytochrome subunit, we believe our 20 Å EM 3D reconstruction model is a good approximation of the size and shape of the RFX RC–LH.
Membrane isolation and AFM of membrane patches
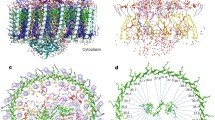
Photosynthetic RFX membranes from lysed cells were fractionated on a discontinuous sucrose density gradient. The red colored band from the gradient yielded a variety of membrane patches, and AFM of several such patches showed the presence of loosely packed proteins of distinct heights (Fig. 4a). Their separation as measured from the highest point to the highest point of the adjacent protein, and overall height as measured from the mica substrate (Fig. 4b) showed that there were at least two types of protein complexes present, the most numerous having a height of 11.1 ± 0.7 nm (SD), n = 106, and the second 13.7 ± 0.4 nm (SD) in height, n = 13. The height of the small proteins is consistent with derivations from single particle analysis of purified RC–LH of 11.0–11.5 nm (Collins et al. 2010) and from the EM 3D reconstruction model of 10.7 nm, where the C subunit is known to be compressed. Moreover, the height of the protrusion relative to the surrounding membrane was measured by AFM to be 3.9 ± 0.5 nm (SD), n = 20, which is compatible with the 4 nm height of the C subunit protrusion measured by Collins et al. (2010) and the 3.2 nm height difference in the 3D EM model. On the basis of these dimensions and the fact that this membrane patch came from a colored band having the absorbance spectrum of the RC–LH complex including the cytochrome, we assign these features as arising from the periplasmic face of the RC–LH complex in situ in the membrane.
Organization of the RFX putative photosynthetic membrane. a 3D representation of medium resolution AFM height data in false color where black is the mica substrate, full z = 18 nm. The patch has numerous protrusions in blue, average height above the mica 11.1 ± 0.7 nm SD, n = 106, assigned as putative RC–LHs. Similar sized protrusions in yellow may also be RC–LHs that are raised off the mica by an underlying piece of membrane, however these were not included in our assessment. b A scan line across three putative RC–LHs in the membrane, shown in blue on the inset, and their separations, highest point to highest point of 13.2 and 7 nm. c A cartoon highlighting the highly asymmetric position of the cytochrome subunit of R. castenholzii with vertical lines denoting the highest point of the complex. Depending upon the orientation of the complexes, the resulting separation would be as large as 14.7 nm or as small as 8.3 nm based on T. tepidum X-ray structure. d A cartoon representation of the organization of 4 RC–LHs based upon their measured separation, highest point to highest point, demonstrating that the complexes do not appear to be closely packed but do have an aligned orientation
There is an elevated section of the membrane which has similar proteins within it, which is likely to be due to an underlying piece of empty lipid membrane (Fig. 4a, yellow area). The overall height of these proteins was not measured; however, the height of the protrusions with respect to the surrounding membrane was measured as 3.9 ± 0.5 nm (SD), n = 12, and thus it seems very likely that these are also RC–LH complexes.
The identity of the larger proteins is more difficult to assign with confidence as there is no 3D structure available for the most likely candidate, ACIII, which functionally replaces the cytochrome bc 1 complex in Chloroflexi; this is addressed in the “Discussion” section.
Analysis of the spacing and packing patterns was done using a line scan and measuring the distances between the putative RC–LHs (smaller proteins) and the putative ACIIIs (larger proteins). A scan line across three putative RC–LHs in the membrane (Fig. 4b, blue line on the inset) reveals their separations, highest point to highest point of 13.2 and 7 nm (Fig. 4b). The average separation distance of 11.5 ± 2.4 nm (SD), n = 72, was determined for the clearly defined examples in the patches. The large fluctuation in average spacing distance is attributed to the tilted and off-center position of the periplasmic cytochrome subunit as it protrudes above the membrane. This notion is explored in Fig. 4c with a cartoon highlighting the highly asymmetric position of the C subunit in the RC–LH complex. Two complexes, based on the T. tepidum RCLH1 structure, are paired to show the maximum distance, 14.7 nm, if the C subunits were as far apart as possible and the minimum distance, 8.3 nm, if the C subunits were as close as possible. If the C subunits were vertically perpendicular to the membrane, we would expect a more consistent separation distance.
After the separation distance was measured, the patches were analyzed for any overall pattern in the arrangement of the smaller and larger proteins within the membrane. The smaller proteins are intermixed with the larger in a loosely packed and chaotic manner with no clear pattern discernible, but the smaller proteins did appear to cluster together. This apparent clustering, which is explored in the cartoon in Fig. 4d, is based upon their measured separation, highest point to highest point. This model demonstrates that the complexes do not appear to be closely packed, as in equivalent RCLH1 only purple bacterial membranes, but do have an aligned orientation. It appears that the RC–LHs are clustered such that their cytochrome subunits are aligned, supported by analysis of the measured average separation of the putative RC–LH complexes, which is 11.5 ± 2.4 nm SD, n = 72. This distance is too small for the known dimensions of the complex, average diameter 13.1 nm, but is compatible with an orientation where the cytochromes are proximal in any ‘pair’ of complexes, as explored in Fig. 4c. It also appears that these clusters of putative RC–LHs are congregated near the larger proteins, putative ACIIIs, which would promote efficiency of the electron transport function within the membrane. While no higher order definitive membrane packing patterns were observed, the number of complexes, putative identities, separation distances, and clustering pattern were determined.
Heme quantitation and membrane protein ratio determination
The ratio of RC–LH to ACIII in the photosynthetic membranes of RFX was determined using SDS-PAGE and TMBZ heme staining. This method was chosen because TMBZ selectively stains c-type hemes and the dominant three c-type heme containing proteins in the membranes of RFX are the tetraheme cytochrome c subunit of the RC, the ActA pentaheme, and ActE monoheme subunits of the ACIII. Therefore, the amount of RC–LH is represented by the cyt c band and the amount of ACIII is shown in the ActA and ActE bands. Based on the molecular weights and previous gels with protein identity confirmation by mass spectrometry (Collins et al. 2010; Gao et al. 2010), we can confidently identify proteins from the gel bands. RC cyt c runs at around 36 kDa, ActA about 25 kDa, and ActE at 20 kDa (Fig. 5a). It was evident from the TMBZ-stained gel that the three subunits we knew to contain c-type hemes are the only subunits with c-type hemes in the membrane at detectable levels. The image of the TMBZ-stained gel was analyzed in the Fiji-ImageJ Gel analysis program yielding intensity plots of each lane. The area under the intensity profile peaks was measured, which corresponds to the amount of protein in each band, and then plotted and found to increase linearly corresponding to the gel loading (Fig. 5b). Then the ratio of the areas was calculated; however, the ratio is approximate since TMBZ signal strength varies with peroxidase activity and each cytochrome has different levels of peroxidase activity (personal communication Robert Kranz). Since the gel was loaded with an increasing linear amount of sample, the calculated area of the intensity from all lanes should also linearly increase. Lanes that were not in the linear region due to overloading were not included in the ratio calculation. The ratio from all linear lanes was calculated and found to range from approximately 3 RC–LH to 1 ACIII for overloaded samples, similar to the RC–LH1 to cyt bc 1 ratio found in purple bacteria (Cartron et al. 2014) up to 10 RC–LH to 1 ACIII for not overloaded samples indicating a further disparity between this FAP and the purple bacteria.
Discussion
EM—RC–LH size, shape, and structure
We characterized the RC–LH and the detergent-derived LH complex by negative stain EM and single particle analysis, which clearly showed the periplasmically exposed bound cytochrome subunit (Fig. 1). The distinct protruding shape of this subunit allows for the determination of the orientation of the complex, namely the periplasmic and cytoplasmic faces as indicated in Fig. 2. The 3D reconstruction of both complexes, at ~20 Å resolution, allows us to superimpose the LH on the RC–LH to better appreciate this size difference, but it must be borne in mind that this is a symmetrized structure that averages out the slightly ovoid shape of the RC–LH and centers the asymmetrically positioned bound cytochrome (Fig. 3).
We have aligned the images in Fig. 2 making an assumption about the orientation of the LH complex based upon our ability to fit the LH 3D reconstruction within the bounds of the RC–LH reconstruction (Fig. 3a). In all three orientations shown in Fig. 2, the LH is smaller than the RC–LH for which there are two probable explanations. One, the loss of the RC is accompanied by damage to the ring upon removal. A feature of the LH-only data is the presence of partial rings (Fig. 1b), which are likely caused by the detergent removal of the RC during solubilization of the RC–LH. There are three classes of LH in the EM data that have clearly open, incomplete, rings. We consider these open rings to be due to partial solubilization of the LH during RC extraction, where 2 or more α1β1Bchl3 subunits have been removed, preventing a closed ring from reforming. The smaller closed rings of the LH could be the result of the loss of a single α1β1Bchl3, which does not prevent a closed ring reforming. The alternative is that there is a compaction of the LH after RC removal without the loss of an α/β pair simply because the RC is not there to force a certain shape or size as a physical mold but also from a loss of intermolecular interactions. This second explanation of the LH appearing smaller in the LH-only complex is the variability of inter-subunit spacings, as observed in the RC–LH1 crystal structures of related purple bacteria. Niwa et al. noted substantive variations in the inter-subunit spacings with a maximum difference of 1.9 Å between the α-polypeptides on the periplasmic face and 3.5 Å on the cytoplasmic face. If these distances were to minimize upon the removal of the RC, then the LH would indeed have a smaller diameter than the RC–LH and would ‘shrink’ on RC removal. On this basis, the bowl shape of the LH-only reconstruction also validates our orientation of the LH in Fig. 2. The partial rings observed in EM cannot be explained by this inter-subunit spacing loss mechanism, so it seems likely that the smaller observed size is the result of a combination of the two proposals, damage during solubilization and compaction.
A comparison of the RC–LH 3D reconstruction and the crystal structure of T. tepidum shows a close match in overall physical dimensions (Fig. 3b), which makes it likely that the LH ring is composed of 16 α1β1bchl3 subunits, in agreement with Collins et al. (2010) who derived a figure of 15 ± 1. The final volume of the EM 3D model of the RC–LH has been adjusted based on the comparison with the X-ray model of the RCLH1 from T. tepidum, from which the H subunit was removed for ease of comparison with the RFX complex. The density threshold levels of the EM 3D models of the RC–LH and LH-only complexes from RFX were adjusted based on their molecular masses (Collins et al. 2010). The RC cytochrome c subunit of RFX was observed by EM to be smaller than the corresponding subunit of T. tepidum; this difference could arise from the larger size of the T. tepidum subunit, which is about 80 amino acid residues longer than its RFX counterpart (Fig. 6c). Additionally, detergent and stain effects can increase the apparent size of 3D EM models (Timmins et al. 1988; Boonstra et al. 1993, 1994); a high-resolution method is needed for precise measurement of size and structure of this complex.
The amino acid alignment and structural dimensions of RFX RC–LH with the solved purple bacterial RC–LH1s allow us to make predictions about the arrangement of the pigments. FAPs and purple bacteria share type II reaction centers with symmetrical pigment arrangements. Crystal structures of purple bacterial RCs have indicated amino acid residues which are responsible for the type II pigment arrangements (Allen et al. 1987; Katona et al. 2003; Niwa et al. 2014). Based on amino acid sequence alignments, we predict the RC pigments of RFX to look like much like those of the purple bacterial systems (Fig. S2B, C). In purple bacteria, the RC pigments are four bacteriochlorophyll a (Bchl a) and two bacteriopheophytins (Bphe) (Deisenhofer et al. 1985; Sundström and van Grondelle 1995). RFX replaces one Bchl a with an additional Bphe (Collins et al. 2009). The other key difference in the RC of RFX versus that of purple bacteria is the lack of the H subunit. In our models, the H subunit has been removed from the purple bacteria to compare. The H subunit does not bind any pigments, but adds protein volume on the cytoplasmic face of the complex, which is absent in RFX.
We expect the RFX LH to differ more from the purple bacterial model than the RC as it has been described as a LH1/LH2 hybrid, binding two types of bacteriochlorophyll with absorbance maxima at 806 and 880 nm; thus the minimal unit may be described as α1β1bchl3. The T. tepidum purple bacterial LH1 system, in common with the archetypal purple bacterial LH1 complex, has a minimal unit consisting of α1β1bchl2, and further contains only one carotenoid per αβ-pair, while Rba. sphaeroides LH1 has two bound carotenoids per minimal unit (Broglie et al. 1980). The RFX LH complex has greater similarity to the archetypal purple bacterial LH2 complex, α1β1bchl3, suggesting that, structurally as well as spectroscopically, the RFX complex is a LH1/LH2 hybrid. In alignments with the purple bacterial LH systems, there are two conserved histidine residues in the β-polypeptide and one histidine in the α-polypeptide; the two closest to the C-terminus of both polypeptides likely form a ligand to the Mg of the bacteriochlorophyll a molecules, as seen in the crystal structures of the LH2 from Rhodopseudomonas acidophila and Phaeospirillum molischianum (REFS) (Fig. 6a, b). Since the pigment binding residues are conserved and there are three Bchl a molecules in the RFX LH system, we predict the pigments in the RFX LH system should have a ring of overlapping Bchl a parallel to the transmembrane helices with 2 Bchl a per αβ-pair as in purple bacterial LH1. A second ring perpendicular to the transmembrane helices of monomeric Bchl a bound in a manner similar to purple bacterial LH2 is also likely and has been indicated by CD spectroscopy (Collins et al. 2009). The prediction of excitonically linked overlapping Bchls and monomeric Bchl a rings is supported by the UV/Vis absorbance spectrum of the complex which has peaks at 880 and 800 nm for the LH and 850 for the RC, as well as linear dichroism spectra that indicate that the 880 and 800 nm pigments have significantly different orientations (Collins et al. 2009). Additional evidence from sequence also points to the LH1/LH2 hybrid nature of the RFX LH complex. When analyzed using BLAST, the RFX LH alpha and beta sequences aligned with sequences from purple bacterial LH1, LH2, and LH3 complexes. Linear dichroism spectroscopy of the WMs indicated that the B800 bacteriochlorophylls are at a large angle to the membrane, or in a similar angle as the carotenoids (Collins et al. 2009). This finding, along with a similar orientation in the analogous B808-866 antenna complex in Chloroflexus aurantiacus, suggests that the function of the 808 nm absorbing bacteriochlorophyll is to transfer energy from the chlorosome baseplate bacteriochlorophyll that has been proposed to be present in the ancestor of both groups of FAP bacteria (Xin et al. 2012).
The notion of the LH1/LH2 hybrid idea can also be examined in terms of kinetics of energy and electron transfer within the complexes. The similarity in predicted pigment arrangements within the RC and LH portions of the RFX and purple bacterial systems could account for the observed similarities in kinetic rates and conversion efficiencies measured in previous studies (Table S1). A notable feature of the purple bacterial RC is that it has 95 % yield of charge separation (Sener et al. 2007). This implies that nearly every time the RC special pair receives excitation energy, charge separation yielding electron movement is achieved. The yield of charge separation is comparable in RFX than in purple bacteria, which could result from the pigment substitution in the RC or any of the other differences between the systems (Collins et al. 2011). Even though it is less than in purple bacteria, the RC conversion efficiency in RFX is high.
The measured trapping time representing LH1 donating excitation energy to the special pair of the RC in both RFX and several purple bacterial species was 60 ps for open RCs and ~200 ps for closed RCs. This indicates the hybrid RFX LH complex is able to donate excitations to the RC at the same rate as a LH1 complex (Pullerits et al. 1994; Pullerits and Sundström 1996; Xin et al. 2012). In purple bacteria that naturally lack an LH2 complex such as Rhodospirillum rubrum, the rate was closer to 40 ps (Visscher et al. 1989). Although RFX does not have a peripheral LH2, the LH2 pigments are part of the RC–LH complex, which may explain why the RFX rates are closer to the purple bacterial systems with LH2 antennas. Bleaching of the special pair, lifetime of the P+H −A state, was also measured to be approximately 200 ps in RFX and purple bacteria (Kirmaier et al. 1985; Kirmaier and Holten 1987; Collins et al. 2011). Despite the substitution of the Bchl for a Bphe, the internal kinetics of the RCs are comparable. Similarly, the tetraheme subunit high potential heme could donate to the special pair on a very similar timescale, 480 ns lifetime of the excited special pair in RFX versus 460 ns in Blc. viridis (Ortega and Mathis 1993; Collins et al. 2011). The RC–LH kinetics indicate that there is not a significant difference in LH1 donation to the RC rates despite the variety of antenna systems in the FAPs and purple bacteria.
After charge separation in the RC, the electron is transferred to a second membrane protein complex that completes the cyclic ETC. The second protein is the cytochrome bc 1 complex in purple bacteria and the ACIII, in FAPs. Both cyt bc 1 and ACIII take electrons from quinol and give them to the soluble acceptor protein that shuttles electrons back to the RC. They also pump protons across the membrane to establish the electrochemical gradient. In order for the RC and whole ETC to operate smoothly, the proton pump/quinol oxidizing enzyme needs to turn over at a rate in line with the RC, contingent on the availability on quinone. The turnover rates of purple bacterial cyt bc 1 complexes, measured as rate constant k, range from 30 to 70 s−1 (Table S1). The turnover of the ACIII was in the middle of the rates measured for two purple bacterial cyt bc 1 complexes indicating that, even though the complexes are completely different, this side of the ETC turns over at nearly equal rates in FAPs and other bacterial systems (Guner et al. 1993; Gao et al. 2009).
Another important consideration in the efficiency of these RC–LH and ACIII systems is the ability of quinone to access the RC through the LH ring. Quinone channels have been observed in purple bacteria in three motifs: PufX, W, and closed. In some dimeric systems with PufX, the complexes are arranged as a ‘S’ shape with an opening in the LH1 ring for quinone diffusion to the cyt bc 1 complex; there is also an opening to allow quinone shuttling between each RC in the dimer (Qian et al. 2005, 2013; Comayras et al. 2005). However, PufX does not always promote dimer formation, such as the monomeric Rba. capsulatus system (Crouch and Jones 2012), but the same opening in the LH1 ring is expected to be present. The closed motif has been observed most recently by Niwa et al. (2014) who noted the presence of holes within the LH1 complex, which they proposed were the quinone diffusion channels with variable dimensions around the ring. The size of the RFX LH complex is closest to T. tepidum and it lacks the PufX polypeptide so even though our EM model is at a lower resolution, we believe the quinone channels probably follow the T. tepidum pattern. The EM models have allowed us to investigate the structure, energy, electron, and quinone flow of the RFX RC–LH.
AFM supramolecular organization
We used AFM and heme quantification to look at the supramolecular organization and electron flow of the major photosynthetic complexes in the membranes of RFX. While it is known that RFX does not have chlorosomes, there is an RC–LH complex that has many similarities to the well-studied purple bacterial RC–LH1 core complex, but it lacks an H subunit on the cytoplasmic face of the complex and has pigment substitutions as discussed above. The RC–LH complex alone would be sufficient to support photosynthetic growth in a manner similar to the purple bacteria such as Rhodospirillum rubrum and Blc. viridis. In AFM images of the latter photosynthetic membranes, large arrays of circular complexes with a protruding C subunit can be seen; however, these are much better resolved than the proteins in membrane pieces derived from RFX and it is clear that the Blc. viridis RC–LH1–Cyt complexes have little space between them (Scheuring et al. 2005, 2007). The RFX membrane shows a looser packing of RC–LH complexes, with an apparently empty lipid bilayer between some of them (Fig. 4). This loose packing has also been observed in the EM images of the related Chloroflexus aurantiacus membranes, which differs from RFX by the presence of a large membrane associated antenna, the chlorosome (Bína et al. 2014). The EM of CFX membranes showed an average RC center-to-center separation distance of 18 nm, larger than observed by AFM in RFX. In the CFX EM, some samples did not remove the chlorosomes and areas under the chlorosomes had tighter packing with 11 RC–LHs per chlorosome (Bína et al. 2014). The absence of the chlorosome or any other type of accessory antenna, either integral membrane or periplasmic, may account for the looser and random packing in the membranes of RFX. However, the resolution is not high enough to exclude the possibility of small proteins with little exposed topology interspersed among the RC–LH’s, which would also account for the loose packing observed, but cannot be imaged by AFM. We would expect many copies of the tethered electron carrier auracyanin, which has been proposed to be the equivalent of cyt c 2 in purple bacteria, to be present in this membrane. The highly flexible nature of the tether of auracyanin, estimated to be ~8.7 nm long, would not be easily detected by AFM but would cause the complexes to be further apart.
The average separation of the putative RC–LH complexes, 11.5 ± 2.4 nm, seems rather small when compared to the known diameter of the complex of 13.0 ± 1 nm (Collins et al. 2010). The measured values are reasonably close. We believe that this small disparity, where complexes appear too close or too far apart, is due to the fact that we are measuring separations on the basis of the highest point of the proteins, i.e., the C subunit, which we observed to be substantially off-center within the LH ring. Two C subunits ‘leaning’ toward each other would lead to a substantially shorter measured separation than a true center-to-center separation, and similarly, the C subunits ‘leaning’ directly away from each other would lead to a much greater perceived distance (Fig. 4c). Allowing for the unconstrained rotation of the largely circular complexes within the membrane could explain all the measured separations, two extremes of which are illustrated in the single scan line data of 13.2 versus 7 nm (Fig. 4). We have tested this hypothesis by modeling the separation, as measured by the highest point to the highest point, in a model made by placing two T. tepidum RC–LH1–Cyt 3D structures side by side in two configurations that place the off-center C subunits proximal and distal to each other for measurement purposes. The separations of 8.4 and 14.7 nm are indicative of the large variation that could be expected for in situ complexes of RFX and are illustrated in the cartoon in Fig. 4c. It should also be borne in mind that the downwards force exerted by the AFM is likely to influence the ‘lean’ of the C subunit by making it more horizontal and this will be reflected in the subsequent separations measured. The model was extended to illustrate how the orientation of four C subunits, seen in Fig. 4d, could match with their measured separations.
With knowledge from the EM model and related X-ray structures, the assignment of the smaller proteins in the membrane patch measured by AFM was assigned as the RC–LH complexes. The assignment of the larger protein is much more speculative, but we suggest that it is the ACIII complex, a functional analog of the cytochrome bc 1 or b 6 f complex in other photosynthetic organisms (for a review see Majumder et al. 2013). Although there is no atomic structure for any ACIII complex, Majumder et al. have proposed a homology-based model that uses the polysulfide reductase (PSR) of Thermus thermophilus, another member of the complex iron–sulfur molybdoenzyme (CISM) family (Jormakka et al. 2008). Taking a measurement of the height of the PSR complex from its PDB file (2VPW) yields a figure of 13.5 nm, which is within the measured height range of the larger proteins within the membrane patch of 13.7 ± 0.4 nm. The orientation of the ACIII complex within the membrane is currently debated but if it was as modeled in Majumder et al., then the ActB1 and ActB2 subunits would protrude into the periplasm and thus be imaged by the AFM (Majumder et al. 2013). Thus, at this time, we tentatively assign these taller proteins as ACIII complexes. We have therefore provisionally assigned the major integral membrane proteins player of the cyclic ETC in RFX.
To explore the validity of the ACIII assignment, the ratio of the RC–LH to the ACIII was determined. In the membrane patch illustrated in Fig. 4a, the ratio of putative RC–LH complexes to the taller putative ACIII complexes is 5.7:1. Heme staining data on WMs from RFX estimates the ratio of RC–LH to ACIII from the WMs closer to 10:1, but as low as 3:1 (Fig. 5). The 5.7 RC–LH to 1ACIII value from AFM is in the middle of the heme staining determined ratios suggesting that the numbers are in range of each other. In Rba. sphaeroides, the ratio of RC–LH1–PufX complexes to cyt bc 1 complexes for a low-light chromatophore has been estimated as 3:1 on the basis of quantitative mass spectrometry (Cartron et al. 2014). The larger ratio in RFX than that for purple bacteria may reflect the limited light-gathering capacity of the RC–LH complexes in comparison with that of Rba. sphaeroides photosystem which is augmented by large numbers of LH2 complexes. The ratio in purple bacterial LH2:RC is 2.7: 1, in a low-light chromatophore (Adams and Hunter 2012; Cartron et al. 2014). However, the RFX samples measured here were not grown in low-light conditions, so it is also possible that the larger ratio in RFX is due to growth in a higher light condition rather than a limited light absorbing capacity of the RC–LH complex. Since the RC–LH contains a LH1/LH2 hybrid antenna complex, the larger ratio in RFX may not be perceived as so large considering that it is doing the job of both the LH1 and the LH2 in one complex. The RFX RC–LH:ACIII ratio is larger than the purple bacterial RC–LH1:cyt bc 1 ratio, but the RFX ratio is not large compared to the combined purple bacterial LH2–RC–LH1:cyt bc 1 ratio. The membrane organization in the species adjusts to environmental conditions and the protein complex systems at the organisms’ disposal.
The enzyme kinetics observed for the ACIII and the cytochrome bc 1 complex are very similar to each other, as well as the kinetics within the RC–LH system (Hall et al. 1987; Gao et al. 2009) (Table S1). Similar kinetic rates of electron transfer are consistent with having a similar ratio of RC–LH/RC–LH1 and ACIII/bc 1 protein complexes. We have used our data to make a simple limited model for the photosynthetic membranes of RFX (Fig. 7).
Proposed supramolecular arrangement of RFX photosynthetic membrane system. View is looking down onto the top of the periplasmic face of the membrane. Teal and gray rings are the LH complex. Dark blue is the cytochrome subunit. Yellow is the M subunit and pink is the M subunit. Blue proteins are auracyanin. Tan is the homology model of ACIII based on polysulfide reductase
Conclusion
A 3D EM reconstruction model of the RFX was generated, which is similar to the X-ray structure of the T. tepidum RC–LH1 complex with the H subunit removed. Superposition of the X-ray structure on the EM model and amino acid sequence alignment yielded clues about pigment arrangement. It also provided size dimensions, particularly heights, which allowed us to assign the RC–LH complex to topographic features in the AFM images of RFX membrane patches. We were also putatively able to assign ACIII complexes from previous homology modeling and RC–LH to ACIII protein ratio was measured. Taken together, the measured membrane packing distances taken from AFM, shape from EM, and system kinetics from previous work indicate how the photosynthetic cyclic ETC operates within the membranes of R. castenholzii. We propose a model for how the RC–LH, ACIII, and auracyanin may arrange themselves in the membrane, as viewed from the periplasm (Fig. 7) and for the operation of this photosynthetic apparatus.
Abbreviations
- ACIII:
-
Alternative complex III
- AFM:
-
Atomic force microscopy
- Bchl:
-
Bacteriochlorophyll
- Bphe:
-
Bacteriopheophytin
- EM:
-
Electron microscopy
- FAP:
-
Filamentous anoxygenic phototrophs
- GSB:
-
Green sulfur bacteria
- RC–LH:
-
Light-harvesting reaction center complex in Roseiflexus castenholzii
- RC–LH1:
-
Light-harvesting reaction center 1 complex in purple bacteria
References
Adams PG, Hunter CN (2012) Adaptation of intracytoplasmic membranes to altered light intensity in Rhodobacter sphaeroides. Biochim Biophys Acta 1817:1616–1627
Allen JP, Feher G, Yeates TO et al (1987) Structure of the reaction center from Rhodobacter sphaeroides R-26: the protein subunits. Proc Natl Acad Sci USA 84:6162–6166
Bahatyrova S, Frese RNR, Siebert CA et al (2004) The native architecture of a photosynthetic membrane. Nature 430:1058–1062
Bína D, Gardian Z, Vácha F, Litvín R (2014) Supramolecular organization of photosynthetic membrane proteins in the chlorosome-containing bacterium Chloroflexus aurantiacus. Photosynth Res 122:13–21
Blankenship RE (2010) Early evolution of photosynthesis. Plant Physiol 154:434–438
Boonstra AF, Visschers RW, Calkoen F et al (1993) Structural characterization of the B800–850 and B875 light-harvesting antenna complexes from Rhodobacter sphaeroides by electron microscopy. Biochim Biophys Acta 1142:181–188
Boonstra AF, Germeroth L, Boekema EJ (1994) Structure of the light harvesting antenna from Rhodospirillum molischianum studied by electron microscopy. Biochim Biophys Acta 1184:227–234
Broglie RM, Hunter CN, Delepelaire P et al (1980) Isolation and characterization of the pigment-protein complexes of Rhodopseudomonas sphaeroides by lithium dodecyl sulfate/polyacrylamide gel electrophoresis. Proc Natl Acad Sci USA 77:87–91
Cao L, Bryant DA, Schepmoes AA et al (2012) Comparison of Chloroflexus aurantiacus strain J-10-fl proteomes of cells grown chemoheterotrophically and photoheterotrophically. Photosynth Res 110:153–168
Cartron ML, Olsen JD, Sener M et al (2014) Integration of energy and electron transfer processes in the photosynthetic membrane of Rhodobacter sphaeroides. Biochim Biophys Acta 1837:1369–1380
Cogdell R, van Grondelle R (2003) The light-harvesting system of purple bacteria. In: Green BR, Parson WW (eds) Light-harvesting antennas in photosynthesis. Advances in Photosynthesis, vol 13. Kluwer Academic Publishers, Dordrecht, pp 169–194
Collins AM, Xin Y, Blankenship RE (2009) Pigment organization in the photosynthetic apparatus of Roseiflexus castenholzii. Biochim Biophys Acta 1787:1050–1056
Collins A, Qian P, Tang Q, Bocian D (2010) Light-harvesting antenna system from the phototrophic bacterium Roseiflexus castenholzii. Biochemistry 49:7524–7531
Collins A, Kirmaier C, Holten D, Blankenship RE (2011) Kinetics and energetics of electron transfer in reaction centers of the photosynthetic bacterium Roseiflexus castenholzii. Biochim Biophys Acta 1807:262–269
Comayras F, Jungas C, Lavergne J (2005) Functional consequences of the organization of the photosynthetic apparatus in Rhodobacter sphaeroides. I. Quinone domains and excitation transfer in chromatophores and reaction center.antenna complexes. J Biol Chem 280:11203–11213
Crouch L, Jones M (2012) Cross-species investigation of the functions of the Rhodobacter PufX polypeptide and the composition of the RC–LH1 core complex. Biochim Biophys Acta 2:336–352
Deisenhofer J, Epp O, Miki K et al (1985) Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3 Å resolution. Nature 318:618–624
Gao X, Xin Y, Blankenship RE (2009) Enzymatic activity of the alternative complex III as a menaquinol:auracyanin oxidoreductase in the electron transfer chain of Chloroflexus aurantiacus. FEBS Lett 583:3275–3279
Gao X, Xin Y, Bell PD et al (2010) Structural analysis of alternative complex III in the photosynthetic electron transfer chain of Chloroflexus aurantiacus. Biochemistry 49:6670–6679
Gao X, Majumder EW, Kang Y et al (2013) Functional analysis and expression of the mono-heme containing cytochrome c subunit of alternative complex III in Chloroflexus aurantiacus. Arch Biochem Biophys 535:197–204
Goodhew CF, Brown KR, Pettigrew GW (1986) Haem staining in gels, a useful tool in the study of bacterial c-type cytochromes. Biochim Biophys Acta 852:288–294
Guner S, Willie A, Millett F et al (1993) The interaction between cytochrome c2 and the cytochrome bc1 complex in the photosynthetic purple bacteria Rhodobacter capsulatus and Rhodopseudomonas viridis. Biochemistry 32:4793–4800
Hall J, Zha X, Durham B et al (1987) Reaction of cytochrome c and c2 with the Rhodobacter sphaeroides reaction center involves the heme crevice domain. Biochemistry 26:4494–4500
Hanada S, Takaichi S, Matsuura K, Nakamura K (2002) Roseiflexus castenholzii gen. nov., sp nov., a thermophilic, filamentous, photosynthetic bacterium that lacks chlorosomes. Int J Syst Evol Microbiol 52:187–193
Hohmann-Marriott MF, Blankenship RE (2011) Evolution of photosynthesis. Annu Rev Plant Biol 62:515–548
Hu X, Ritz T (2002) Photosynthetic apparatus of purple bacteria. Q Rev Biophys 35:1–62
Jormakka M, Yokoyama K, Yano T et al (2008) Molecular mechanism of energy conservation in polysulfide respiration. Nat Struct Mol Biol 15:730–737
Katona G, Andréasson U, Landau EM et al (2003) Lipidic cubic phase crystal structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.35 Å resolution. J Mol Biol 331:681–692
Kirmaier C, Holten D (1987) Primary photochemistry of reaction centers from the photosynthetic purple bacteria. Photosynth Res 13:225–260
Kirmaier C, Holten D, Parson WW (1985) Temperature and detection-wavelength dependence of the picosecond electron-transfer kinetics measured in Rhodopseudomonas sphaeroides reaction centers. Resolution of new spectral and kinetic components in the primary charge-separation process. Biochim Biophys Acta 810:33–48
Liu H, Zhang H, Niedzwiedzki DM et al (2013) Phycobilisomes supply excitations to both photosystems in a megacomplex in cyanobacteria. Science 342:1104–1107
Majumder ELW, King JD, Blankenship RE (2013) Alternative Complex III from phototrophic bacteria and its electron acceptor auracyanin. Biochim Biophys Acta 11:1383–1391
Niedzwiedzki DM, Collins AM, LaFountain AM et al (2010) Spectroscopic studies of carotenoid-to-bacteriochlorophyll energy transfer in RC-LH photosynthetic complex from Roseiflexus castenholzii. J Phys Chem B 114:8723–8734
Niwa S, Yu L, Takeda K et al (2014) Structure of the LH1-RC complex from Thermochromatium tepidum at 3.0 Å. Nature 508:228–232
Ortega JM, Mathis P (1993) Electron transfer from the tetraheme cytochrome to the special pair in isolated reaction centers of Rhodopseudomonas viridis. Biochemistry 32:1141–1151
Pettersen EF, Goddard TD, Huang CC et al (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Pierson BK, Castenholz RW (1974) A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus, gen. and sp. nov. Arch Microbiol 100:5–24
Pugh R, McGlynn P, Jones M, Hunter C (1998) The LH1–RC core complex of Rhodobacter sphaeroides: interaction between components, time-dependent assembly, and topology of the PufX protein. Biochim Biophys Acta 1366:301–316
Pullerits T, Sundström V (1996) Photosynthetic light-harvesting pigment-protein complexes: toward understanding how and why. Acc Chem Res 8:381–389
Pullerits T, Visscher KJ, Hess S et al (1994) Energy transfer in the inhomogeneously broadened core antenna of purple bacteria: a simultaneous fit of low-intensity picosecond absorption and fluorescence kinetics. Biophys J 66:236–248
Qian P, Hunter C, Bullough P (2005) The 8.5 Å projection structure of the core RC–LH1–PufX dimer of Rhodobacter sphaeroides. J Mol Biol 349:948–960
Qian P, Bullough P, Hunter C (2008) Three-dimensional reconstruction of a membrane-bending complex the RC-LH1-PufX core dimer of Rhodobacter sphaeroides. J Biol Chem 283:14002–14011
Qian P, Papiz MZ, Jackson PJ et al (2013) Three-dimensional structure of the Rhodobacter sphaeroides RC-LH1-PufX complex: dimerization and quinone channels promoted by PufX. Biochemistry 43:7575–7585
Roszak AW, Howard TD, Southall J et al (2003) Crystal structure of the RC-LH1 core complex from Rhodopseudomonas palustris. Science 302:1969–1972
Scheuring S, Sturgis JN (2009) Atomic force microscopy of the bacterial photosynthetic apparatus: plain pictures of an elaborate machinery. Photosynth Res 102:197–211
Scheuring S, Lévy D, Rigaud J-L (2005) Watching the components of photosynthetic bacterial membranes and their in situ organisation by atomic force microscopy. Biochim Biophys Acta 1712:109–127
Scheuring S, Boudier T, Sturgis J (2007) From high-resolution AFM topographs to atomic models of supramolecular assemblies. J Struct Biol 159:268–276
Sener MK, Olsen JD, Hunter CN, Schulten K (2007) Atomic-level structural and functional model of a bacterial photosynthetic membrane vesicle. Proc Natl Acad Sci USA 104:15723–15728
Sturgis JNJ, Niederman RRA (2008) Atomic force microscopy reveals multiple patterns of antenna organization in purple bacteria: implications for energy transduction mechanisms and membrane modeling. Photosynth Res 95:269–278
Sturgis JN, Tucker JD, Olsen JD et al (2009) Atomic force microscopy studies of native photosynthetic membranes. Biochemistry 48:3679–3698
Sundström V, van Grondelle R (1995) Kinetics of excitation transfer and trapping in purple bacteria. Anoxyg Photosynth Bact 2:349–372
Takaichi S, Maoka T, Yamada M et al (2001) Absence of carotenes and presence of a tertiary methoxy group in a carotenoid from a thermophilic filamentous photosynthetic bacterium Roseiflexus castenholzii. Plant Cell Physiol 42:1355–1362
Tang G, Peng L, Baldwin PR et al (2007) EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46
Tang K-H, Barry K, Chertkov O et al (2011) Complete genome sequence of the filamentous anoxygenic phototrophic bacterium Chloroflexus aurantiacus. BMC Genomics 12:334
Timmins PA, Leonhard M, Weltzien HU et al (1988) A physical characterization of some detergents of potential use for membrane protein crystallization. FEBS Lett 238:361–368
Tsukatani Y, Nakayama N, Shimada K et al (2009) Characterization of a blue-copper protein, auracyanin, of the filamentous anoxygenic phototrophic bacterium Roseiflexus castenholzii. Arch Biochem Biophys 490:57–62
Tucker JD, Siebert CA, Escalante M et al (2010) Membrane invagination in Rhodobacter sphaeroides is initiated at curved regions of the cytoplasmic membrane, then forms both budded and fully detached spherical vesicles. Mol Microbiol 76:833–847
Van der Meer MTJ, Klatt CG, Wood J et al (2010) Cultivation and genomic, nutritional, and lipid biomarker characterization of Roseiflexus strains closely related to predominant in situ populations inhabiting Yellowstone hot spring microbial mats. J Bacteriol 192:3033–3042
Van ver Meer MTJ, Schouten S, Hanada S et al (2002) Alkane-1,2-diol-based glycosides and fatty glycosides and wax esters in Roseiflexus castenholzii and hot spring microbial mats. Arch Microbiol 178:229–237
Visscher KJ, Bergström H, Sundström V et al (1989) Temperature dependence of energy transfer from the long wavelength antenna BChl-896 to the reaction center in Rhodospirillum rubrum, Rhodobacter sphaeroides (w.t. and M21 mutant) from 77 to 177 K, studied by picosecond absorption spectroscopy. Photosynth Res 22:211–217
Xin Y, Pan J, Collins A et al (2012) Excitation energy transfer and trapping dynamics in the core complex of the filamentous photosynthetic bacterium Roseiflexus castenholzii. Photosynth Res 111:149–156
Yamada M, Zhang H, Hanada S et al (2005) Structural and spectroscopic properties of a reaction center complex from the chlorosome-lacking filamentous anoxygenic phototrophic bacterium Roseiflexus castenholzii. J Bacteriol 187:1702–1709
Acknowledgments
This material is based upon work supported as part of the Photosynthetic Antenna Research Center (PARC), an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Award Number DE-SC 0001035; EL-WM was supported by an Olin Fellowship for Women and a PEO Scholar Award.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Majumder, E.LW., Olsen, J.D., Qian, P. et al. Supramolecular organization of photosynthetic complexes in membranes of Roseiflexus castenholzii . Photosynth Res 127, 117–130 (2016). https://doi.org/10.1007/s11120-015-0179-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-015-0179-9