Abstract
Annual loss of productivity of the important crop legume chickpea has received prime scientific concern at recent times. Vascular wilt caused by fungal pathogen Fusarium oxysporum f. sp. ciceris race 1 (Foc1) accounts for major share of yield loss of chickpea. Control of this disease remains a challenge due to the lack of appropriate breeding programs to manage fast pathogen mutability. Previous studies with this pathogen have highlighted the role of reactive oxygen species (ROS) as chemical signal in enkindling downstream systemic resistance response instead of activating site specific defense. But the role of salicylic acid in modulating resistance is still unexplored. Present study explains the probable function of salicylic acid (SA) in coordination with ROS. The external SA application reveals the restoration of relative water content of infected susceptible chickpea plants. The qRT-PCR based expression study of key SA biosynthetic genes indicate that the SA biogenesis takes place by the activity of phenylalanine ammonia lyase (PAL) that activates other SA responsive genes and TGA transcription factors to induce an active defense against Foc1. Finally, detection of SA by LC MS/MS along with the accumulation of transcripts of SA marker genes, PR1 and PR5, strengthens the involvement of SA in translocation of distant systemic signals in chickpea-Foc1 interaction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant immunity lacks mobile defender cells but the well orchestrated signal transduction mechanism makes them potent combatant against a wide array of pathogens. Additionally, plants show the phenomenon of SAR (systemic acquired resistance) wherein the encounter of a plant organ with the pathogen results in local hypersensitive response followed by transduction of signal to the nearby plant parts so that the secondary infections could be ceased. Recent findings not only strengthened this long distance signaling events in plant (SAR), but also established the development of a short term memory against the infestation of a particular type of pathogen, known as “priming” (Aranega-Bou et al. 2014). These systemic responses largely depend upon hormonal cross talk and interactions of small molecules which ultimately promote a broad spectrum resistance phenomenon in plants (Shah et al. 2014).
Small molecules are generally chemical substances synthesized by plants to execute signaling events. Pipecolic acids (PiP), dihydroabietinal (DA), azelaic acid (AzA) etc. are reported recently to be the important small molecules acting as systemic defense inducers (Dempsey and Klessig 2012). SFD1/GLY1 (suppressor of fatty acid desaturase 1 alias GLY1)-derived glycerol 3 phosphate protein (G3P) and defective in induced resistance 1 (DIR1) are also found to function in coordination to impart SAR (Shah et al. 2014). Besides, another important small molecule which has long been known as key regulator of SAR response in plants is salicylic acid (SA). It is a plant stress hormone that imparts resistance by modulating the defense pathways in varied ways (Kumar 2014).
SA biosynthesis is grossly dependent on the expression of phenylalanine ammonia lyase (PAL), which is a part of phenylpropanoid pathway. PAL provides precursors for lignin biosynthesis and several other secondary metabolites that play critical role in host defense signaling (Sticher et al. 1997). SA also induces reactive oxygen species (ROS) generation and subsequent oxidative burst which could further induce several defense signaling pathways (Choi et al. 2017).
SA responses are associated with induction of pathogenesis-related (PR) proteins (Selitrennikoff 2001). Among several PRs, PR1 is the marker that is induced during SA response in most of the plants, also known as salicylic acid marker (Meldau et al. 2014; Lemos et al. 2016). However, other PR proteins like chitinase (in cucumber) or PR5 are also found to be accumulated during SAR on pathogen infection. For example, Puccinia triticina, a leaf rust pathogen is shown to induce characteristic expression of PR5 gene upon infection in wheat leaf tissues (Li et al. 2015).
Studies involving SA mediated signaling during pathogen infections are rarely studied on chickpea plants. Chickpea is one such important crop legume in India that meets (10 to 15%) annual yield loss due to wilting caused by Fusarium oxysporum f. sp. ciceris race 1 (Foc1), and in severe condition, it escalates up to 90% loss in productivity. Foc1 enters the plant through breaches of underground root system and colonizes themselves in the xylem vessels of the host plant causing complete chocking of water flow through roots. Several works have been carried out on this plant-pathogen interaction to unveil the hidden clues behind the defense pathway. It has been established that reactive oxygen species (ROS) is important defense signaling molecule in this host pathogen interplay (Gupta et al. 2013; Gupta et al. 2017). Another study had confirmed the connection of oxidative burst with the subtle induction of pathogenesis-related proteins in chickpea upon Foc1 endeavor (Chatterjee et al. 2014). Although, induction of pathogenesis-related protein (PR proteins), particularly PR1 directs clear indication towards long distance signaling via salicylic acid pathway but detailed work on systemic defense response is largely missing in chickpea-Foc1 interaction.
The present study focus on expression analyses of some salicylic acid responsive transcripts after Foc1 invasion through real time PCR in both compatible and incompatible interaction. Transcript accumulations of SA biosynthetic genes and temporal expressional patterns of some SA responsive pathogenesis-related proteins were also examined on a time scale upon infection. Some physiological parameters like relative water content (RWC) and percentage of disease incidence were studied with and without external SA application in susceptible and resistant chickpea lines. Finally, the endogenous SA was detected through LC MS/MS. Thus, entire study demonstrates the probable role of salicylic acid in mediating defense in chickpea upon Foc1 infection in control and SA-treated plants of both susceptible and resistant lines.
Materials and Methods
Maintenance of the Plant and Pathogen
Experiments were performed with seeds of two different lines of chickpea (Cicer arietinum), JG62 (wilt susceptible) and WR315 (wilt resistant), obtained from Indian Institute of Pulses Research (IIPR), Kanpur, Uttar Pradesh, India. Seeds of both cultivars were sown in small plastic pots filled with sterilized sand and synthetic soil at a ratio of 1:1. The plantlets obtained were maintained under natural optimum greenhouse conditions at 22 to 28 °C and 35 to 40% relative humidity and photoperiod of 16:8 h day and night (Haware and Nene 1982). Watering was done on alternate days by sprinkling.
F. oxysporum f. sp. ciceris race 1 (Foc1) culture was obtained from ICRISAT and subsequently grown on half-strength potato dextrose agar (PDA) plates (Himedia Laboratories, Mumbai, India) (Summerell et al. 2003) and incubated in complete darkness at 25 to 28 °C for 12 to 14 days. The fungal plate with mycelial mat was then flooded with sterile water (10 ml) and gently scrapped with a sterile spatula. The spore suspension was collected and stored at − 80 °C with 30% sterile glycerol for further use.
In planta Fungal Infection
Sterilized seeds of both JG62 and WR315 were sown in autoclaved sand-soil mixture (1:1) and allowed to germinate in optimum conditions required for chickpea. Twelve- to fifteen-day-old seedlings were infected with Foc1 as described by Gupta et al. (2009). Mock-inoculated plants served as controls. Both control and infected plants were maintained under uniform greenhouse conditions. Root and shoot tissues were harvested from both control and infected (1DPI, 2DPI, 3DPI, 4DPI, and 7DPI) plants of wilt susceptible (JG62) and wilt resistant (WR315) chickpea lines for the entire study.
External Treatment with Salicylic Acid
The well-grown chickpea seedlings (14 days) of both JG62 and WR315 were grouped into two batches (each batch comprises twenty plants belonging to each time points of both the varieties). One group of plants were subjected to external foliar spray with 200 μM of salicylic acid prior to the infection procedure where as another group no external application of SA has been performed (Mandal et al. 2009). Mock inoculated control plants were also treated with similar manner.
Determination of Percentage of Disease Incidence (PDI)
The percentage of disease incidence were calculated from external morphological changes, e.g., yellowing of leaves, drooping of shoot tissues, and morphology of root system etc. in both salicylic acid-treated and non-treated group of susceptible and resistant chickpea lines following the formula of percentage of disease incidence (PDI) = (total number of infected plants/total number of plants assessed) × 100 (Larkin and Fravel 1998).
Determination of Relative Water Content
Relative water content of control and infected chickpea plants (both SA untreated and treated of both lines JG62 and WR315) were measured weighing method followed by Barrs and Kozlowski 1968 with modifications. The entire plants were taken for the study. Firstly, fresh weights of the entire plants were measured and recorded (FW) then the plantlet roots were allowed to soak water by emerging the plants into the water for about 6 h. After 6 h of complete soaking, the weights of the plantlets were again recorded as turgid weight (TW). The plants were then subsequently oven dried at 70 °C for 72 h. The dried plants were finally weighed and recorded as dry weight (DW). RWC was calculated following the formula, RWC = (FW−DW)/(TW−DW).
RNA Isolation, cDNA Preparation and qRT-PCR
Roots of uninfected and infected plants of both varieties (JG62 and WR315) were collected at 1DPI, 2DPI, 3DPI, 4DPI, and 7DPI and flash frozen in liquid nitrogen. Total RNA was isolated using TRI reagent kit (Sigma-Aldrich) according to the manufacturer’s guidelines. RNA was quantified using Nanodrop Spectrophotometer at 260 nm wavelength. Qualitative assessment was done by running 5 μg of total RNA in 1.2% formaldehyde gels.
The 5 μg mRNA was used and first strand mixture prepared using first strand cDNA synthesis kit (Fermentas, USA) according to the manufacturer’s guidelines. Prepared cDNA was quantified in Nanodrop Spectrophotometer at 260 nm wavelength of light. Then cDNA samples were resolved in 1.2% agarose gel. The prepared cDNA was stored at − 80 °C for further use.
To identify the systemic defense response in chickpea on Foc1 infection several salicylic acid responsive genes were selected and categorized. The genes known to be modulated during SA-induced response in different plant-pathogen interactions were searched in databases, e.g., Chickpea Trancriptome Database (CTDB, http://www.nipgr.res.in/ctdb.html), National Centre for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov), the Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org), and Medicago truncatula DFCI Gene Index [http://knowpulse2.usask.ca/portal/project/Medicago-truncatula-DFCI-Gene-Index-(version-11)]. However, the matches found in the CTDB (http://www.nipgr.res.in/ctdb.html) were given the most priority in selecting out the genes. All the selected genes were categorized under five major groups: (a) genes involved in salicylic acid biosynthesis, (b) genes involved in conversion of free SA to SA conjugate, (c) genes involved in SA signal transduction pathway, (d) SA responsive transcription factors, and (e) SA marker genes. The details of each of these categories were discussed in Table 1.
Quantitative real time PCR was performed on a BioRad iCycler (http://www.biorad.com/) using SyBr Green technology. Reaction mix (20 μl) containing SyBr green, qPCR Super Mix (2×) (Bio Rad), 25 ng cDNA, and 0.3 mM of forward and reverse primers was taken for PCR. The following cycle conditions were used: 95 °C for 5 min, followed by 40 cycles at 95 °C for 30 s, 50–55 °C for 30 s, and 72 °C for 30 s (Gupta et al. 2010). Primer sequences are provided in the S1 Table. Melt curve analyses were done to determine the primer specificities. Variations in cDNAs of the samples were normalized using actin as an internal control. The relative fold changes were calculated using the 2-δδCt method normalized against mock-inoculated control plants (Livak and Schmittgen 2001). All experiments were conducted in triplicates. Standard deviation and standard error were calculated for each transcript. Unpaired student’s t test was performed and p value (< 0.05) calculated for entire data set (Table S3).
Isolation and LC-MS/MS Analyses of SA from Chickpea Plants
SA was extracted from shoot and root tissues from infected and uninfected control plants of both susceptible (JG62) and resistant (WR315) chickpea plants. The isolation was performed following the protocol summarized by Pan et al. 2010 with some suitable modifications. Shoot as well as root tissues of uninfected control and infected (2DPI) chickpea plants were collected in 2 g aliquots, quick frozen, and crushed in a mortar and pestle in presence of liquid nitrogen. The powder was suspended with the extraction solvent containing 2-propanol/water/concentrated HCl (2:1:0.002). The slurry was transferred into a 250 ml conical flask and allowed to continuous agitation in a rotary shaker at 100 rpm, 4 °C for overnight. On the next day, equal volume of dichloromethane was added to each flask and allowed to shake again for 2 h at 4 °C. The slurry was then transferred to a centrifuge tube and centrifugation done at 13000 g, 4 °C for 10 min. After centrifugation, lower organic phase was collected and dried in the continuous flow of N2 gas in a nitrogen evaporator. The samples were re-dissolved in 0.5 ml of HPLC grade methanol followed by centrifugation at 15000 g for 10 min at 4 °C. The upper clear supernatant was collected and used further for detection of SA by liquid chromatography-mass spectrometry (LC-MS/MS).
The extracted phytohormone was subjected to LC-MS/MS system integrated with a Shimadzu UFLCXR HPLC connected to an API 5500 Qtrap mass analyzer (AB Sciex, Toronto, Canada). Phytohormone detection and analysis was performed in multiple reactions monitoring (MRM) mode with positive and negative polarity of electro spray ionization (ESI). Molecular ions were detected on [M + H] + in positive ionization mode and [M−H] - in negative ionization mode. Salicylic acid were analyzed in negative ionization mode with a continuous scan time of 60 ms. The chromatographic separation was performed using the mobile phase composed of (solution A) methanol:water:formic acid (99:900:1 v/v/v) and (solution B) methanol: water: formic acid (900:99:1 v/v/v). The oven temperature was set at 25 °C with the continuous flow rate of 0.4 ml min−1. The detected SA concentration was expressed as nano gram SA (ng)/ml solution.
Statistical Analysis
All the experiments were conducted in triplicates and statistical errors and standard deviation (SD) were calculated. The graph represented as mean value ± SD. The statistical analyses were performed through Excel Stat and GraphPad Prism (version 5.0) software. Significance of each data was tested by one-way ANOVA followed by Tukey’s multiple comparison test as a part of post-hoc analysis (S2 Table). Bland-Altman method of comparison was done for LC MS/MS data. Average and percentage (%) difference were calculated between control and infected (2DPI) plants. Finally, bias of SD within the data sets was also measured with 95% limits of agreement (S2 Table). The significant differences among quantitative real time PCR data set was evaluated by unpaired student’s t test at 5% probability level (p < 0.05) (S3 Table).
Results
SA Treatment Minimizes the Disease Incidence in Susceptible (JG62) Plants
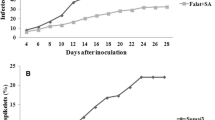
The susceptible (JG62) plants exhibit gradual morphological manifestation of disease severity from 2DPI to 12 DPI as compared to control plants (Fig. 1a). The clear chlorosis and degeneration of root system have been observed in susceptible chickpea plants at 12DPI (Fig. 1b, c) whereas resistant (WR315) lines were devoid of such instances and most of the plants exhibit healthy growth even after 12DPI (Fig. 1d). The JG62 plants when treated with external foliar spray with SA were shown to regain health. The SA untreated infected JG62 plants were completely drooped down at 7DPI (Fig. 1e); however, when treated with external SA, many of them exhibit sturdy nature (Fig. 1f, g). The PDI showed to be high in SA-untreated JG62 [JG62 (−SA)] plants in each time points with the progression of the disease but the same was significantly reduced when treated with external SA [(JG62 (+SA)]. Similarly, in WR315 plants SA application [WR315 (+SA)] reduces PDI as compared to SA untreated WR315 [WR315 (−SA)] plants that exhibit some disease symptoms at 4DPI, but the reduction of disease incidence is not as pronounced as in case of JG62 plants exposed to SA (Fig. 1h).
External morphology of susceptible (JG62) and resistant (WR315) plants. a External morphological changes due to Foc1 infection in JG62 plants in different time points. b Close view of yellowing of leaves in JG62 plants at 12DPI. c Close view of altered root morphology in JG62 plants at 12DPI. d External morphology of WR315 plants in control and at 12DPI. e Drooped down JG62 plant at 7DPI (f) SA-treated JG62 plant at 7DPI. g Root morphology of SA treated JG62 plant at 7DPI. h Percentage of disease incidence in JG62 and WR315 plants in both SA treated and untreated conditions at different time points
SA Treatment Restores Relative Water Content in Infected JG62 Plants
Relative water content was tested to understand the percentage amount of retained water within the plant body. It has been observed that RWC is gradually lowered with the progression of the disease in case of susceptible (JG62) plants. The RWC was also found to be reducing in resistant plants but the level was not as significant as susceptible ones. The susceptible plants which were treated externally with SA [JG62 (+SA) were shown to restore the RWC significantly as compared to JG62 (−SA) (Fig. 2a, b, c).
Quantitative Real Time PCR (qRT PCR)-Based Gene Expression Analyses of Selected Genes
Transcriptional reprogramming is a prevalent consequence of any plant microbe interaction. In chickpea-Fusarium interaction, it has been established previously that reactive oxygen species acts as a signaling molecule to induce defense responses in chickpea by modulating transcription factors and cellular transporters (Gupta et al. 2013). Additionally, it was also evident that root infection by Fusarium oxysporum f. sp. ciceris causes severe effect on photosynthetic genes in chickpea (Bhar et al. 2017). These two findings set the path to find out the key long distance signaling event operating in this host pathogen interplay. Salicylic acid is a customary biotic stress hormone that is well known for its systemic defense response. In chickpea-Fusarium interplay, the role of this important defense hormone is largely illusive. Phenylalanine ammonia lyase and isochorismate synthase (ICS) are the two major SA biosynthetic genes in plants. SA glucosyl transferase (SAGT) and SA methyl transferase (SAMT) are the genes involved in conversion of free SA to its glucoside or methyl derivatives. This conversion is very much important for SA mediated systemic defense response pathway. The ratio of free and conjugated form of SA actually controls the signaling event properly. Enhanced disease susceptibility 1 (EDS1), phytoalexin deficient 4 (PAD4), ethylene insensitive 2 (EIN2), and non-repressor of pathogenesis-related protein 1 (NPR1) are the principal genes in SA signaling pathway involving various metabolic pathways. TGA transcription factor 1 (TGA1), TGA transcription factor 3 (TGA3), and TGA transcription factor 6 (TGA6) are well known for their transcriptional control of different genes in SA-induced conditions, particularly activation of SA marker genes pathogenesis-related 1 (PR1) and pathogenesis-related 5 (PR5) by binding with TGA binding sites. The expression analyses of these genes will definitely give an insight into systemic defense signaling pathway in this plant-pathogen interplay.
Expression of Key Genes Involved in Salicylic Acid Biosynthesis
In shoot tissues of susceptible (JG62) plants, PAL expression level was found to be downregulated throughout the time points after infection, but resistant plants showed expressional inductions. In resistant plants, PAL expression gradually increases from 1DPI to 7DPI in stepwise pattern. ICS expression steeply downregulated for susceptible plants throughout the post infection time points. Resistant plants show gradual upregulation till 3DPI in which it shows highest expressional peak. Then the expression drops to some extent at 4DPI and suddenly fall down to basal level at 7DPI (Fig. 3 a, b).
Relative expressions of genes involved in salicylic acid biosynthesis. a, c represent the expression of phenyl alanine ammonia lyase (PAL) in shoot and root tissues of susceptible (JG62) and resistant (WR315) plants. b, d represent the expression of isochorismate synthase (ICS) in shoot and root tissues of susceptible (JG62) and resistant (WR315) plants. The relative fold changes are normalized against mock-inoculated control plants and bar represents standard error (n = 3, p < 0.05)
Root tissues of susceptible (JG62) chickpea plants exhibit random expression for PAL gene. The gene was slightly upregulated at 2DPI and 7DPI but shows downregulation at 1DPI, 3DPI, and 4DPI. In resistant plants, expression of PAL was thoroughly upregulated throughout the time points. The expression was gradually geared up from 2DPI to 7DPI, with expressional induction at 7DPI. ICS expression did not follow a particular pattern for both susceptible and resistant plants. In susceptible plants, ICS induced largely at 1DPI then it sharply falls at 2DPI and gradually recovered till 4DPI and at 7DPI it again impelled slightly. In resistant plants, ICS was downregulated at 1DPI, which then induced at 2DPI and increased till 3DPI with highest expressional value. Then at 4DPI time point the expression of ICS fell down for resistant plant that peaked up again at 7DPI (Fig. 3c, d).
Expression of Genes Involved in Conversion of Free Salicylic Acid to Salicylic Acid Conjugate
In shoot tissues, SAGT expression showed positive induction in both susceptible and resistant plants, but expressions in resistant plants were quiet higher than that of susceptible plants. Susceptible plants show an expressional peak at 2DPI which then gradually fall till 7DPI. In resistant plants, SAGT expression gradually increases with the progression of disease with the expressional height at 7DPI. In case of susceptible plant, SAMT is significantly downregulated throughout the progression of the disease. On the contrary, resistant plants exhibit a higher expressional induction of SAMT throughout the time points. SAMT expression maintains more or less steady state induction throughout the post infection stages except at 2DPI where it steps down slightly (Fig. 4a, c).
Relative expressions of genes involved in conversion of free SA to SA conjugate. a, b represent the expression of SA glucosyl transferase (SAGT) in shoot and root tissues of susceptible (JG62) and resistant (WR315) plants. c, d represent the expression of SA methyl transferase (SAMT) in shoot and root tissues of susceptible (JG62) and resistant (WR315) plants. The relative fold changes are normalized against mock-inoculated control plants and bar represents standard error (n = 3, p < 0.05)
In case of roots SAGT, the pattern was largely same as SAMT, at least in case of susceptible plants. In resistant plants at 1DPI post-inoculation time point, the degree of SAGT was very low but was gradually increased and shows slight expressional induction at 3DPI. The expression then further increases and exhibits a moderate upregulation at 7DPI. The expression of SAMT in root tissues of both susceptible (JG62) and resistant (WR315) chickpea plants was highly downregulated, but the level of expression in susceptible (JG62) plants was reduced to great extent as compared to resistant plants. In resistant (WR315) plant, although the expression level was largely downregulated, it was further recovered at the later time points (4DPI and 7DPI) (Fig. 4b, d).
Genes Involved in Salicylic Acid Signal Transduction Pathway
In shoot tissues of susceptible plant, all the genes involved in SA signaling exhibit sharp downregulation throughout the time points except at 2DPI. At 2DPI, all the genes exhibit upregulation with the highest induction of EIN 2. NPR 1 and EDS 1 showed moderate expression and PAD 4 induced slightly. On the contrary, all the genes were steeply upregulated in resistant (WR315) plants at all the time points except 7DPI, where NPR 1 exhibit downregulation. NPR 1 shows highest expressional induction at 3DPI and 4DPI. PAD 4, EDS 1, and EIN 2 induced at 3DPI and 4DPI. PAD 4 and EDS 1 maintained their expression at these two time points but gradually fell down with the progression of disease. EIN 2 expression fell gradually from 3DPI to 7DPI (Fig. 5a, b).
Relative expressions of genes involved in SA signal transduction pathway. a, b represent the expression of non-repressor of pathogenesis-related 1 (NPR1), phytoalexin deficient 4 (PAD4), enhanced disease susceptibility 1 (EDS1), and ethylene insensitive 2 (EIN2) in shoot tissues of susceptible (JG62) and resistant (WR315) plants. c, d represent the expression of non-repressor of pathogenesis-related 1 (NPR1), phytoalexin deficient 4 (PAD4), enhanced disease susceptibility 1 (EDS1), and ethylene insensitive 2 (EIN2) in root tissues of susceptible (JG62) and resistant (WR315) plants. The relative fold changes are normalized against mock-inoculated control plants and bar represents standard error (n = 3, p < 0.05)
In roots of susceptible JG62 plants, NPR 1, PAD 4, EDS 1, and EIN 2 exhibited a bit mixed expressional pattern. NPR 1 possessed a continuous upregulation throughout the post infection time points, with a highest expression at 1DPI. The expression of this gene showed a sudden drop at 2DPI which was maintained at moderate stage till 4DPI, then the expression was again boosted up at 7DPI. PAD 4 exhibited largely downregulation throughout the infection time points except 2DPI and 4DPI where expression was slightly geared up. EDS 1 showed expressional undulations with the highest expressions at 1DPI and 7DPI. At 2DPI, EDS 1 level was sharply fallen and then gradually recovered till 4DPI. EIN 2 was downregulated at 1DPI, which induced at 2DPI, fell down at 3DPI to 4DPI, and then again upregulated at 7DPI. In resistant WR 315 plants, most of the genes in this category exhibited upregulation except EIN 2. NPR 1 level was highest at 2DPI and 7DPI. From 3DPI to 4DPI, NPR 1 showed moderate expression but EDS 1 although expressed moderately at 3DPI sharply downregulated at 4DPI. PAD 4 exhibited a basal expression at 1DPI but the level of expression geared up at 2DPI and maintained throughout the post-infection time points (Fig. 5c, d).
Expression Pattern of SA Responsive Transcription Factors
In shoots, TGA 1 and TGA 6 were downregulated throughout the post infection time points in case of susceptible chickpea line, but resistant plants showed upregulation of these two transcription factors. Although, TGA 1 was slightly downregulated at 1DPI but it gradually upregulated with the progression of disease. TGA 6 expression was upregulated in case of resistant plants in all the time points which gradually increased with the progression of the disease. TGA 3 exhibited a mixed expressional pattern where initially slight expressional induction was noticed in case of susceptible plants, but it gradually downregulated in the later stages of infection. TGA 3 was faintly downregulated at 1DPI but gradually increased up to 3DPI and then again fell down with the later time of infection (Fig. 6 a, c, e).
Relative expressions of SA responsive transcription factors. a, c, e represent the expression of TGA1, TGA3, and TGA6 in shoot tissues of susceptible (JG62) and resistant (WR315) chickpea plants. b, d, f represent the expression of TGA1, TGA3, and TGA6 in root tissues of susceptible (JG62) and resistant (WR315) plants. The relative fold changes are normalized against mock-inoculated control plants and bar represents standard error (n = 3, p < 0.05)
In roots, the pattern of expression of TGA 1 transcription factor was susceptible and resistant plants was converse. In the initial time points of infection TGA 1 expression in susceptible plants was upregulated that gradually fell with the progression of the disease. Contrarily, resistant plants TGA 1 was downregulated at initial time points that gradually boosted up with the progression of the disease and finally exhibited a peak expression at 7DPI. TGA 3 was mostly downregulated in susceptible plants except at 7DPI at which point of infection the expression was slightly upregulated. In resistant plants, the expression of TGA 3 transcription factor increased gradually in a stepwise fashion that peaked up at 7DPI. The overall pattern of expression of TGA 6 was upregulated in both susceptible and resistant plants. In susceptible (JG62) plants, the TGA 6 expression was comparatively higher than that of resistant plants except at 2DPI where the expression fell down in case of susceptible plants. In resistant (WR315) plants, TGA 6 exhibited steady state pattern throughout the infection time points except at 4DPI, where the expression of TGA 6 fell to some extent (Fig. 6b, d, f).
Expression of SA Marker Genes
In susceptible shoot tissues, PR 1 was slightly upregulated at the initial time points but gradually downregulated later. PR 5 was found to be downregulated throughout the time points. In resistant plants, both PR 1 and PR 5 were found to be upregulated throughout the infection time points with the gradual increment of expressional peak at 7DPI (Fig. 7a, c).
Relative expressions of SA marker genes. a, c represent the expressions of pathogenesis-related 1 (PR1) and pathogenesis-related 5 (PR5) genes in shoot tissues of susceptible (JG62) and resistant (WR315) chickpea plants. b, d represent the expressions of pathogenesis-related 1 (PR1) and pathogenesis-related 5 (PR5) genes in root tissues of susceptible (JG62) and resistant (WR315) plants. The relative fold changes are normalized against mock-inoculated control plants and bar represents standard error (n = 3, p < 0.05)
In roots, PR 1 is induced more in resistant plants as compared to susceptible one throughout the post infection time points. In susceptible chickpea lines, initially, PR 1 was induced up to 3DPI with the peak expression at 2DPI. Then the expression of PR 1 falls down at 4DPI and 7DPI. Resistant plants exhibited gradual induction of expression with the progression of the disease with the peak expression at 7DPI. PR 5 was also largely downregulated in susceptible plants but gradual expressional induction was noted in resistant chickpea plants after Foc1 infection (Fig. 7b, d).
LC-MS/MS Analysis of Salicylic Acid
The presence of salicylic acid has been detected in both susceptible (JG62) and resistant (WR315) plants but SA concentration is found to be higher in resistant plants (WR315) as compared to the susceptible (JG62) plants. Both shoot and root tissues of JG62 and WR315 exhibit presence of SA by chromatographic detection system. In susceptible plants, the level of SA does not increases significantly after infection (2DPI) as compared to the uninfected control plants. Resistant plants show significant accumulations of SA after infection (2DPI) with Foc1 in both shoot as well as root tissues. It has been observed that the concentration of SA in shoot tissues is quiet higher than that of root tissues of resistant chickpea plants (S4 Table) (Fig. 8a, b).
Discussion
Systemic response is an important part of the long distance defense signaling in plant microbe interaction and is directly connected with the cellular redox state of the host tissues. On the other hand, infection associated ROS generation is an indispensable phenomenon in most of the pathogen endeavor. ROS thus formed is mainly utilized in HR-mediated programmed cell death events in a localized fashion. This may also take part in induction of signaling by activating signaling molecules, i.e., salicylic acid, jasmonic acid etc. Our previous studies demonstrated that chickpea-Fusarium interaction leads to the redox alterations in root tissues that also modulate the cellular transportations and transcriptional reprogramming (Gupta et al. 2013; Gupta et al. 2017). The possibility of interconnection of this oxidative burst with salicylic acid pathway is again established through root proteome analysis of chickpea lines infected with Foc1 (Chatterjee et al. 2014). The procedure of migration of defense signals from root to shoot tissues in this particular plant-pathogen interaction is highly obscured till date. The present study demonstrates the possibility of involvement of classical SA mediated signaling cascade in trafficking root to shoot defense signals.
The occurrence of studies on systemic acquired resistance (SAR) in chickpea upon Fusarium oxysporum infection is scarce. Foc1 infection leads to dramatic morphological changes in chickpea. The susceptible (JG62) plants exhibit tremendous water loss due to wilting, flaccidity in shoot system, as well as total disintegration of root system gradually. External SA treatment is capable to stall these morphological degenerative processes and maintains the normal growth of the susceptible plant even after 7 DPI. This clearly suggests the involvement of SA in resistance phenomenon in chickpea. It has been shown that SA treatment can induce several catalase isoforms in susceptible chickpea lines as compared to the resistant backgrounds (Gayatridevi et al. 2013). In the present study, two principal SA biosynthetic genes phenylalanine ammonia lyase (PAL) and isochorismate synthase were found to be induced in the shoot and root tissues of resistant (WR315) chickpea lines. This clearly indicates that Foc1 may boost up the SA biosynthesis in resistant plants to activate signaling against fungal pathogen. The overall downregulation of these genes in susceptible (JG62) plants contributes to the failure of efficiency of this host plant in aggregating signaling cascade against Foc1. The cell wall proteins from Fusarium were shown to induce PAL in chickpea (Saikia et al. 2006). Additionally, ICS was found to be induced in chickpea plants upon infection with Meloidogyne incognita (Meher et al. 2015). It has been demonstrated that Fusarium oxysporum f. sp. lycopersici can also induce PAL in tomato plant upon infection (Manikandan and Raguchander 2014). The induction of PAL may also indicate the activation of phenyl propanoid biosynthesis pathway which is a source of a wide range of antimicrobial secondary metabolites (Fock-Bastide et al. 2014). The liquid chromatography-mass spectrometry (LC-MS) analysis further confirms that SA is accumulated in resistant (WR315) chickpea plants after Foc1 infection. The progressive induction of PAL in resistant plants confers that the accumulation of SA is probably due to the action of PAL over expression.
SA produced in the host plant in response to infection can activate downstream signaling in free form but it can be converted into inactive or less active conjugate forms. The principal inactive form of SA is SA O-beta glucoside (SAG) that can be stored within the cell for future utilization (Hennig et al. 1993). This conversion is mediated by SA glucosyltransferase gene (Umemura et al. 2009). The high level of expression of SAGT in shoot tissues of resistant chickpea plants conferring higher degree of SAG accumulation in shoot than root tissues after infection. Another important derivative of SA is methyl salicylate (MeSA), which is volatile in nature and help in priming of uninfected regions of host plant during pathogenesis. SA to MeSA conversion is mediated by SA methyl transferase gene (Tieman et al. 2010). The expression analysis of SAMT reveals its consistently high degree of accumulation in shoot tissues of resistant chickpea plants. This may indicate that Foc1 infection in root tissues may activate MeSA-mediated priming in shoot tissues of resistant chickpea plants. Susceptible plants lack this protective mechanism as both SAGT and SAMT were found to be downregulated after Foc1 infection in this particular case.
More prominently, it has been shown that the genes involved in SA signal transduction pathways were modulated in response to Foc1 infection. EDS 1 and PAD 4 are two very important signaling agents mainly function upstream of SA biosynthesis pathway. In this chickpea—Fusarium interaction, EDS 1 is found to be induced in both shoot and root tissues of infected susceptible and resistant chickpea plants, but in case of resistant chickpea lines, the expression is quiet consistent with the progression of the disease. PAD 4 expression pattern follows the same pattern exhibited by EDS 1. This may indicate the positive correlation of these two proteins in SA signaling pathways. PAD4 expression progressively increases with the progression of the disease in case of resistant chickpea lines. In susceptible chickpea plants, PAD 4 is principally downregulated in both shoot and root tissues after Foc1 infection. The similar expression pattern of EDS 1 and PAD 4 demonstrates that these two proteins may function coordinately to induce SA biosynthesis. It has been reported that PAD 4 is an important interaction partner for EDS 1 to induce both HR as well as auto activation of SA biosynthesis (Dempsey et al. 2011). The similar pattern of accumulation of EDS1 and PAD4 has been reported in Arabidopsis in response to wilt causing pathogen Fusarium oxysporum (Berrocal-Lobo and Molina 2008). EIN 2 is directly related to ethylene signaling events, but it also interacts with SA, JA signaling loop. SA is known to suppress JA-mediated signaling and EIN2 is coordinately associated with JA signaling cascade. EIN2 is also involved in HR-mediated cell death in a NPR1 independent pathway (Kunkel and Brooks 2002; Shah 2003). In the present study, low to moderate level of expression of EIN2 in both susceptible (JG62) and resistant (WR315) chickpea plants indicates that EIN2-mediated HR may not be operative in this case. NPR1 is the key molecular switch in SA-mediated resistance response. The consistent activation of NPR1 in resistant (WR315) chickpea plants in both shoot and root tissues may indicate the involvement of NPR1 dependent defense signaling in chickpea-Fusarium interaction. NPR1 is the principal regulator of PR1 and other pathogenesis-related proteins. It contains a BTB/POZ (broad-complex, tramtrac, bric-à-brac/poxvirus, zinc finger) domain with an ankyrin-repeat domain and its activation depends upon oligomer to monomer conversion upon infection (Mukhtar et al. 2009). It activates PR1 and other pathogenesis-related proteins by the help of TGA transcription factors. TGA transcription factors binds to the TGA box (TGACGTCA) of several SA responsive genes for the transcriptional reprogramming and activation of pathogenesis-related proteins (Gatz 2013).
The TGA transcription factors (TGA1, TGA3, and TGA6) are not induced in susceptible (JG62) chickpea plants whereas in resistant chickpea (WR315) plants all the TGA transcription factors are found to be induced effectively. In resistant lines, the induction of TGA TFs in shoot tissues are more prominent than that of root tissues. This may indicate that in resistant chickpea plants, SA induces the activation of TGA transcription factors that in turn activates NPR1. This hypothesis is further strengthened by the progressive induction of two SA marker genes PR1 and PR5 with the succession of the disease in resistant chickpea plants.
The overall downregulation of SA biosynthetic genes, NPR1, TGA transcription factors as well as marker genes PR1 and PR5 clearly indicates that susceptible lines cannot instigate SA refereed defense signaling efficiently to impart resistance against Foc1. Contrarily, resistant chickpea plants exhibit higher degree of internal SA accumulation by the transcriptional activity of PAL gene. The free SA is used to activate TGA transcription factors that in turn instigate pathogenesis-related protein (PR1 and PR5) by NPR1 dependent manner. In order to shed light on molecular switch of PR1 and PR5 regulation, in future, the study should focus towards cis-acting elements identification for TGA transcription factors in SA responsive gene pool. The investigation on trans-acting elements regulating pathogenesis proteins may also provide further insight into this complex interaction network.
References
Aranega-Bou P, de la O Leyva M, Finiti I, García-Agustín P et al (2014) Priming of plant resistance by natural compounds. Hexanoic acid as a model. Front Plant Sci 5:488. https://doi.org/10.3389/fpls.2014.00488
Barrs HD, Kozlowski TT (1968) Determination of water deficits in plant tissues. Water Deficits Plant Growth 1:235–368
Berrocal-Lobo M, Molina A (2008) Arabidopsis defense response against Fusarium oxysporum. Trends Plant Sci 13(3):145–150. https://doi.org/10.1016/j.tplants.2007.12.004
Bhar A, Gupta S, Chatterjee M, Sen S, Das S (2017) Differential expressions of photosynthetic genes provide clues to the resistance mechanism during Fusarium oxysporum f. sp. ciceri race 1 (Foc1) infection in chickpea (Cicer arietinum L.). Eur J Plant Pathol 148(3):533–549
Chatterjee M, Gupta S, Bhar A, Chakraborti D, Basu D, Das S (2014) Analysis of root proteome unravels differential molecular responses during compatible and incompatible interaction between chickpea (Cicer arietinum L.) and Fusarium oxysporum f. sp. ciceri Race1 (Foc1). BMC Genomics 15(1):949. https://doi.org/10.1186/1471-2164-15-949
Choi WG, Miller G, Wallace I, Harper J, Mittler R, Gilroy S (2017) Orchestrating rapid long-distance signaling in plants with Ca2+, ROS and electrical signals. Plant J 90(4):698–707. https://doi.org/10.1111/tpj.13492
Dempsey AD, Klessig DF (2012) SOS–too many signals for systemic acquired resistance? Trends Plant Sci 17(9):538–545. https://doi.org/10.1016/j.tplants.2012.05.011
Dempsey DMA, Vlot AC, Wildermuth MC et al (2011) Salicylic acid biosynthesis and metabolism. Arabidopsis Book 9:e0156. https://doi.org/10.1199/tab.0156
Fock-Bastide I, Palama TL, Bory S, Lécolier A, Noirot M, Joët T (2014) Expression profiles of key phenylpropanoid genes during Vanilla planifolia pod development reveal a positive correlation between PAL gene expression and vanillin biosynthesis. Plant Physiol Biochem 74:304–314. https://doi.org/10.1016/j.plaphy.2013.11.026
Gatz C (2013) From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. Mol Plant-Microbe Interact 26(2):151–159. https://doi.org/10.1094/MPMI-04-12-0078-IA
Gayatridevi S, Jayalakshmi SK, Mulimani VH, Sreeramulu K (2013) Salicylic acid and salicylic acid sensitive and insensitive catalases in different genotypes of chickpea against Fusarium oxysporum f. sp. ciceri. Physiol Mol Biol Plants 19(4):529–536. https://doi.org/10.1007/s12298-013-0184-4
Gupta S, Chakraborti D, Rangi RK, Basu D, Das S (2009) A molecular insight into the early events of chickpea (Cicer arietinum) and Fusarium oxysporum f. sp. ciceri (race 1) interaction through cDNA-AFLP analysis. Phytopathology 99(11):1245–1257. https://doi.org/10.1094/PHYTO-99-11-1245
Gupta S, Chakraborti D, Sengupta A, Basu D, Das S (2010) Primary metabolism of chickpea is the initial target of wound inducing early sensed Fusarium oxysporum f. sp. ciceri race I. PLoS One 5(2):e9030. https://doi.org/10.1371/journal.pone.0009030
Gupta S, Bhar A, Chatterjee M, Das S (2013) Fusarium oxysporum f. sp. ciceri race 1 induced redox state alterations are coupled to downstream defense signaling in root tissues of chickpea (Cicer arietinum L.) PLoS One 8(9):e73163. https://doi.org/10.1371/journal.pone.0073163
Gupta S, Bhar A, Chatterjee M, Ghosh A, Das S (2017) Transcriptomic dissection reveals wide spread differential expression in chickpea during early time points of Fusarium oxysporum f. sp. ciceri race 1 attack. PLoS One 12(5):e0178164. https://doi.org/10.1371/journal.pone.0178164
Haware MP, Nene YL (1982) Races of Fusarium oxysporum f. sp. ciceri. Plant Dis 6:809–810
Hennig J, Malamy J, Grynkiewicz G, Indulski J, Klessig DF (1993) Interconversion of the salicylic acid signal and its glucoside in tobacco. Plant J 4(4):593–600. https://doi.org/10.1046/j.1365-313X.1993.04040593.x
Kumar D (2014) Salicylic acid signaling in disease resistance. Plant Sci 228:127–134. https://doi.org/10.1016/j.plantsci.2014.04.014
Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5(4):325–331. https://doi.org/10.1016/S1369-5266(02)00275-3
Larkin RP, Fravel DR (1998) Efficacy of various fungal and bacterial biocontrol organisms for control of Fusarium wilt of tomato. Plant Dis 82(9):1022–1028. https://doi.org/10.1094/PDIS.1998.82.9.1022
Lemos M, Xiao Y, Bjornson M, Wang JZ, Hicks D, Souza A, Wang CQ, Yang P, Ma S, Dinesh-Kumar S, Dehesh K (2016) The plastidial retrograde signal methyl erythritol cyclopyrophosphate is a regulator of salicylic acid and jasmonic acid crosstalk. J Exp Bot 67(5):1557–1566. https://doi.org/10.1093/jxb/erv550
Li XY, Gao L, Zhang WH, Liu JK, Zhang YJ, Wang HY, Liu DQ (2015) Characteristic expression of wheat PR5 gene in response to infection by the leaf rust pathogen, Puccinia triticina. J Plant Interact 10(1):132–141. https://doi.org/10.1080/17429145.2015.1036140
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Mandal S, Mallick N, Mitra A (2009) Salicylic acid-induced resistance to Fusarium oxysporum f. sp. lycopersici in tomato. Plant Physiol Biochem 47(7):642–649. https://doi.org/10.1016/j.plaphy.2009.03.001
Manikandan R, Raguchander T (2014) Fusarium oxysporum f. sp. lycopersici retardation through induction of defensive response in tomato plants using a liquid formulation of Pseudomonas fluorescens (Pf1). Eur J Plant Pathol 140(3):469–480. https://doi.org/10.1007/s10658-014-0481-y
Meher HC, Gajbhiye VT, Singh G, Chawla G (2015) Altered metabolomic profile of selected metabolites and improved resistance of Cicer arietinum (L.) against Meloidogyne incognita (Kofoid & White) Chitwood following seed soaking with salicylic acid, benzothiadiazole or nicotinic acid. Acta Physiol Plant 37(7):1–12. https://doi.org/10.1007/s11738-015-1888-6
Meldau S, Kästner J, von Knorre D, Baldwin IT (2014) Salicylic acid-dependent gene expression is activated by locomotion mucus of different molluscan herbivores. Commun Integr Biol 7(3):e28728. https://doi.org/10.4161/cib.28728
Mukhtar MS, Nishimura MT, Dangl J (2009) NPR1 in plant defense: it’s not over’til it’s turned over. Cell 137(5):804–806. https://doi.org/10.1016/j.cell.2009.05.010
Pan X, Welti R, Wang X (2010) Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nat Protoc 5(6):986–992. https://doi.org/10.1038/nprot.2010.37
Saikia R, Yadav M, Singh BP et al (2006) Induction of resistance in chickpea by cell wall protein of Fusarium oxysporum f. sp. ciceri and Macrophomina phaseolina. Curr Sci 91:1543–1546
Selitrennikoff CP (2001) Antifungal proteins. Appl Environ Microbiol 67(7):2883–2894. https://doi.org/10.1128/AEM.67.7.2883-2894.2001
Shah J (2003) The salicylic acid loop in plant defense. Curr Opin Plant Biol 6(4):365–371. https://doi.org/10.1016/S1369-5266(03)00058-X
Shah J, Chaturvedi R, Chowdhury Z, Venables B, Petros RA (2014) Signaling by small metabolites in systemic acquired resistance. Plant J 79(4):645–658. https://doi.org/10.1111/tpj.12464
Sticher L, Mauch-Mani B, Métraux AJ (1997) Systemic acquired resistance. Annu Rev Phytopathol 35(1):235–270. https://doi.org/10.1146/annurev.phyto.35.1.235
Summerell BA, Salleh B, Leslie JF (2003) A utilitarian approach to Fusarium identification. Plant Dis 87(2):117–128. https://doi.org/10.1094/PDIS.2003.87.2.117
Tieman D, Zeigler M, Schmelz E, Taylor MG, Rushing S, Jones JB, Klee HJ (2010) Functional analysis of a tomato salicylic acid methyl transferase and its role in synthesis of the flavor volatile methyl salicylate. Plant J 62(1):113–123. https://doi.org/10.1111/j.1365-313X.2010.04128.x
Umemura K, Satou J, Iwata M, Uozumi N, Koga J, Kawano T, Koshiba T, Anzai H, Mitomi M (2009) Contribution of salicylic acid glucosyltransferase, OsSGT1, to chemically induced disease resistance in rice plants. Plant J 57(3):463–472. https://doi.org/10.1111/j.1365-313X.2008.03697.x
Acknowledgments
Authors thank Dr. S.C. Pande (ICRISAT, Patancheru) for providing fungal culture and Dr. S.K. Chaturvedi (IIPR, Kanpur) for providing chickpea seeds. Authors are also thankful to Dr. Kaushik Bannerjee, National Research Centre for Grapes, Solapur, Pune, India, for performing the LC MS/MS of the sample. Unwearied assistance of Mr. Swarnava Das for physiological experiments is greatly acknowledged. Mr. Sudipta Basu is duly acknowledged for seed multiplication. Finally, authors acknowledge the Director, Bose Institute for infrastructural facilities.
Funding
This work was supported by the grant provided to A.Bhar by Council of Scientific and Industrial Research, India (09/015(0378) /2009-EMR-1) and to M.Chatterjee by Department of Biotechnology, Government of India (BT/01/COE/06/03/2006). The funding organizations had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Supplementary Table S1
(DOC 51 kb)
Supplementary Table S2
(XLS 645 kb)
Supplementary Table S3
(DOC 63 kb)
Supplementary Table S4
(DOCX 11 kb)
Rights and permissions
About this article
Cite this article
Bhar, A., Chatterjee, M., Gupta, S. et al. Salicylic Acid Regulates Systemic Defense Signaling in Chickpea During Fusarium oxysporum f. sp. ciceri Race 1 Infection. Plant Mol Biol Rep 36, 162–175 (2018). https://doi.org/10.1007/s11105-018-1067-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-018-1067-1