Abstract
The dehydration-responsive element-binding (DREB) transcription factors play important roles in regulation of plant responses to abiotic stresses. In the present study, ZmDBF3, a novel DREB transcription factor gene from maize (Zea mays L.), was cloned and characterized. Sequence analyses revealed that ZmDBF3 is classified into A-4 group. It was demonstrated that ZmDBF3 was induced in by salt, drought, cold, and high temperature, as well as by signaling molecules abscisic acid (ABA), but no significant changes were observed under salicylic acid (SA) and methyl jasmonate (MeJA) conditions. The results of transient expression assays and transcriptional activity analysis revealed that ZmDBF3 is a nuclear protein with transcriptional activity. Overexpression of ZmDBF3 in yeast (Saccharomyces cerevisiae) exhibited increased survival rate under NaCl, KCl, Na2CO3, NaHCO3, PEG6000, freezing, and sorbitol treatment, compared with the control. Furthermore, ectopic expression of ZmDBF3 in Arabidopsis significantly enhanced tolerance to salt, drought, and freezing tolerance. Taken together, the findings indicated that the ZmDBF3 is a novel member of DREB transcription factor which may act as a regulatory factor involved in multiple stress response pathways.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Abiotic stresses, especially drought and high salinity, are major limiting factors that retard plant growth and severely decrease crop productivity. To survive, plants have to evolve complex mechanisms to adapt to ever-changing environmental conditions. Drought stress induces a range of physiological and biochemical responses in plants, including stomatal closure, inhibition of photosynthesis, and respiration activation (Shinozaki and Yamaguchi-Shinozaki 2007). Additionally, plants respond to abiotic stresses at the cellular and molecular levels, including stress perception, signal transduction to cellular components, gene expression, and metabolic changes (Agarwal et al. 2006; Shinozaki and Yamaguchi-Shinozaki 2007).
Transcription factors have been proved to mediate the function of multiple stress response genes in a coordinated manner, which are considered as attractive targets for application through genetic engineering. The dehydration-responsive element-binding (DREB) proteins are the major transcription factors that induce a set of abiotic stress-inducible gene expression in the ABA-independent pathway (Yamaguchi-Shinozaki and Shinozaki 2006). DREBs are parts of APETALA2/ethylene-responsive factor (AP2/ERF) superfamily, specially bind the cis-element of dehydration responsive element (DRE, A/GCCGAC), in the promoter region of many stress-inducible genes (Yamaguchi-Shinozaki and Shinozaki 1994; Jiang et al. 1996). Two groups of DREB genes were discovered in the Arabidopsis, DREB1s and DREB2s (Agarwal et al. 2006). Expression of the Arabidopsis DREB1 genes is induced by cold, but not by dehydration or high-salinity. By contrast, the DREB2 genes can be induced by dehydration, high-salinity, and heat stresses, but not by cold stress (Lata and Prasad 2011). Hence, DREB1s and DREB2s are involved in two separate signal transduction pathways under low temperature and dehydration, respectively. However, DREB1D, DREB1E, and DREB1F are also induced by osmotic stress, suggesting the existence of cross talk between the DREB1 and the DREB2 pathways (Haake et al. 2002; Nakashima et al. 2009).
Numeral studies demonstrated that overexpression of special DREB genes in transgenic plants using constitutive or inducible promoters resulted in plants more tolerant to drought, salt, heat, and freezing stresses (Lata and Prasad 2011). For example, transgenic Arabidopsis plants overexpressing DREB1B under control of the cauliflower mosaic virus (CaMV) 35S promoter showed strong tolerance to freezing stress (Jaglo-Ottosen et al. 1998). Similarly, transgenic Brassica napus or tobacco plants overexpressing the Arabidopsis DREB1/CBF genes increased the freezing tolerance and induced the expression of cold-related genes (Jaglo-Ottosen et al. 2001; Kasuga et al. 2004). The overexpression of AtDREB1A driven by the stress-inducible rd29A promoter in transgenic wheat also showed improved tolerance to drought stress (Pellegrineschi et al. 2004). Besides in Arabidopsis, DREB transcription factors have been identified in other various plant species, such as rice (Oryza sativa), soybean (Glycine max), sorghum (Sorghum bicolor), hot pepper (Capsicum annum), tobacco (Nicotiana tabacum), and chickpea (Cicer arietinum) (Lata and Prasad 2011). These observations suggested that the DREB regulation can be used to improve the tolerance of various kinds of agriculturally important crops to multiple abiotic stresses by gene transfer.
Maize (Zea mays L.), one of the most important crops in the world, is frequently impacted by drought, high salinity, and low temperature. Therefore, how to improve abiotic stress tolerance in maize is a priority target in many breeding programs. Previously, two DREB genes (ZmDREB1A and ZmDREB2A) belonging to the DREB1 and DREB2 subgroups, respectively, were identified and characterized (Qin et al. 2004; Qin et al. 2007). ZmDREB1A was accumulated in response to cold stress, while transcripts of ZmDREB2A were increased by cold, dehydration, salt, and heat stresses in maize seedlings (Qin et al. 2007; Qin et al. 2004). Moreover, constitutive or stress-inducible expression of ZmDREB2A resulted in improved drought stress tolerance in plants (Qin et al. 2007). However, until now, much less is known about functions of the other DREB transcription factors in maize. In the present study, we isolated ZmDBF3 gene from maize and examined its expression pattern in response to various abiotic stresses and signal molecules. Furthermore, the role of ZmDBF3 in tolerance to abiotic stresses was examined by ectopic expression in both yeast cells and Arabidopsis.
Materials and Methods
Plant Material, Growth Conditions and Treatments
Maize (Z. mays L. cv. B73) was employed to isolate the ZmDBF3 gene and to evaluate its expression patterns under various treatments. Seeds were germinated in pots containing vermiculite and cultured in growth chambers (16-h light/8-h dark cycle with a light intensity of 200 μmol m−2 s−1 at 25 °C). Similar sized seedlings at the same stage (8 days old) were selected and subjected to the following treatments. For salt, drought, abscisic acid (ABA), salicylic acid (SA), and methyl jasmonate (MeJA) treatments, the roots of the seedlings were immersed in Hoagland’s solutions containing 250 mM NaCl, 20 % polyethylene glycol 6000 (PEG 6000) and 100 μM ABA, 2 mM SA, and 100 μM MeJA, respectively. Cold treatment was conducted by exposing the seedlings to 4 °C in a chamber with a 16-h light/8-h dark regime for 24 h. For high-temperature stress, seedlings were kept in a chamber at 40 °C for 0.5, 1, 3, 4, 5, and 6 h. Leaves from plants in each treatment were collected at special time points, immediately frozen in liquid nitrogen and stored at −80 °C for RNA extraction.
Amplification and Sequence Analysis of ZmDBF3
Genomic DNA was extracted by the cetyltrimethylammonium bromide (CTAB) method. The full-length coding sequence of ZmDBF3 was amplified with Pfu DNA polymerase (TransGen, Beijing, China) from the DNA by gene-specific primers (forward ATGAGCTTGCTTGTGACTG; reverse TCTTGCTCGTCAGTGCTCC) based on the sequence from NCBI database (NM_001157828). The products were purified and cloned into pMD18-T vector (Takara, Dalian, China) for sequencing. The isoelectric point (pI) and molecular weight (MW) of ZmDBF3 were predicted with ExPASy (http://expasy.org/tools/pi_tool.html) for computation. Multiple sequence alignment was performed by DNAMAN software. Phylogenetic analysis was generated by neighbor-joining method and displayed using MEGA4.0 based on the similarity of amino acid sequences (Tamura et al. 2007).
Subcellular Localization of ZmDBF3 Protein
Primers (ZmDBF3-F: CGAGCTCATGAGCTTGCTTGTGACTG, the underlined indicates Sac I restriction site, ZmDBF3-R: CACCATGGTGGCGACCGGTGGGTGCTCCCAGAGCAGCAAC) and primers eGFP-F: AGCACCCACCGGTCGCCACCATGGTGAGCAAGGGCGAG, eGFP-R: 5′-TCCCCCGGGTTACTTGTACAGCTCGTC, the underlined indicates Sma I site) were used to amplify ORF of ZmDBF3 without the termination codon and eGFP, respectively. The products were linked by overlapping PCR method, and the recombinant fragment was digested by Sac I/Sma I and inserted into the corresponding sites of pCHF-1301 vector (Li et al. 2014) and verified by sequencing. Then, the resultant DNA was transformed into Agrobacterium EHA105. Transient expression of the ZmDBF3::eGFP fusion construct and eGFP control was performed by introducing the resultant DNAs into onion epidermal cells using Agrobacterium-mediated genetic transformation method. Transformed cells were cultured on Murashige and Skoog (MS) medium for 16–48 h and observed under a confocal microscope (Olympus, Japan).
Transcriptional Activation Assay
The complete coding sequence of ZmDBF3 was amplified by PCR with primers (ZmDBF3-F: CCGGAATTCTCTTGCTCGTCAGTGCTCC (EcoR I), ZmDBF3-R: CGCGGATCCTCTTGCTCGTCAGTGCTCC (BamH I)) and cloned into the pGBKT7 vector. The pGBKT7-ZmDBF3 construct, pGAL4 plasmid (as s positive control), and the negative control (pGBKT7 vector) were transformed into yeast strain AH109. Transformed cells were confirmed by PCR and then plated on SD/-Trp or 10 mM 3-AT of SD/-Trp-His-Ade medium. Transcription activation was evaluated according to the growth status of yeast cells after incubating plates at 30 °C for 3 days with 5-bromo-4-chloro-3-indolyl-b-d-galacto- pyranoside (X-gal).
Abiotic Stress Analysis of ZmDBF3 in Yeast Cells
The full-length sequence of ZmDBF3 was amplified and directionally cloned into the yeast expression vector pYES2 (named pYES2-ZmDBF3), which contained the Ura3 selection marker driven by a GAL1 promoter. The pYES2-ZmDBF3 construct and pYES2 (the negative control) were transformed to the yeast strain INVSc1 (His-, Leu-, Trp-, and Ura-) according to the pYES2 vector kit instructions (Invitrogen). The positive recombinant INVSc1 strains harboring pYES2-DREB1B and pYES2 were cultured in SC-U liquid medium containing 2 % glucose at 30 °C for 12–16 h until the OD600 reached 0.4. Stress tolerance assays were performed according to the methods described previously (Gao et al. 2011).
Plasmid Construction and Arabidopsis Transformation
The full-length coding sequence of ZmDBF3 was amplified by PCR using primers CGAGCTCATGAGCTTGCTTGTGACTG (Sac I) and TCCCCCGGGTCTTGCTCGTCAGTGCTCC (Sma I). The PCR products were cloned into the pCHF1301 vector driven by the CaMV 35S promoter (Li et al. 2014). The recombinant plasmid was introduced into the EHA105. Arabidopsis plants transformation was performed by the floral dip method (Clough and Bent 1998). Transgenic Arabidopsis was selected on MS medium containing 30 μg/l hygromycin and confirmed by PCR analysis. T3 generation plants were used for stress tolerance analysis in this study.
Analysis of Drought, Cold, and Salt Tolerance in Transgenic Arabidopsis
In all experiments, Arabidopsis seeds were surface-sterilized with 10 % NaOCl for 3–6 min and germinated on MS agar medium with 1 % (w/v) sucrose and 0.6 % (w/v) agar. The growth conditions of ZmDBF3 transgenic plants (T3 generation) and the control (Col-0) were the same as described above. For salt and drought stress on plates, the Arabidopsis seeds were germinated on MS agar for 5 days and then transferred to fresh MS medium containing different concentrations of NaCl (100, 200, and 300 mM) and mannitol (200, 300, and 400 mM), respectively. The plates were positioned vertically on shelves to assist the evaluation of root growth rates. The survival rates were recorded for another 5 days after transferring. For the salt tolerance test at the adult stage, 3-week-old plants were irrigated with 350 mM NaCl solution. When the soil was completely saturated with salt solution, free NaCl solution was removed and the plants were cultured under normal conditions. Photos were taken on the fifth and tenth day after salt treatment. For drought tolerance analysis, 3-week-old plants were cultured without watering for 2 weeks. For freezing stress treatment, 3-week-old ZmDBF3 transgenic and control plants were exposed −6 °C for 3 h and then cultured under normal conditions described as above. All abiotic stress tolerance experiments were conducted in triplicate.
qRT-PCR
Total RNAs from maize leaves were isolated using RNAiso Plus reagent (Takara) according to the manufacturer’s instructions. Genomic DNA digestion in total RNA and reverse transcription were carried out using PrimeScript® RT reagent Kit With gDNA Eraser kit (Takara). qPCR was performed in a total volume of 20 μl, 1 μl of diluted complementary DNA (cDNA), 200 nM for each primer, 0.4 μl of ROX Reference Dye II (50×), and 10 μl of 2× SYBR Green PCR Master Mix (Takara) on an ABI 7500 Real Time System (PE Applied Biosystems, USA). The qPCR program included a preliminary step of 30 s at 95 °C, followed by 40 cycles of 95 °C for 5 s and 58 °C for 40 s. ZmGAPDH (GenBank accession number NM_001112119) was used as internal control for normalization of different cDNA samples, respectively. Relative gene expression was calculated according to a 2−ΔΔCT method (Livak and Schmittgen 2001). The primer sequences are as follows: ZmGAPDH-L CCGTGTTCCTACTGTGGATG, ZmGAPDH-R TGTCACCAAGGAAGTCGGTA, ZmDBF3-L GGCTGCGATGGAGATGGAGA, and ZmDBF3-R GGGCATTGTCGGCGTTGTT.
Results
Isolation and Structure Analysis of ZmDBF3
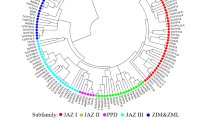
Using a gene-specific primer pair designed based on ZmDREB1B (accession number NM_001157828) nucleotide sequence, a 783-bp fragment containing full length of the gene was successfully obtained by PCR method from B73 maize DNA samples since there is no intron predicted in this gene. Sequence analysis revealed that ZmDREB1B consisted of 783-bp open reading frame (ORF) encoding 260 amino acids. The deduced amino acids sequence of ZmDREB1B had a calculated molecular weight of 28 kDa with a pI of 5.11 (http://web.expasy.org/compute_pi/). To analyze the phylogenetic relationship of ZmDREB1B with the group DREB/CBF proteins from other plants, a phylogenetic tree was constructed based on the amino acid sequences of these proteins. As shown in Fig. 1a, ZmDBF2, StDREB1B, AtTINY, and AtTINY2 were in the same group, which belongs to subgroup A-4. Moreover, ZmDREB1B showed high homolog to ZmDBF2; therefore, this gene was named as ZmDBF3 instead. Comparison of amino acids sequences of these proteins indicated that the ZmDBF3 protein contains a conserved DNA-binding domain (AP2/ERF domain) which is composed of 64 amino acid residues (73–136), with conserved valine (V) and glutamic acid (E) residues at the 14th and 19th positions (Fig. 1b).
Sequence analysis of ZmDREB1B. a Phylogenetic analysis of ZmDBF3 with AP2/ERF transcription factors from other plant species. The phylogenetic analysis was conducted by MEGA 4.0 software. The appended proteins are as follows: ZmDREB1A (AF450481), HvCBF3 (AF298230), ZmDREB2A (NP_001105876), HvDRF1 (AY223807), HvCBF1 (AF418204), HvCBF2 (AF442489), TaDREB1 (DQ195068), TaCBF1 (AF376136), OsDREB1A (AF300970), ZmDBF1 (AAM80486), ZmDBF2 (AF493799), ZmABI4 (AY125490), GhDBP1 (AY174160), AtDREB1A (AB007787), AtDREB1B (AB007788), AtDREB1C (AB007789), AtDREB2A (AB007790), GmDREBa (AAT12423), GmDREBb (AAQ57226), GmDREBc (AAP83131), OsDREB2A (AF300971), PeDREB2 (ABY19375), AtABI4 (A0MES8), GmDREB2 (ABB36645), AtRAP2.10 (Q9SW63), AtRAP2.1 (Q8LC30), AtRAP2 (AAP04063.1), AtRAP2.4 (NP_177931), GhDBP2 (AAT39542), OsDREB3 (NP_001048142), AtTINY (Q39127), AtTINY2 (AY940160.1), LeCBF1 (AY034473), BnCBF7 (AF499032), BnCBF17 (AF499034), GhDREB1A (AY321150), OsDREB1B (AF300972), HvBCBF3 (AF298231), AtDREB2B (NM_111939.2), AtRAP2.9 (NM_179009.1), ScCBF1 (AF370730), GhDBP3 (DQ224382), StDREB1B (JN125862.1), and AtDREB2C (At2g40340). b Multiple alignments of the amino acid sequences of DREB proteins. The AP2/ERF domain is underlined, and two conserved residues of V14 and E19 are indicated over asterisks. Identical sequences are shaded in black
Nuclear Localization and Transcriptional Activity of ZmDBF3 Protein
To investigate the subcellular localization of ZmDBF3 in plant cells, we conducted transient expression assays with 35S::eGFP and a chimeric 35S::ZmDBF3-eGFP construct (Fig. 2a) in onion epidermal cells. As shown in Fig. 2b, GFP fluorescence distributed evenly in the nucleus and the cytoplasm with the control plasmid 35S-eGFP, while the CaMV 35S::ZmDBF3-eGFP fusion protein was targeted to the nucleus. These results clearly indicate that ZmDBF3 is a nuclear localized protein.
Subcellular localization and transcriptional activity analysis of ZmDBF3. a Schematic structures of 35S::eGFP and 35S::ZmDBF3-eGFP vectors. b Subcellular localization of ZmDBF3 protein in onion epidermal cells. The transformed cells with 35S::eGFP and 35S::ZmDBF3-eGFP were observed under a confocal microscope after inoculation for 36 h. The photographs were taken in dark field for green fluorescence, bright light to illustrate the morphology of the cells and in overlay. The bar represents 100 μm. c Transcriptional activity analysis of ZmDBF3. The full-length ORF of ZmDBF3 was fused to the GAL4 DNA-binding domain in the vector pGBKT7, and the construct was transformed into yeast strain AH109 (Invitrogen). pGBKT7 and pGAL4 plasmids were used as negative and positive control, respectively
To test whether ZmDBF3 has transcriptional activity, the entire coding region was fused to the GAL4 DNA-binding domain in the vector pGBKT7, and the construct was transformed into yeast. Colony-lift filter assays showed that the reporter gene LacZ was expressed in the yeast cells transformed with the pGAL4 or pGBKT7-ZmDBF3 constructs (Fig. 2c), indicating that the ZmDBF3 has transcriptional activity in yeast.
Expression Pattern of ZmDBF3 Under Abiotic Stresses and Signaling Molecules
The transcript profiling of ZmDBF3 in response to abiotic stresses and signaling molecules was investigated by qRT-PCR analysis. As shown in Fig. 3a, the expression of ZmDBF3 was enhanced about 22-fold at 1 h and then declined following PEG treatment. Salt stress led to a significant increase of ZmDBF3 transcript level within 1 h and reached a maximum at 6 h and then decreased at 24 h (Fig. 3b). After ABA treatment, the transcript level of ZmDBF3 was strongly induced at 4 h time-point and continuously accumulated up to 24 h of treatment (Fig. 3c). Similar to cold treatment, ZmDBF3 mRNA accumulated quickly in response to high temperature and reached maximum at 5 h of stress treatment (Fig. 3d, e). However, no significant changes of ZmDBF3 were detected at each tested time-point as compared with non-treated control, when subjected to MeJA or SA (Fig. 3f, g). Above results indicated that ZmDBF3 may play important roles in abiotic stress to help plants adapting to adverse environmental conditions.
Transcript accumulation of ZmDBF3 in maize leaves in response to different abiotic stresses and signaling molecules as determined by qRT-PCR. Maize seedlings were treated with 20 % PEG 6000 (a), 250 mM NaCl (b), 100 μM ABA (c), 40 °C (d), 4 °C(e), 100 μM MeJA (f), and 2 mM SA (g). Total RNA was isolated from leaves at indicated times after different treatments. Bars indicate mean relative expression values and three replicates
Overexpression of ZmDBF3 in S. cerevisiae INVSc1 Cells Improved Tolerance to Multiple Abiotic Stresses
ZmDBF3 was introduced into yeast cells to analyze the possible function of this protein under different abiotic stresses. The growth performance of yeast cells transformed with pYES2 (control) or pYES2-ZmDBF3 was tested under various abiotic stress conditions. As shown in Fig. 4a, the growth of ZmDBF3 transgenic yeast cells was inhibited relative to control under stress-free conditions, when diluted to 10−5, indicating that ZmDBF3 expression retards the growth of yeast cells. However, ZmDBF3-overexpressing yeast cells exhibited increased survival rate compared to the control, when subjected to NaCl, KCl, 6 % Na2CO3, and NaHCO3 treatment (Fig. 4b–d, f, g). No significant difference in growth levels between the ZmDBF3 transgenic and control yeast cells under 8 % Na2CO3, 40 % PEG6000, sorbitol, and freezing (Fig. 4e, h–j). These results indicated that ZmDBF3 might be involved in the yeast multiple abiotic stress tolerance.
Stress tolerance analysis of ZmDBF3 in yeast cells. The transformed yeast INVSc1 harboring ZmDBF3 (pYES2-ZmDBF3) and INVSc1 harboring empty pYES2 were diluted into different ratio and treated with various stress conditions, and their survival rates were compared. a Non-stress, b 5 M NaCl, c 5 M KCl, d 6 % Na2CO3, e 8 % Na2CO3, f 6 % NaHCO3, g 8 % NaHCO3, h 40 % PEG6000, i 5 M sorbitol, and j −20 °C
Ectopic Expression of ZmDBF3 in Arabidopsis Resulted in Enhanced Salt and Drought Tolerance
To investigate the function of ZmDBF3 in plants, transgenic Arabidopsis plants expressing the ZmDBF3 under the control of the CaMV 35S promoter were obtained. Five-day-old Col-0 and transgenic seedlings were transferred to the MS plates containing different concentrations of NaCl and mannitol after germination on MS medium. As shown in Fig. 5a, b, no significant difference between the transgenic plants and control was observed on MS medium containing 100 mM NaCl or 200 mM mannitol. However, when supplemented with 200 mM NaCl, over 70 % Col-0 plants showed etiolation or death, whereas the transgenic plants exhibited significantly higher survival rate and longer primary roots relative to control (Fig. 5a, c, d). Similarly, ZmDBF3-overexpressing lines exhibited longer roots than Col-0 on MS plates containing 300 mM mannitol (Fig. 5b, d), indicating overexpression of ZmDBF3 has decreased NaCl and mannitol sensitivity. On MS containing 300 mM NaCl or 400 mM mannitol, both transgenic lines and control displayed sign of death or severely inhibited root length (Fig. 5a, b), suggesting that the concentration of NaCl is beyond the reach of tolerance.
Salt and drought tolerance assay of transgenic Arabidopsis plants overexpressing ZmDBF3. a, b Phenotype of transgenic lines (OE15, OE8, and OE3) and control (Col-0) on MS medium containing different concentrations of NaCl (100, 200, or 300 mM) and mannitol (200, 300, or 400 mM), respectively. Survival rates of ZmDBF3 transgenic lines (OE3, OE8, and OE15) and Col-0 on MS medium containing 200 mM NaCl (c). d Primary root length of ZmDBF3-overexpressing lines (OE3, OE8, and OE15) and Col-0 on MS medium containing 200 mM NaCl or 300 mM mannitol. Data are shown as mean ± SD of three replicates. The statistical significance was determined by Duncan’s multiple comparison tests. Different letters above bars indicate significant differences (P < 0.05) among different genotypes in the same treatment. e, f Performance of Col-0 and ZmDBF3-overexpressing seedlings under salt and drought stresses. Survival rates of ZmDBF3 transgenic lines and Col-0 under 350 mM NaCl and drought stresses. Data are shown as mean ± SD of three replicates
To further investigate the role of ZmDBF3 in improvement of plant salt and drought tolerance at adult stage, 3-week-old soil-grown plants were irrigated with 350 mM NaCl or without irrigation for 2 weeks, respectively. The survival percentage of transgenic plants was 47–73 % under salt stress, which was significantly higher than that of Col-0 (7 %) (Fig. 5e, g). After drought treatment for 2 weeks, Col-0 plants became severely wilted, whereas ZmDBF3 overexpression lines appeared relatively healthy (Fig. 5f, h). After re-watering for 3 days, only 20 % Col-0 plants survived, whereas the survival rates of overexpressing lines ranged from 25 to 80 %. These results indicated that the transgenic plants are more tolerant to salt and drought than the Col-0 plants.
Overexpression of ZmDBF3 in Arabidopsis Increases Freezing Tolerance
To evaluate the contribution of ZmDBF3 to plant tolerance to low temperature, 3-week-old ZmDBF3-overexpressing lines and Col-0 seedlings were exposed to −6 °C for 6 h. After 1 day of recovery at normal conditions, over 80 % of the Col-0 seedlings were dead; conversely, several of those from the transgenic lines (OE3, OE8, and OE15) were green and regrew normally after recovery (Fig. 6a). The survival percentages of the three transgenic lines ranged from 43 to 46 % (Fig. 6b), significantly higher than the control (19 %) under the freezing condition, indicating that overexpression of ZmDBF3 in Arabidopsis improves tolerance to freezing stress.
Discussion
Plant growth and yield are severely decreased by abiotic stresses. Identification of the transcription factors that regulate responses to abiotic stress is considered as a key target for improving stress tolerance in crop plant species. DREBs play a vitally important role in abiotic stress tolerance through binding to the DRE/CRT element in the promoter regions of a large number of stress-responsive genes in plants (Yamaguchi-Shinozaki and Shinozaki 2006). In the present study, we isolated ZmDBF3, a novel member of DREB transcription factor family gene from maize, and studied the function of this gene by overexpression in yeast cells and Arabidopsis.
Identification and classification of genes are significant for the functional analysis of a gene family. According to the conserved AP2/ERF domain, the subfamily of DREB transcription factors can be further divided into six subgroups from A-1 to A-6 (Sakuma et al. 2002). Although much information has elucidated the regulatory function of the A-1 (DREB1/CBF) and A-2 (DREB2) subfamily in Arabidopsis, there is still less learn about the species roles of DREBs belonging to A-3 to A-6. Phylogenetic analysis suggested that ZmDBF3 is classified into A-4 group (Fig. 1a). Each DREB protein contains a highly conserved AP2/ERF domain with 14th valine (V14) and the 19th glutamic acid (E19), whereas alanine and aspartic acid are conserved in the corresponding positions of the other ERF proteins (Sakuma et al. 2002). The conserved valine and glutamic acid residues in the AP2/ERF domain play an important role in binding to the target DNA sequences (Liu et al. 1998; Sakuma et al. 2002; Sakuma et al. 2006). It was also reported that 14th residue is more conserved and important than the 19th residue in the DRE-binding activity of DREB (Cao et al. 2001; Sun et al. 2014). The analysis of ZmDBF3 protein revealed that it contains a conserved ERF/AP2 domain of 64 amino acids with conserved V14 and E19 residues (Fig. 1b), which was consistent with the characteristics of DREB1 subclass. In addition, the results of transient expression assays and transcriptional activity analysis demonstrated that ZmDBF3 is a nuclear localized protein with transcriptional activity (Fig. 2a–c). Above data indicated that ZmDBF3 is one new member of the A-4 subfamliy of DREB1 transcription factors.
Previous studies showed that DREB1s and DREB2s are the largest groups that involved in ABA-independent pathways (Liu et al. 1998). Generally, DREB1 genes are induced by cold, whereas DREB2 genes are induced by dehydration, high salinity, and heat stresses in Arabidopsis (Lata and Prasad 2011). However, many DREB1 genes isolated from various plant species exhibited different expression pattern following abiotic stresses induction. CaDREBLP1 from hot pepper was rapidly induced by dehydration and salt, but not by cold stress (Hong and Kim 2005). PpDBF1 from Physcomitrella patens and HvDREB1 from Hordeum vulgare were significantly induced by salt, cold, and drought, respectively (Liu et al. 2007; Xu et al. 2009). In this study, ZmDBF3 was induced by drought, salt, cold, and high temperature, indicating that ZmDBF3 is an abiotic stress-responsive gene and may contribute to plant stress tolerance. ABA is an important hormone signal to improve plants to overcome various environment stresses (Fujita et al. 2011). The ABA-inducible expression pattern of ZmDBF3 suggests that ZmDBF3 may be involved in the plant response to ABA (Fig. 3c). Consistent with our results, previous studies indicated that ABA plays a role in the regulation of ZmDBF activity and the existence of an ABA-dependent pathway for the regulation of genes through the C-repeat/DRE element (Kizis D and Pagès 2002). As important signaling molecules, JA and SA play important roles in wounding and defense responses against pathogens, respectively. In our study, it was also showed that ZmDBF3 was not affected by MeJA or SA treatment (Fig. 3f, g), suggesting that it may not be involved in wounding or biotic stress response, although previous study has showed that expression of some DREB1 genes is correlated with wounding (Dubouzet et al. 2003; Hong and Kim 2005).
Gene expression in yeast cells is a good model system for screening the candidate gene or studying the mechanisms underlying stress tolerance. For example, expression of ThVHAc1 in yeast increased salt, drought, ultraviolet (UV), oxidative, heavy metal, cold, and high temperature tolerance (Gao et al. 2011). The transgenic yeast expressing GhBCP1 and GhBCP4 significantly increased cell growth rate under Cu2+, Zn2+, and high salinity stresses (Ruan et al. 2011). Using this expression system, we also have successfully identified and characterized a heavy metal-associated protein gene, AcHMA1, from Atriplex canescens, in metal stress tolerance (Sun et al. 2014). In this study, ZmDBF3-overexpression yeast cells exhibited increased growth rate compared with the control under NaCl, KCl, Na2CO3, NaHCO3, PEG6000, and freezing stresses (Fig. 4a–j), suggesting that this gene is sufficient to enhance abiotic stress tolerance in yeast.
To further investigate the stress tolerance of ZmDBF3, we obtained ZmDBF3 transgenic overexpressing lines of Arabidopsis. In most cases, DREB transgenic plants under constitutive promoter showed dwarfism or abnormal phenotype, which limits the practical use of DREB genes (Agarwal et al. 2006). The retard growth phenotype is thought to be related to interference in gibberellic acid (GA) metabolism (Achard et al. 2008). In contrast to the phenotype in yeast cells (Fig. 4a), ZmDBF3-overexpressing Arabidopsis plants exhibited normal growth rate compared with Col-0 under non-stressed conditions (Fig. 5a, b). The results also indicated the existence of difference between two expression systems. Similar results were obtained from AtCBF3 overexpression in diverse plant species (Gilmour et al. 2000; Oh et al. 2005). Overexpression of AtCBF3 in Arabidopsis plants caused severe growth retardation (Gilmour et al. 2000); however, AtCBF3 transgenic plants exhibited growth retardation when expressed in rice (Oh et al. 2005). Overexpression of the TINY or HARDY gene, two genes belong to A-4 DREB subgroup, resulted in stunted growth and thick leaves (Wilson et al. 1996; Karaba et al. 2007), suggesting that distinctive DREB genes may have various functions, although sharing high sequence similarity. Therefore, selecting proper DREB transcription factors or using stress-inducible promoters may be good choices to minimize the negative effects of DREB on plant growth and development.
Previous studies have showed the role of DREB transcription factors in tolerance to abiotic stress. Transgenic tobacco plants overexpressing PpDBF1 showed higher tolerance to salt, drought, and cold stresses (Liu et al. 2007). Similarly, overexpression of DREB1A under the control of the CaMV 35S promoter also increased the tolerance to drought, high-salt, and freezing stresses (Liu et al. 1998; Kasuga et al. 1999; Gilmour et al. 2000). It was also reported that Arabidopsis plants overexpressing AtDREB2A did not improve stress tolerance, suggesting that a posttranslational modification is necessary for the function of AtDREB2A (Liu et al. 1998; Sakuma et al. 2006). In this study, ZmDBF3 transgenic Arabidopsis plants showed reduced salt and drought sensitivity, and enhanced tolerance to drought and salt (Fig. 5a–h). Moreover, overexpression of ZmDBF3 in Arabidopsis also exhibited enhanced tolerance to freezing stress (Fig. 6a, b). In addition, we also noted that the three transgenic lines showed different stress tolerance under stress conditions. The overexpressing line OE15 showed stronger tolerance to drought and high-salt than the others (Fig. 5e–h). The difference may be due to the various expression levels of transgenic lines or the different insertion site in plant genome.
In conclusion, we identified a novel maize DREB A-4 group gene, ZmDBF3. The results clearly showed that ZmDBF3 was upregulated at the transcription level by cold, high salinity, cold, and ABA but was not affected by exogenous JA and SA. Furthermore, overexpression of ZmDBF3 in yeast cells and Arabidopsis enhanced tolerance to salt, drought, and freezing stresses, suggesting that ZmDBF3 may be a positive regulator of multiple abiotic stress tolerance. Therefore, in the future, ZmDBF3 might be a candidate gene for genetic engineering in plants to improve stress tolerance.
References
Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P (2008) The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20:2117–2129
Agarwal PK, Agarwal P, Reddy MK, Sopory SK (2006) Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep 25:1263–1274
Cao Z, Li J, Chen F, Li Y, Zhou H, Liu Q (2001) Effect of two conserved amino acid residues on DREB1A function. Biochem 66:623–627
Clough S, Bent A (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Dubouzet J, Sakuma Y, Ito Y, Kasuga M, Dubouzet E, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt-, and cold-responsive gene expression. Plant J 33:751–763
Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K (2011) ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res 124:509–525
Gao C, Wang Y, Jiang B, Liu G, Yu L, Wei Z, Yang C (2011) A novel vacuolar membrane H + -ATPase c subunit gene (ThVHAc1) from Tamarix hispida confers tolerance to several abiotic stresses in Saccharomyces cerevisiae. Mol Biol Rep 38:957–963
Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 124:1854–1865
Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang J (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130:639–648
Hong JP, Kim WT (2005) Isolation and functional characterization of the Ca-DREBLP1 gene encoding a dehydration-responsive element binding-factor-like protein 1 in hot pepper (Capsicum annuum L. cv. Pukang). Planta 220:875–888
Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces cor genes and enhances freezing tolerance. Science 280:104–106
Jaglo-Ottosen KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ, Deits T, Thomashow MF (2001) Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol 127:910–917
Jiang C, Lu B, Singh J (1996) Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol Biol 30:679–684
Karaba A, Dixit S, Greco R, Aharoni A, Trijatmiko K, Marsch-Martinez N, Krishnan A, Nataraja K, Udayakumar M, Pereira A (2007) Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proc Natl Acad Sci U S A 104:15270–15275
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17:287–291
Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2004) A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol 45:346–350
Kizis D, Pagès M (2002) Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. Plant J 30:679–689
Lata C, Prasad M (2011) Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot 62:4731–4748
Li J, Yu G, Sun X, Jia C, Du Q, Li Q, Pan H (2014) Modification of vectors for functional genomic analysis in plants. Genet Mol Res 13:7815–7825
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:391–406
Liu N, Zhong N, Wang G, Li L, Liu X, He Y, Xia G (2007) Cloning and functional characterization of PpDBF1 gene encoding a DRE-binding transcription factor from Physcomitrella patens. Planta 226:827–838
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408
Nakashima K, Ito Y, Yamaguchi-Shinozaki K (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149:88–95
Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M, Kim YK, Nahm BH, Kim JK (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138:341–351
Pellegrineschi A, Reynolds M, Pacheco M, Brito RM, Almeraya R, Yamaguchi-Shinozaki K, Hoisington D (2004) Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome 47:493–500
Qin F, Sakuma Y, Li J, Liu Q, Li Y, Shinozaki K, Yamaguchi-Shinozaki K (2004) Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant Cell Physiol 45:1042–1052
Qin F, Kakimoto M, Sakuma Y, Maruyama K, Osakabe Y, Tran L, Shinozaki K, Yamaguchi-Shinozaki K (2007) Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J 50:54–69
Ruan X, Luo F, Li D, Zhang J, Liu Z, Xu W, Huang G, Li X (2011) Cotton BCP genes encoding putative blue copper-binding proteins are functionally expressed in fiber development and involved in response to high-salinity and heavy metal stresses. Physiol Plant 141:71–83
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration and cold-inducible gene expression. Biochem Biophys Res Commun 290:998–1009
Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18:1292–1309
Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress tolerance and response. J Exp Bot 58:221–227
Sun X, Yu G, Li J, Jia P, Zhang J, Jia C, Zhang Y, Pan H (2014) A heavy metal-associated protein (AcHMA1) from the halophyte, Atriplex canescens (Pursh) Nutt., confers tolerance to iron and other abiotic stresses when expressed in Saccharomyces cerevisiae. Int J Mol Sci 15:14891–14906
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Wilson K, Long D, Swinburne J, Coupland G (1996) A dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8:659–671
Xu Z, Ni Z, Li Z, Li L, Chen M, Gao D, Yu X, Liu P, Ma Y (2009) Isolation and functional characterization of HvDREB1: a gene encoding a dehydration-responsive element binding protein in Hordeum vulgare. J Plant Res 122:121–130
Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6:251–264
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Acknowledgments
This study was supported by grants of the 12th Five-Year Plan Project of Science and Technology Support, China (2012BAD19B04, 2014BAD14B02), by the Research and Development of Industrial Technology Special at Jilin Provincial Development and Reform Commission (2013C001), and by the Ministry of Agriculture Key Project of GM Cultivation of New Varieties (2013ZX08004004), P.R. China.
The Ethical Statement
We all have read the Instructions for Authors carefully and fully comply with the Ethical Standards.
-
1.
The manuscript has not been submitted to more than one journal for simultaneous consideration.
-
2.
The manuscript has not been published previously (partly or in full).
-
3.
A single study is not split up into several parts to increase the quantity of submissions and submitted to various journals or to one journal over time.
-
4.
No data have been fabricated or manipulated (including images) to support our conclusions.
-
5.
No data, text, or theories by others are presented as if they were the author’s own (“plagiarism”). Proper acknowledgements to other works have been given (this includes material that is closely copied (near verbatim), summarized, and/or paraphrased), quotation marks are used for verbatim copying of material, and permissions are secured for material that is copyrighted.
-
6.
Consent to submit has been received explicitly from all coauthors, as well as from the responsible authorities—tacitly or explicitly—at the institute/organization where the work has been carried out, before the work is submitted.
-
7.
Authors whose names appear on the submission have contributed sufficiently to the scientific work and therefore share collective responsibility and accountability for the results.
Conceived and designed the experiments: Hongyu Pan, Wei Zhou, Chengguo Jia; Performed the experiments: Wei Zhou, Chengguo Jia; Analyzed the data: Wei Zhou, Gang Yu, Jinliang Liu, Xianghui Zhang; Wrote the paper: Chengguo Jia, Wei Zhou, Hongyu Pan; Contributed reagents/materials/analysis tools: Others.
-
8.
We will not change authorship or the order of authors after acceptance of this manuscript.
-
9.
We will provide relevant documentation or data in order to verify the validity of the results if needed.
Compliance with Ethical Standards
ᅟ
Conflict of Interest
The authors declare that they have no conflict of interest.
Research without involving human participants and/or animals.
Author information
Authors and Affiliations
Corresponding author
Additional information
Wei Zhou and Cheng-Guo Jia contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhou, W., Jia, CG., Wu, X. et al. ZmDBF3, a Novel Transcription Factor from Maize (Zea mays L.), Is Involved in Multiple Abiotic Stress Tolerance. Plant Mol Biol Rep 34, 353–364 (2016). https://doi.org/10.1007/s11105-015-0926-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-015-0926-2