Abstract
Broccoli (Brassica oleracea L. var. italica) is an important vegetable crop all over the world. However, rapid post-harvest senescence in harvested floral heads reduces its value. Mutation in GIGANTEA (GI) caused delay of flowering and increased tolerance level to H2O2-induced oxidative stress in Arabidopsis. BoGI, a GI orthologue, was isolated and characterized from B. oleracea. BoGI mRNA is expressed throughout development and can be detected in leaves, stem, root, and flowers. Further analysis indicated that the expression of BoGI is modulated by the circadian clock. To investigate the senescence flowering-associated mechanism regulated by BoGI gene and the agricultural application of BoGI in controlling flowering time and floret yellowing for B. oleracea, constructs containing antisense cDNA of BoGI driven by 35S or a flower-specific AP1 promoter were transformed into B. oleracea and the transgenic plants were generated. The flowering time and the senescence of the detached leaves were significantly delayed in transgenic 35S::BoGI antisense plants. Reverse transcriptase polymerase chain reaction (RT-PCR) analysis showed that clear reduction of BoGI expression was observed in these 35S::BoGI antisense plants compared to that in wild-type plants. Furthermore, post-harvest yellow and flower senescence was delayed in AP1::BoGI antisense plants. These findings indicate that BoGI could be involved in regulation of flowering time, leaf, floret, and flower senescence in broccoli.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The transition from vegetative rosette leaf to reproductive inflorescence development has been extensively studied in Arabidopsis. This process involves regulation by multiple environmental and endogenous inputs and corresponding genes which have been identified and characterized (Koornneef et al. 1991; Amasino 1996; Levy and Dean 1998; Reeves and Coupland 2000; Araki 2001; Simpson and Dean 2002). Flowering genes such as FLOWERING LOCUS T (FT) has been thought to be a floral integrator gene, CONSTANS (CO), and GIGANTEA (GI) were involved in the photoperiod flowering pathway, whereas FCA, LUMINIDEPENDENS (LD), and FLOWERING LOCUS D (FLD) have been thought to function in the autonomous flowering pathway to promote flowering (Levy and Dean 1998; Chou and Yang 1998; Reeves and Coupland 2000; Araki 2001; Simpson and Dean 2002). The Arabidopsis GIGANTEA (GI) gene is an earliest acting conserved gene encoding a protein of 1173 amino acids (Koornneef et al. 1991; Fowler et al. 1999; Park et al. 1999; Mizoguchi et al. 2005). GI protein has no sequence similarity to any known proteins in the database (Park et al. 1999; Fowler et al. 1999) and appears to be absent from genomes of Chlamydomonas (microorganism) and animals (Mittag et al. 2005). The GI was originally discovered as a late-flowering mutant (Rédei 1962; Koornneef et al. 1991; Araki and Komeda 1993) and contains novel nuclear localization signals (NLSs) (Huq et al. 2000). GI positively regulates the expression of flowering time genes and acts upstream of CO and FT (Kardailsky et al. 1999; Suárez-López et al. 2001).
In Arabidopsis, gi mutants flowered later and produced more leaves than wild-type plants in long-day (LD) conditions, but not in short-day (SD) conditions (Koornneef et al. 1991). Ectopic expression of Arabidopsis antisense GI gene fragment also caused late flowering in transgenic radish (Curtis et al. 2002). This data indicated that downregulation of the GI gene could delay bolting in a heterologous plant. Arabidopsis GI gene is also important in controlling circadian rhythms (Park et al. 1999; Fowler et al. 1999; Yanovsky and Kay 2002; Edwards et al. 2005; Gould et al. 2006; Kim et al. 2007; Sawa et al. 2007) and phytochrome signaling (Huq et al. 2000). In wild-type Arabidopsis, GI expression is regulated by circadian clock with the highest expression at 8 to 12 h of the light period and the lowest expression at dawn (Park et al. 1999; Fowler et al. 1999). GI has been thought to have a role in early phytochrome signaling by transcriptional regulation of CCA1 and LHY since the expression patterns of these genes are altered in gi mutants (Park et al. 1999; Fowler et al. 1999).
In addition to regulate flowering, it has been shown that GI might also play a role in response to oxidative stress, cold responses, and salt tolerance (Huq et al. 2000; Cao et al. 2005, 2006, 2007; Kim et al. 2013). Mutation in GI significantly increased tolerance level to paraquat or H2O2-induced oxidative stress (Kurepa et al. 1998). GI has been thought to play a complex role in regulating plant development in diverse ways. It is interesting to explore whether mutation in GI can also cause tolerance to other biotic and abiotic stresses in plants and to investigate the possible mechanism of GI regulation.
Broccoli, Brassica oleracea L. var. italica, is a floral vegetable that is consumed worldwide. It contains rich amount of vitamins A and C and isothiocyanine, especially glucoraphanin and its derivative sulforaphane. Broccoli is used for the prevention of breast cancer (Fahey et al. 1997, 2002). Increasing demand for broccoli will necessitate improvement of its agronomic characteristics, such as disease resistance and environmental tolerance. In recent years, several reports have pointed out the merits and biological impacts of transformation technologies (Potrykus 1991; Walden and Wingender 1995; Christou 1996; Birch 1997). Indeed, the genetics of broccoli were recently improved via transgenic breeding (Chen et al. 2008). Antisense approaches regulate fruit ripening, increase floral longevity, and slow post-harvest yellowing have been reported (Smith et al. 1988; Oeller et al. 1991; Gray et al. 1992; Ayub et al. 1996; Aida et al. 1998; Henzi et al. 1999; Giovannoni 2004; Higgins et al. 2006). Broccoli has been transformed with antisenescence genes such as isopentenyl transferase (ipt) and broccoli mutated ethylene response sensor gene (boers) to slow post-harvest yellowing, and transgenic lines with these genes have been developed (Chen et al. 2001, 2004). In this study, we describe that BoGI, the GI orthologue from broccoli, was involved in leaf senescence, flowering time, post-harvest yellowing and flower senescence in broccoli.
Materials and Methods
Plant Materials
Broccoli (B. oleracea var. italica cv. Green King) seeds were obtained from the Know-You Seed Company (Kaohsiung, Taiwan). The seeds were surface-sterilized for 2 min with 70 % EtOH, then 20 min for 1 % sodium hypochlorite, followed by five rinses with sterile distilled water. Seeds were then germinated under aseptic conditions on a hormone-free germination medium in Magenta boxes at 25 °C under 16-h light/8-h dark photoperiod with light intensity of 35 μmol m−2 s−1. This germination medium containing half strength MS salts (Murashige and Skoog 1962) plus B5 (Gamborg et al. 1968) vitamins (MSB5) supplemented with 15 g l−1 sucrose and 8.0 g l−1 agar buffered to pH 5.7. Explants of cotyledon, hypocotyl, and epicotyl from 7-day-old seedlings were used for transformation.
Determination of Antibiotic Sensitivity
A dose-response assay was conducted to determine the optimal concentrations of kanamycin and carbenicillin in the selection medium. Different concentrations of kanamycin (0, 25, 50, 75, and 100 mg l−1) were added to the callus induction medium (MSB5, 30 g l−1 sucrose supplemented with 2.0 μM benzylaminopurine (BAP), 0.5 μM naphthaleneacetic acid (NAA), and 8.0 g l−1 agar). As a control, uninfected explants were cultured on selection medium. Explants were cultured on each selection medium for 5 weeks at 25 °C under 16/8-h photoperiod. This experiment was performed with three replications. After 3 weeks, the number of explants producing callus was calculated. In addition, bacterial cells of A. tumefaciens were cultured on MSB5 medium containing different concentrations of carbenicillin (100–400 mg l−1) to determine the appropriate concentration for inhibiting bacterial growth.
The Cloning of BoGI and the Construction of BoGI Antisense Constructs
A 2.4-kb cDNA fragment containing the sequence from exon 9 to 3′-UTR of BoGI (Fig. 1a) was cloned from broccoli (B. oleracea var. italica cv. Green King) using polymerase chain reaction (PCR) strategy. The 1.7 kb Arabidopsis AP1 promoter fragment was obtained by PCR using primers AP1p 5′ (5′-GTCTTCAAGGCCACAAGCTTAG-3′) and AP1p 3′ (5′-CTCTAAAGGATCCAAACAAAACAAA-3′). The primers contained the generated HindIII (5′-AAGCTT-3′, underlined) or BamHI recognition site (5′-GGATCC-3′, underlined) to facilitate the cloning of this cDNA into PBI121 in which 35S promoter was replaced. A 1.5-kb antisense DNA fragment encoding entire exon 9 was obtained by PCR using primers BoGI-9-1 (5′-AGGATCCGCACTATATGGG-3′) and BoGI-9-2 (5′-GGATCCCAGGAGAAGCGATC-3′). Both specific 5′ and 3′ primers contained the generated BamHI recognition site (5′-GGATCC-3′, underlined) to facilitate the cloning of this cDNA. This antisense fragment was cloned into binary vector PBI121 under the control of cauliflower mosaic virus (CaMV) 35S promoter (35S::BoGI-anti) or flower specific AP1 promoter (AP1::BoGI-anti).
Sequence information for the BoGI protein. a The genomic structure of cloned partial BoGI. This 2.4-kb cDNA fragment of partial BoGI contained the sequence from entire exon 9 to 13 and the 3′-UTR of BoGI. A 1.5-kb antisense DNA fragment encoding entire exon 9 was obtained by PCR using primers BoGI-9-1 and BoGI-9-2. b Sequence comparison of BoGI (B. oleracea), AtGI (Arabidopsis thaliana), and OzGI (rice). The 2.4-kb cDNA fragment of partial BoGI encodes 745 amino acids which showed 91 and 63 % sequence identity to AtGI and OzGI, respectively. The boxed regions represent nuclear localization signals (NLSs) sequence. The underlined regions represent putative transmembrane domains. Amino acid residues identical to AtGI are indicated as dots. To improve the alignment, dashes were introduced into the sequence
Plant Transformation
The Agrobacterium tumefaciens strain LBA 4404 harboring the binary vector pBI121, with 35S::BoGI-anti or AP1::BoGI-anti, was used for broccoli transformation. Hypocotyl explants were cultured on callus induction medium for pre-cultivation (0–4 days). The explants were dipped into an overnight suspension of Agrobacterium. The bacterial concentration was controlled at 0.8–1.0 OD600 and diluted to tenfold. After overnight of co-incubation, explants were transferred in medium containing half MSB5 salts, 20 g l−1 sucrose, and 8.0 g l−1 agar at 25 °C under a 16/8-h (light/dark) photoperiod with light intensity of 45 μmol m−2 s−1 for 2 days. Explants were washed under shaking in half MSB5 liquid medium containing 300 mg l−1 carbenicillin for 2 days with medium replaced daily. After this washing, explants were transferred to selection medium containing MSB5, 2.0 μM BAP, 0.5 μM NAA, 300 mg l−1 carbenicillin, and 75 mg l−1 kanamycin for callus induction. After 3 weeks of culture, well-developed callus was produced from the cut ends of the explants and transferred to MSB5 medium with 30 g l−1 sucrose, 8.0 g l−1 agar, and 2.0 μM BAP for induction of transgenic shoots. After 4 weeks of culture, multiple shoots developing from the callus were separated and transferred to rooting medium containing half MSB5 media supplemented with 20 g l−1 sucrose, 2.0 g l−1 gelrite, 1.0 μM IBA, and 75 mg l−1 kanamycin. The rooted plantlets were washed in sterile distilled water to remove traces of medium and then transferred to plastic pots (5 cm diameter) containing sterile soil, sand, and vermiculite mixture (3:1:1) for 2–3 weeks under 35 μmol m−2 s−1 with 16/8-h photoperiod before transfer to 30 cm pots. The hardened plants were grown to maturity to collect seeds from the T0 plants and raised T1 plants which were also observed under greenhouse condition. The plants (transgenic and non-transgenic) were separately allowed to undergo self-pollination and seeds were collected.
Polymerase Chain Reaction and Reverse Transcriptase Polymerase Chain Reaction Analysis
Total DNA or RNA was isolated from leaves of different putative transgenic plants and non-transformed plants using the Trizol method according to the instructions of the manufacturer. For cDNA synthesis, total RNA (1 μg) was reverse-transcribed in a 20-μl reaction mixture using the BcaBEST™ RNA PCR system (TaKaRa Shuzo Co., Shiga, Japan). A 5.0-μl of cDNA sample from RT reaction was used for PCR. Primers specific for BoGI: BoGI-9-1 (5′-AGGATCCGCACTATATGGG-3′) and BoGI-9-2 (5′-GGATCCCAGGAGAAGCGATC-3′); BoGIin-1 (5′-AACCGTAAAAGTAGGAACGTCAAGG-3′) and BoGIin-2 (5′-AGCAGATCTTTGATACCCTTCTCTG-3′). Primers specific for detection of the presence of BoGI antisense fragment from genomic DNA of 35S::BoGI antisense and AP1::BoGI antisense plants: BoGI-9-2 (in BoGI) (5′-GGATCCCAGGAGAAGCGATC-3′) and GUS-2 (in T-DNA) (5′-AGTTCAGTTCGTTGTTCACACA-3′). The fragment was amplified under the following conditions of initial hot start at 95 °C for 5 min, then 35 cycles of denaturation (95 °C; 1 min), annealing (56 °C; 1 min), and extension (72 °C; 1), followed by a final extension of 20 min at 72 °C. The reverse transcriptase polymerase chain reaction (RT-PCR) products were separated on a 1 % agarose gel by electrophoresis and photographed using a Kodak EDAS 290 Electrophoresis documentation system.
Phenotypic Analysis of Leaves, Floral Heads, and Flowers
Leaves from lower parts of mature plants without any symptom of yellow were detached, and floret heads were harvested 7–15 days after the beginning of bolting. The detached leaves and a branch from each floret head were placed in a plastic container, covered with a transparent plastic film, and exposed under light (14 h light/10 h dark) at 25 °C in the controlled room or an incubator for 12 days. Flower senescence analyses were observed under greenhouse condition.
Inheritance Analysis
Seeds (25 seeds from each plant) collected from five transgenic plants (T0) were sterilized and germinated on MSB5 basal media supplemented with 75 mg l−1 kanamycin. The plates were incubated under the conditions described for in vitro culture. After 1 week, kanamycin sensitive seedlings germinated but bleached quickly, whereas resistant seedlings were green and formed true leaves and roots. The transgenic T1 plants were analyzed for the presence of the transgenic BoGI gene by the PCR assay.
Statistical Analysis
All the experiments were set up in a completely randomized design, and experiments were performed in triplicates and each experiment was repeated twice. The data was expressed as means ± standard error. One-way ANOVA analysis followed by Duncan’s test was used to determine significant (P < 0.05) differences. All the statistical analyses were done by using SPSS software version 9.0 (SPSS Inc. USA).
Results
Isolation of BoGI cDNA from B. oleracea
To investigate the function of GI gene in broccoli, the identification and functional analysis of broccoli GI homologues is necessary. For this purpose, a partial sequence of broccoli GI homologue of (BoGI) (Fig. 1a) was cloned and characterized. This 2.4 kb cDNA fragment contained the sequence from exon 9 to 3′-UTR of BoGI and encodes 745 amino acids (Fig. 1b). The BoGI showed 91 and 63 % sequence identity to Arabidopsis AtGI and rice OzGI, respectively (Fig. 1b). Similar to GI, a region contained four nuclear localization signal (NLS) sites was also identified for BoGI (Fig. 1b).
Gene Expression of BoGI
To further explore the gene expression patterns, the detection of BoGI expression is necessary. As shown in Fig. 2a, BoGI mRNA is expressed throughout development and can be detected in leaves, stem, root, and flowers. In addition to high sequence homology to GI in Arabidopsis, the expression of BoGI was also regulated by light. The expression level of BoGI mRNA varied under LD conditions, with the level highest at the 8 to 12 h of the light period (Fig. 2b, c) similar to that observed for GI in Arabidopsis (Fowler et al. 1999). This result suggested that GI homologue of broccoli was regulated by mechanisms similar to that in Arabidopsis.
Detection of expression of BoGI by RT-PCR analysis. a Total RNA isolated at 8 h of the light period from root, stem, cotyledon of a 10-day-old seedlings, and stem, leaf, and flower bud of a 6-month-old B. oleracea var. italica were used as templates to detect the expression of BoGI. rRNA stained in an EtBr gel was used to show the amount of RNA used for each RT-PCR reactions. b To examine whether the expression of BoGI was influenced by light, total RNA was isolated and analyzed from leaf of 25-day-old B. oleracea var. italica plants every 6 h. Sample collection started at the beginning of the light period (time 0) and continued every 6 h for 24 h in LD conditions. Each experiment was repeated three times with similar results. rRNA stained in an EtBr gel was used to show the amount of RNA used for each RT-PCR reactions. The related strength of the BoGI expression for each transgenic plant were listed below. c A statistical result from b which clearly showed that the expression level of BoGI mRNA varied under LD conditions, with the level highest at the 8 to 12 h of the light period and lowest at dawn
Genetic Transformation of Broccoli
Hypocotyl has a better survival rate than cotyledon or epicotyl (Table 1). Similar results were reported for B. oleracea by Chen et al. (2001). After 4 weeks of culture, 90 % callus induction was attained in explants cultured on callus induction medium but lacking kanamycin (Tables 1 and 2). On medium containing kanamycin, the percentage of explants with callus (36 %) was obtained at 25 mg l−1. At 75 mg l−1, 100 % of explants bleached and died (Table 2). To analyze the effect of 35S::BoGI antisense and AP1::BoGI antisense in B. oleracea, Agrobacterium-mediated genetic transformation was performed. In broccoli, the reported kanamycin concentrations employed for nptII gene selection were 50 mg l−1 (De Block et al. 1989) and 25 mg l−1 (Metz et al. 1995). Chen et al. (2001) reported that 75 mg l−1 kanamycin for selection, over 60 % of survival plantlets was positive for both the nptII and the ipt genes. We have used 75 mg l−1 kanamycin for the transformation experiments.
The effect of pre-culture on the frequency of transformation in broccoli was examined using hypocotyl explants cultured on MSB5 medium supplemented with 2.0 μM BAP and 0.5 μM NAA for 3 days. Similar reports have also addressed the adaptation of pre-culture for the success of transformation (McHughen et al. 1989; Sangwan et al. 1992; Metz et al. 1995; Villemont et al. 1997), and it was shown that putative transformation competent cells could be increased by pretreatment (Sangwan et al. 1992). After 2 days of co-cultivation with 35S::BoGI antisense, the explants were transferred onto the callus induction medium consisted of MSB5 containing 2.0 μM BAP, 0.5 μM NAA, 300 mg l−1 carbenicillin, and 75 mg l−1 kanamycin. This was in accordance with other Brassica species (Metz et al. 1995; Paul and Sikdar 1999; Cao et al. 2008). Callus initiation was observed in selective medium within 4 weeks of culture, while non-transformed control explants on selective medium turned yellow and did not produce calli. Transgenic callus were transferred to MSB5 containing 2.0 μM BAP for induction of transgenic shoots. Shoots that survived this selection stage were transgenic and were transferred to rooting medium containing half MSB5 media supplemented with 20 g l−1 sucrose, 2.0 g l−1 gelrite, 1.0 μM IBA, and 75 mg l−1 kanamycin. The rooted plantlets were transferred to pots, acclimated for 2 weeks in the culture room, and were moved to the greenhouse; similar to that for the non-transgenic rooted plants generated from the tissue culture. Progeny analysis for most transgenic T0 plants in 75 mg l−1 kanamycin medium showed a 3:1 ratio for kanamycin resistant/susceptible phenotype, indicating a likely single gene insertion.
Delay of Flowering and Leaf Senescence was Observed in 35S::BoGI Antisense of B. oleracea
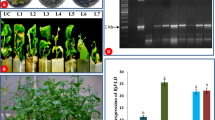
The construct contained antisense of BoGI, a GI homologue of B. oleracea, driven by the CaMV 35S promoter was transformed into broccoli, and ten transgenic plants (T1) were obtained and analyzed. These 35S::BoGI antisense transformants showed similar late-flowering phenotype that differed strongly from non-transformed broccoli plants. The flowering time for these 35S::BoGI antisense plants was significantly delayed (Fig. 3a). Compared to non-transformed wild-type broccoli that flowered at about 100 days after sowing, 35S::BoGI antisense plants flowered at about 180 days after sowing (Fig. 4a). When the detached leaves from these 35S::BoGI antisense plants were examined, their senescence was significantly delayed (Fig. 3b–e). Detached leaves from non-transformed wild-type broccoli were senescent within 3 days by showing the dehydration of the leaf tissue and diminution of the chloroplasts resulting in the yellowish color of the leaves (Fig. 3c). By contrast, leaves from 35S::BoGI antisense plant were not senescent even after 5 days of detachment (Fig. 3c). At 7 days after detachment, wild-type leaves were completely senescent, whereas only slight yellowing was observed in 35S::BoGI antisense leaves (Fig. 3d). At 12 days after detachment, wild-type leaves were completely dry (Fig. 3e, f), whereas petiole and part of the leaf were still green in 35S::BoGI antisense leaves (Fig. 3e, g). This result indicated that BoGI regulates flowering and leaf senescence in broccoli.
Phenotypic analysis of transgenic (T1) B. oleracea var. italica ectopically expressing antisense of BoGI. a A 35S::BoGI antisense broccoli plant (middle) flowered significantly later than wild-type (left) and AP1::BoGI antisense plant (right). At 180 days after germination, wild-type and AP1::BoGI antisense plants already flowered and set siliques and seeds, whereas 35S::BoGI antisense plant remained in vegetative stage without flowering. b–e Detached leaves from wild-type broccoli (left) were senescent within 3 days by showing the dehydration of the leaf tissue and diminution of the chloroplasts resulting in the yellowish color of the leaves. By contrast, leaves from 35S::BoGI antisense plant (right) was not senescent after 5 days of detachment. b 1 day, c 5 days, d 7 days, and e 12 days after detachment. f Close-up of the wild-type leaf petiole in e. In this stage, leaf tissue was completely dried. g Close-up of the 35S::BoGI antisense leaf petiole in e. In this stage, some leaf tissue still remained green. h The floral head of an AP1::BoGI antisense broccoli plant (top) was not senescent 7 days after detachment, whereas the floral head of a wt broccoli plant (bottom) was completely senescent and turned to yellow in the same stage
Comparison of flowering time in non-transgenic and BoGI antisense transgenic plants (T1) of B. oleracea var. italica. a Flowering time of wild type, AP1::BoGI antisense, and 35S::BoGI antisense transgenic broccoli under long-day condition. The genotype tested is shown along the horizontal axis and the average from 10 (T1) anti-BoGI transformants (AP1:: and 35S::), and non-transgenic plants were compared. Vertical bar represented SD. b Comparison of flower senescence time in non-transgenic and transgenic plants of AP1::BoGI antisense and 35S::BoGI antisense. The data measured survival days of 10 mature flowers from flowering until fading. Vertical bar represented SD
The presence of the BoGI antisense gene in transgenic 35S::BoGI antisense plants was further confirmed by the PCR assay (Fig. 5a). To explore whether the phenotype correlated to BoGI expression in 35S::BoGI antisense plants, RT-PCR analysis was performed. As shown in Fig. 5b, a clear reduction of BoGI expression was observed in these 35S::BoGI antisense plants compared to that in wild-type plants. This result indicated that the phenotypes generated in 35S::BoGI antisense transgenic broccoli were due to the repression of the BoGI expression.
PCR and RT-PCR assay for transgenic (T1) B. oleracea var. italica ectopically expressing antisense of BoGI. a Detection of the presence of the DNA fragment for BoGI antisense gene in two 35S::BoGI antisense transgenic (T1) Brassica plants (1–2) by PCR. A 1.5-kb fragment was amplified in these two transgenic plants, whereas no band can be amplified in wild-type (WT) plant. b Detection of the BoGI expression in four 35S::BoGI antisense transgenic (T1) Brassica plants (1–4) and in a non-transgenic wild-type (WT) plant through RT-PCR analysis. rRNA stained in an EtBr gel was used to show the amount of RNA used for each RT-PCR reactions. c Detection of the presence of the DNA fragment for BoGI antisense gene in two AP1::BoGI antisense transgenic (T1) Brassica plants (1–2) by PCR. A 1.5-kb fragment was amplified in these two transgenic plants, whereas no band can be amplified in wild-type (WT) plant
Delay of Floret Yellowing was Observed in AP1::BoGI Antisense of B. oleracea
To provide more evidence that GI function was related with senescence, construct containing antisense of BoGI driven by the flower specific AP1 promoter was transformed into broccoli and ten transgenic plants (T1) were obtained and analyzed. The flowering time for these AP1::BoGI antisense plants was similar to the non-transformed plants (Figs. 3a and 4a). When the post-harvested florets from these AP1::BoGI antisense plants were examined, their yellowing was significantly delayed (Fig. 3h). Post-harvested florets from non-transformed wild-type broccoli were yellow within 3 days. By contrast, florets from AP1::BoGI antisense plant were not yellow even after 7 days of harvested florets (Fig. 3h). At 7 days after harvested florets, wild-type florets were completely yellowing, whereas no yellowing was observed in AP1::BoGI antisense florets (Fig. 3h). When the flower senescence time was measured in plants from flowering until fading, the flower senescence for these AP1::BoGI antisense plants was also significantly delayed (Fig. 4b). Compared to non-transformed wild-type broccoli flowers that were senescent at about 15 days after flowering, AP1::BoGI antisense flowers faded at about 25–30 days after flowering (Fig. 4b). This result indicated that BoGI also regulates floret and flower senescence in broccoli. The presence of the BoGI antisense gene in transgenic AP1::BoGI antisense plants was further confirmed by the PCR assay (Fig. 5c).
Discussion
In an attempt to investigate the roles for the GI gene in regulating plant development beside flowering, GI orthologue BoGI was cloned and characterized from B. oleracea. BoGI contains several characteristics that were conserved among GI orthologues. For example, all four putative NLS regions identified in GI protein (Fowler et al. 1999; Huq et al. 2000) were found in the BoGI proteins (Fig. 1b). Furthermore, similar to GI (Park et al. 1999; Fowler et al. 1999), BoGI mRNA is also regulated by circadian clock with the level highest at 8 to 12 h of the light period and lowest at dawn (Fig. 2b, c). Based on the sequence, structure, and expression similarity, it is clear that BoGI is B. oleracea GI orthologue.
Leaf senescence exhibits chloroplast degradation and declined photosynthetic activity that finally results in cell death, which could be regarded as a form of programmed cell death (Noodén and Leopold 1978; Gan and Amasino 1997; Lim et al. 2003; van Doorn and Woltering 2004). The terminal phase of leaf senescence was indicated by loss of antioxidant capacity and increased release of reactive oxygen species (ROS) in plants (Leshem 1988; Buchanan-Wollaston 1997; Zimmermann and Zentgraf 2005). Since gi mutants significantly increased tolerance level to H2O2-induced oxidative stress (Kurepa et al. 1998), the senescence of the leaf was therefore analyzed in antisense BoGI plants of B. oleracea in this study. Ectopic expression of GI homologue BoGI antisense did not only cause the late flowering but also the delay of leaf senescence phenotypes in transgenic broccoli. Therefore, in addition to promote flowering time, the ability for GI homologues to regulate leaf senescence in plants was confirmed. Since we only tested the flowering time and leaf senescence under long-day conditions (14 h light/10 h dark), whether the delay of the flowering time and leaf senescence in 35S::BoGI antisense transgenic broccoli was similar under short-day conditions remains to be investigated. Interestingly, the correlation between flowering time and leaf senescence has been reported. For example, the development of early flowering and leaf senescence under long-day condition was observed in vtcl and vtc2 mutants (Barth et al. 2006). Delay of flowering and senescence was also reported in HDA6 mutant of axe1-5 (Wu et al. 2008). These data further support the idea that a complex mechanism regulating both flowering and leaf senescence.
More recently, researchers have been studying early gene changes in an attempt to understand the regulation of post-harvest senescence (Coupe et al. 2003; Eason et al. 2005; Gapper et al. 2005a, b; Page et al. 2001; Pogson et al. 1995; Pramanik et al. 2005). External application of cytokinins is known to retard yellowing in broccoli (Rushing 1990; Clarke et al. 1994; Tian et al. 1995). Rushing (1990) reported that post-harvest broccoli florets deteriorated rapidly and became unmarketable in 3 to 4 days, while florets treated with benzyladenine or zeatin lasted yellow retardation for 5 to 6 days. Harvested broccoli deteriorates quickly due to relatively high respiration and susceptibility to wilting when stored at room temperature (Gillies and Toivonen 1995). Wang (1977) studied the length of broccoli shelf life and observed that florets showed significant yellowing after 2–3 days at 20 °C and essentially complete yellowing in 4 days. The yellowing is caused by sepal chlorophyll degradation, but the pedicel and stem did not turn yellow after harvest (Corcuff et al. 1996). In this study, we found that the floret of AP1::BoGI antisense transgenic broccoli did not turn yellow even after 7 days of being harvested. By contrast, wild-type florets were completely yellowing at 7 days after being harvested. Furthermore, the flower senescence of the mature flowers in these AP1::BoGI antisense plants was significantly delayed compared to non-transformed wild-type and 35S::BoGI antisense broccoli flowers. 35S::BoGI antisense flowers faded at about 20 days after flowering, slightly later than that in non-transformed wild-type plant (about 15 days) (Fig. 4b). Thus, we have demonstrated that the expression of an anti-GI homologue gene fragment in broccoli could cause effects in delaying flowering time, leaf senescence, and post-harvest yellow retardation.
References
Aida R, Yoshida T, Ichimura K, Goto R, Shibata M (1998) Extension of flower longevity in transgenic torenia plants incorporating ACC oxidase transgene. Plant Sci 138:91–101
Amasino RM (1996) Control of flowering time in plants. Curr Opin Genet Dev 6:480–487
Araki T (2001) Transition from vegetative to reproductive phase. Curr Opin Plant Biol 4:63–68
Araki T, Komeda Y (1993) Analysis of the role of the late flowering locus, GI, in the flowering of Arabidopsis thaliana. Plant J 3:231–239
Ayub R, Guis M, Amor MB, Gillot L, Roustan JP, Latche A, Bouzayen M, Pech JC (1996) Expression of ACC oxidase antisense gene inhibits ripening of cantaloupe melon fruits. Nat Biotechnol 14:862–866
Barth C, Tullio MD, Conklin PL (2006) The role of ascorbic acid in the control of flowering time and the onset of senescence. J Exp Bot 57:1657–1665
Birch RG (1997) Plant transformation: problems and strategies for practical application. Annu Rev Plant Physiol Plant Mol Biol 48:297–326
Buchanan-Wollaston V (1997) The molecular biology of leaf senescence. J Exp Bot 48:181–199
Cao SQ, Ye M, Jiang ST (2005) Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep 24:683–690
Cao SQ, Jiang ST, Zhang RX (2006) The role of GIGANTEA gene in mediating the oxidative stress response and in Arabidopsis. Plant Growth Regul 48:261–270
Cao SQ, Song YQ, Su L (2007) Freezing sensitivity in the gigantean mutant of Arabidopsis is associated with sugar deficiency. Biol Plant 51:359–362
Cao J, Shelton AM, Earle ED (2008) Sequential transformation to pyramid two Bt genes in vegetable Indian mustard (Brassica juncea L.) and its potential for control of diamondback moth larvae. Plant Cell Rep 27:479–487
Chen LFO, Hwang JY, Charng YY, Sun CW, Yang SF (2001) Transformation of broccoli (Brassica oleracea var. italica) with isopentenyltransferase gene via Agrobacterium tumefaciens for postharvest yellowing retardation. Mol Breed 7:243–257
Chen LFO, Huang JY, Wang YH, Chen YT, Shaw JF (2004) Ethylene insensitivity and postharvest yellowing retardation in mutant ethylene response sensor (boers) gene transformed broccoli (Brassica oleracea var. italica.). Mol Breed 14:199–213
Chen LFO, Lin CH, Kelkar SM, Chang YM, Shaw JF (2008) Transgenic broccoli (Brassica oleracea var. italica) with antisense chlorophyllase (BoCLH1) delays postharvest yellowing. Plant Sci 174:25–31
Chou ML, Yang CH (1998) FLD interacts with genes that affect different developmental phase transitions to regulate Arabidopsis shoot development. Plant J 15:231–242
Christou P (1996) Transformation technology. Trends Biotechnol 1:423–431
Clarke SF, Jameson PE, Downs C (1994) The influence of 6-benzyl-aminopurine on post-harvest senescence of floral tissues of broccoli Brassica oleracea var. italica. Plant Growth Regul 14:21–27
Corcuff R, Arul J, Hamza F, Castaigne F, Makhlouf J (1996) Storage of broccoli florets in ethanol vapor enriched atmosphere. Postharvest Biol Technol 7:219–229
Coupe SA, Sinclair BK, Watson LM, Heyes JA, Eason JR (2003) Identification of dehydration-responsive cysteine proteases during post-harvest senescence of broccoli florets. J Exp Bot 54:1045–1056
Curtis IS, Nam HG, Yun JY, Seo KH (2002) Expression of an antisense GIGANTEA (GI) gene fragment in transgenic radish causes delayed bolting and flowering. Transgenic Res 11:249–256
De Block M, De Brouwer D, Tenning P (1989) Transformation of Brassica napus and Brassica oleracea using Agrobacterium tumefaciens and the expression bar and new genes in transgenic plants. Plant Physiol 91:694–701
Eason JR, Ryan DJ, Watson LM, Hedderley D, Christey MC, Braun RH, Coupe SA (2005) Suppression of the cysteine protease, aleurain, delays floret senescence in Brassica oleracea. Plant Mol Biol 57:645–657
Edwards KD, Lynn JR, Gyula P, Nagy F, Millar AJ (2005) Natural allelic variation in the temperature compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics 170:387–400
Fahey JW, Zhang Y, Talalay P (1997) Broccoli sprouts: an exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc Natl Acad Sci U S A 94:10367–10372
Fahey JW, Haristoy X, Dolan PM, Kensler TW, Scholtus I, Stephenson KK, Talalay P, Lozniewski A (2002) Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyreneinduced stomach tumors. Proc Natl Acad Sci U S A 99:7610–7615
Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18:4679–4688
Gamborg O, Miller R, Ojima K (1968) Nutrients requirements of suspension cultures of soybean root cells. Exp Cell Res 50:151–158
Gan S, Amasino RM (1997) Making sense of senescence. Molecular genetic regulation and manipulation of leaf senescence. Plant Physiol 113:313–319
Gapper NE, Coupe CA, McKenzie MJ, Sinclair BK, Lill RE, Jameson PE (2005a) Regulation of harvest-induced senescence in broccoli (Brassica oleracea var. italica) by cytokinin, ethylene, and sucrose. J Plant Growth Regul 24:153–165
Gapper NE, Coupe SA, McKenzie MJ, Scott RW, Christey MC, Lill RE, McManus MT, Jameson PE (2005b) Senescence-associated down-regulation of 1-aminocyclopropane-1-carboxylate (ACC) oxidase delays harvest-induced senescence in broccoli. Funct Plant Biol 32:891–901
Gillies SL, Toivonen PMA (1995) Cooling method influences the postharvest quality of broccoli. HortSci 30:313–315
Giovannoni JJ (2004) Genetic regulation of fruit development and ripening. Plant Cell 16:170–180
Gould PD, Locke JCW, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, Hall A (2006) The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18:1177–1187
Gray J, Picton S, Shabbeer J, Schuch W, Frierson D (1992) Molecular biology of fruit ripening and its manipulation with antisense genes. Plant Mol Biol 19:69–87
Henzi MX, Christey MC, McNeil DL, Davies KM (1999) Agrobacterium rhizogenes-mediated transformation of broccoli (Brassica oleracea L. var. italica) with an antisense 1-aminocyclopropane-1-carboxylic acid oxidase gene. Plant Sci 143:55–62
Higgins JD, Newbury HJ, Barbara DJ, Muthumeenakshi S, Puddephat IJ (2006) The production of marker-free genetically engineered broccoli with sense and antisense ACC synthase 1 and ACC oxidase 1 and 2 to extend shelf-life. Mol Breed 17:7–20
Huq E, Tepperman JM, Quail PH (2000) GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc Natl Acad Sci U S A 97:9789–9794
Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286:1962–1965
Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449:356–360
Kim WY, Ali Z, Park HJ, Park SJ, Cha JY, Perez-Hormaeche J, Quintero FJ, Shin G, Kim MR, Qiang Z, Ning L, Park HC, Lee SY, Bressan RA, Pardo JM, Bohnert HJ, Yun DJ (2013) Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat Commun 4:1352–1364
Koornneef M, Hanhart CJ, van der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229:57–66
Kurepa J, Smalle J, Van Montagu M, Inze D (1998) Effects of sucrose supply on growth and paraquat tolerance of the late-flowering gi- 3 mutant. Plant Growth Regul 26:91–96
Leshem YY (1988) Plant senescence processes and free radicals. Free Rad Biol Med 5:39–49
Levy YY, Dean C (1998) The transition to flowering. Plant Cell 10:1973–1990
Lim PO, Woo HR, Nam HG (2003) Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci 8:272–278
McHughen A, Jordan M, Feist G (1989) A preculture period prior to Agrobacterium inoculation increase production of transgenic plants. J Plant Physiol 135:245–248
Metz TD, Dixit R, Earle ED (1995) Agrobacterium tumefaciens-mediated transformation of broccoli (Brassica oleracea var. italica) and cabbage (B. oleracea var. capitata). Plant Cell Rep 15:287–292
Mittag M, Kiaulehn S, Johnson CH (2005) The circadian clock in Chlamydomonas reinhardtii. What is it for? What is it similar to? Plant Physiol 137:399–409
Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, Coupland G (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17:2255–2270
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Noodén LD, Leopold AC (1978) Phytohormones and the endogenous regulation of senescence and abscission. In: Letham DS, Goodwin PB, Higgins TJV (eds) Phytohormones and related compounds: A comprehensive treatise, vol II. Elsevier, Amsterdam, pp 329–369
Oeller PA, Ming-Wong L, Taylor L, Pike DA, Theologis A (1991) Reversible inhibition of fruit senescence by antisense RNA. Science 254:437–439
Page T, Griffiths G, Buchanan-Wollaston V (2001) Molecular and biochemical characterization of postharvest senescence in broccoli. Plant Physiol 125:718–727
Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285:1579–1582
Paul S, Sikdar SR (1999) Expression of npt II marker and gus reporter genes and their inheritance in subsequent generations of transgenic Brassica developed through Agrobacterium mediated gene transfer. Curr Sci 76:1569–1573
Pogson BJ, Downs CG, Davies KM (1995) Differential expression of two 1-aminocyclopropane-1-carboxylic acid oxidase genes in broccoli after harvest. Plant Physiol 108:651–657
Potrykus I (1991) Gene transfer to plants: assessment of published approaches and results. Annu Rev Plant Physiol Plant Mol Biol 432:205–225
Pramanik BK, Matsui T, Suzuki H, Kosugi Y (2005) A sucrose synthase gene from broccoli: cDNA cloning sequencing and its expression during storage. Biotechnology 4:288–295
Rédei GP (1962) Supervital mutants of Arabidopsis. Genetics 47:443–460
Reeves PH, Coupland G (2000) Response of plant development to environment: control of flowering by daylength and temperature. Curr Opin Plant Biol 3:37–42
Rushing JW (1990) Cytokinins affect respiration, ethylene production, and chlorophyll retention of packaged broccoli florets. HortSci 25:88–90
Sangwan RS, Bourgeois Y, Brown S, Vasseur G, Sangwan-Norreel B (1992) Characterization of competent cells and early events of Agrobacterium-mediated genetic transformation in Arabidopsis thaliana. Planta 188:439–456
Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318:261–265
Simpson GG, Dean C (2002) Arabidopsis, the Rosetta stone of flowering time? Science 296:285–289
Smith CJS, Watson CF, Ray J, Bird CR, Morris PC, Schuch W, Grierson D (1988) Antisense RNA inhibition of polygalacturonase gene expression in transgenic tomatoes. Nature 334:724–726
Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410:1116–1120
Tian MS, Davies L, Downs CG, Liu XF, Lill RE (1995) Effects of floret maturity, cytokinin and ethylene on broccoli yellowing after harvest. Postharvest Biol Tech 6:29–40
van Doorn WG, Woltering EJ (2004) Senescence and programmed cell death: substance or semantics? J Exp Bot 55:2147–2153
Villemont E, Dubois F, Sangwan RS, Vasseur G, Bourgeois Y, Sangwan-Norreel B (1997) Role of the host cell cycle in the Agrobacterium-mediated genetic transformation of Petunia: evidence of an S-phase control mechanism for T-DNA transfer. Planta 201:160–172
Walden R, Wingender R (1995) Gene-transfer and plantregeneration techniques. Trends Biotechnol 13:324–331
Wang CY (1977) Effect of aminoethoxy analog of rhizobitoxine and sodium benzoate on senescence of broccoli. HortSci 12:54–56
Wu K, Zhang L, Zhou C, Yu CW, Chaikam V (2008) HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot 59:225–234
Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419:308–312
Zimmermann P, Zentgraf U (2005) The correlation between oxidative stress and leaf senescence during plant development. Cell Mol Biol Lett 10:515–553
Acknowledgments
This work was supported by grants to C-H Y from the Ministry of Science and Technology, Taiwan, ROC, grant numbers: NSC95-2317-B-005-006 and NSC96-2317-B-005-019. This work was also supported in part by the Ministry of Education, Taiwan, ROC, under the ATU plan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Muthu Thiruvengadam and Ching-Fang Shih contributed equally to this work.
Rights and permissions
About this article
Cite this article
Thiruvengadam, M., Shih, CF. & Yang, CH. Expression of An Antisense Brassica oleracea GIGANTEA (BoGI) Gene in Transgenic Broccoli Causes Delayed Flowering, Leaf Senescence, and Post-Harvest Yellowing Retardation. Plant Mol Biol Rep 33, 1499–1509 (2015). https://doi.org/10.1007/s11105-015-0852-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-015-0852-3