Abstract
Pathogenesis-related (PR) proteins encoded by plant defense genes play key roles in plant disease-resistance responses, specialized in systemic-acquired resistance. However, their roles in the response of wheat to fungal infection are still not well known. Our earlier studies have reported that a full-length TcLr19PR1 gene (818 bp) was isolated from wheat infected by leaf rust (Puccinia triticina). Here, we showed that TcLr19PR1 was induced earlier and its expression level was higher in the incompatible interaction between seedling stage of wheat and P. triticina than in compatible interactions. TcLr19PR1 was strongly induced after P. triticina inoculation and ABA and SA treatments, in which the expression level of TcLr19PR1 significantly increased and reached the maximum at 12 and 72 h, respectively. Furthermore, the transgenic T1-stable TcLr19PR1 lines generated in the susceptible wheat cultivar Zhengzhou 5389 background exhibited certain extent disease resistance against P. triticina infection by preventing disease development. In addition, Western blotting was conducted to confirm that TcLr19PR1 protein was induced by P. triticina infection in positive transgenic plants at the protein expression level. These findings suggest that TcLr19PR1 gene plays an important role in wheat development and resistance to leaf rust pathogen attack.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Leaf rust, caused by Puccinia triticina, is one of the most destructive diseases of common wheat all over the world (Komer 1996). Its epidemic commonly causes both yield losses and production with lower quality, and it occurs more frequently and in more worldwide regions as compared with stem rust of wheat (P. graminis f. sp. tritici) or stripe rust of wheat (Puccinia striiformis f. sp. tritici) (Bolton et al. 2008). Development of genetic resistance to rust has been approved as the most efficient, cost-effective, and environment-friendly approach to prevent the losses caused by rust epidemics (Singh et al. 2004). However, new wheat cultivars lose their resistance due to the occurrence of new virulent races. Therefore, it is important to elucidate the molecular mechanism of resistance to leaf rust and to use wheat cultivars with durable resistance.
Plants show a series of complex physiological and biochemical changes in response to pathogen attack in co-evolution process. Recognition of a pathogen often triggers a localized resistance reaction, known as the hypersensitive response (HR) which is characterized by rapid cell death around the site of infection, so that the growth of pathogen is inhibited and a series of defense responses are stimulated causing plant systemic-acquired resistance (SAR) (Hammond-Kosack and Jones 1996; Ryals et al. 1996).
The SAR is a "whole-plant" resistance response that occurs following an earlier localized exposure to a pathogen. The resistance conferred is long-lasting and effective against a broad spectrum of pathogens, including viruses, bacteria, fungi, and oomycetes (Durrant and Dong 2004). SAR can be activated in many plants by pathogens, either as part of the HR or as a symptom of disease. SAR is characterized by the increased expression of large number pathogenesis-related genes (PR genes) (Yoti et al. 2013; Uwe. 2006).
In host-pathogen interactions, various novel proteins are induced which are collectively referred to as “pathogenesis-related proteins” (PRs) (Hamamouch et al. 2011), does not only accumulate locally in the infected leaf but also induced systemically, which defined as proteins coded by the host plant but induced specifically in pathological or related situations (Hamamouch et al. 2011; Van Loon et al. 2006). Induction of PRs has been found in many plant species belonging to various families, PR1 to PR17 (Sels et al. 2008; Liu and Ekramoddoullah 2006), which suggests a general role for these proteins in resistant reaction and in adaptation to biotic stress conditions. Many PRs have direct antibacterial activity (Reddy. 2013). In different families of PRs, PR1 is one of the most important proteins whose function is unclear. PR1 gene induced by pathogenic bacteria, salicylic acid (SA), or ethylene (ET) is often used as a molecular marker gene of SAR (Van Loon and Van 1999), and the research on the proteins encoded by PR1 is a “hot spot” in the area of resistance of plant.
PR1 genes were first discovered in tobacco (Nicotiana tabacum L.) (Antoniw et al. 1980), then many PR1 genes were identified from many monocotyledonous and dicotyledonous plants (Mitsuhara et al. 2008; Li et al. 2011). Most PR1 proteins with molecular weights of 14~17 kD are alkaline. PR1 family is found most abundantly in PRs. PR1 family is highly conserved in structure and shows a high level of similarity in some domains. Their amino acid sequences share a certain degree of homology. PR1 proteins have the same functions in eukaryotic cells (Aglika. 2005) and participate in plant responses to various biotic and abiotic stresses. Some research results showed that PR1 proteins mediated resistance of host plant to fungi. Yang et al. (2013) found that PR1 was upregulated in Paeonia suffruticosa (‘Luhehong’) infected by Cylindrocladium canadense at 24 h postinoculation (hpi), which suggested that PR1 was closely associate with P. suffruticosa resistance to C. canadense. Sujon et al. (2005) found that highly expressed PR1 gene in Phytophthora nicotianae can enhance plant resistance to tobacco mosaic virus.
Although many PR1 genes were cloned and reported to be involved in plant disease-resistance responses and SAR, fewer studies targeted to the function of PR gene. In our earlier studies, a PRl gene designated TcLr19PR1 with an open reading frame (ORF) of 495 bp encoding 164-amino acids from wheat near-isogenic lines TcLr19 infected by leaf rust (P. triticina) was obtained and located on chromosome 7D by nulli-tetrasomic lines of “Chinese Spring,” which is in common with the location of wheat-leaf rust-resistance gene Lr19. Semiquantitative PCR showed that the expression of TcLr19PR1 gene was induced by leaf rust pathogen (Wang et al. 2012). In the present study, the expression of TcLr19PR1 gene induced by pathogen, salicylic acid (SA) and abscisic acid (ABA) was measured temporally and spatially in wheat tissues to explore the relationship between TcLr19PR1 gene and leaf rust disease resistance. To further understand the gene’s function, the constructed vector pAHC-TcLr19PR1 was transformed into immature embryo-derived calli of a susceptible wheat cultivar ZhengZhou 5389 through particle bombardment, and molecular detection and disease-resistance identification of transgenic wheat with TcLr19PR1 gene were studied. These results will provide systematic data for further study on the function and the mechanism of TcLr19PR1 in defending adversity stress.
Materials and Methods
Plant Materials and Inoculation
Wheat (Triticum aestivum L.) near isogenic line TcLr19, Thatcher, and leaf rust (P. triticina) race 07-10-421-3 (FHJT) were the biological materials used for expression analyses. The susceptible wheat cultivar ZhengZhou 5389 was used for generating transgenic lines. Based on the gene-for-gene model, wheat TcLr19 and leaf rust race 07-10-421-3 showed the incompatible reaction (type). However, Thatcher and leaf rust race 07-10-421-3 showed the compatible reaction (type 4) according to Roelfs standard (1985). The plants were grown in a 15-cm diameter pot in the greenhouse for about 7 days when primary leaves were fully expanded, leaf rust urediospores collected from the susceptible wheat Zhengzhou 5389 were applied with a brush to the surface of the primary leaves of TcLr19; sterile water was inoculated to wheat leaves as control. After inoculation, the plants were kept at 100 % relative humidity in the dark for 24 h at 20 °C, and then moved to the greenhouse. Wheat leaves were harvested at 0, 6, 12, 24, 36, 48, 60, 72, 96, and 120 hpi and immediately frozen in liquid nitrogen and stored at −80 °C prior to extraction of total RNA. Following the same inoculation method, leaves of T1 generate transgenic plants and ZhengZhou 5389 were harvested at 0, 12, 24, 48, 72, 96, and 120 hpi and stored at −80 °C for extraction of total RNA. Young leaves, young stems, and young roots collected from noninfected TcLr19 were used for spatial expression analyses.
SA/ABA Treatments
Fifty micromolars SA or ABA solution was sprayed on the leaf surface of seedlings with one mature leaf and one new leaf until the solution drops down. Wheat leaves were harvested at 0, 12, 24, 48, 72, and 96 h after SA or ABA solution treatment and immediately frozen in liquid nitrogen and stored at −80 °C for total RNA extraction.
Temporal and Spatial Expression Profiles of TcLr19PR1 Gene
Total RNA was extracted from infected wheat leaves at different time points from 0 to 120 hpi and noninoculated wheat leaves, stems, and roots according to the protocol of the Triozol reagent. Subsequently, they were treated with amplification-grade DNAse I (TaKaRa, Japan) to remove any residual DNA, and reverse-transcript PCR was performed using SuperScript II reverse transcriptase, oligo(dT)12–18, and RNAse OUT (Invitrogen). Synthesized complementary DNA (cDNA) was treated with RNAse H and used as the template for quantitative reverse transcription PCR (qRT-PCR). Amplification, detection, and data analyses of qRT-PCR were performed with the iCycler IQ real-time detection system (Bio-Rad, Amsterdam, Netherlands). A pair of TcLr19PR1-specific primers (qTcLr19PR1-F: 5′-CAATAACCTCGGCGTCTTCATC-3′; qTcLr19PR1-R: 5′-ATTTACTCGCTCGGTCCCTC-3′) was designed and synthesized based on the sequence obtained from RT-PCR. Wheat glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as a normalization factor for equal RNA concentrations. A pair of primers for GAPDH gene (qGAPDH-F: 5′-CTGCCTTGCTCGTCTTGCTAA-3′; qGAPDH-R: 5′-CTTGATGGAAGGACCATCAAC-3′) was designed based on the deposited wheat GAPDH sequence (GeneBank ID: AF251217). PCR reaction system was as follows: 12.5 μL 2× TransStartTM Green qPCR SuperMix (TransGen Biotech), 2.0 μL cDNA, 0.5 μL of each primer (10 μM) in a total volume of 25 μL. PCR was performed according to the following amplification procedure: an initial denaturation step at 95 °C for 2 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. Quantification of the target gene was assessed by relative standard curves. The 2−ΔΔCT (Thomas and Kenneth 2008) method was employed to quantify the relative gene expression. Three independent biological replicates were performed for each sample.
Vectors Construction
TcLr19PR1 was amplified from TcLr19 DNA by PCR using the primer pairs (TcLr19PR1-F-SmaI: 5′-TCCCCCGGGCGAAACGAGAATG-3′; TcLr19PR1-R-SacI: 5′-CGAGCTCGTGTGCCATAGAAAGGCTCAT-3′). The amplified fragment was cloned to the pEGM-T vector following the manufacture’s protocol. Subsequently, the obtained construct pEGM-T:: TcLr19PR1 was digested with SmaI and SacI. Meanwhile, the binary expression pAHC25, driven by the strong ubi promoter, was digested with SmaI and SacI as well. The digested TcLr19PR1fragment was inserted into the digested pAHC25 vector. Consequently, the vector pAHC25::TcLr19PR1 was generated.
Particle Bombardment-Mediated Wheat Transformation
The generated vector pAHC::TcLr19PR1 was transformed into immature embryo-derived calli of a susceptible wheat cultivar ZhengZhou 5389 using particle bombardment, following the method described by Wang et al. (2006). After several rounds of regeneration, calli were transferred to the differentiation medium for 1–2 weeks when adventitious buds were formed. When bud seedlings reach 3 cm, they were transferred to rooting and strengthening medium. Subsequently, the containers with open lid in which seedling has above 7 cm in height and over 5 cm in root length are moved to greenhouse for adaptation for 3 days. After that, the seedlings were transferred to soil. The advantages of cultivar ‘ZhengZhou 5389’ that was used for transformation are because it does not have leaf rust resistance gene Lr19 and is efficiently transformed by particle bombardment.
Screening of T0 and T1 Transgenic Plants and Histological Observation
To test the transgenic plants, a single leaf from each plant was harvested. Subsequently, DNA and RNA were extracted using a modified CTAB method (Saghai-Maroof et al. 1984) and RNeasy kits, respectively. PCR screening for the presence of the transgenic gene was performed using the right primer of the bar gene and the left primer of TcLr19PR1 gene (TcLr19PR1-F: 5′-TCCCCCGGGCGAAACGAGAATG-3′; Bar-R: 5′-AAACCCACGTCATGCCAGTTC-3′). Fourteen-day old transformed plantlets (T0 and T1) and ZhengZhou 5389 (approximately three- to four-leaf stage) were inoculated with leaf rust race (FHJT) following the method described earlier. Incidence of leaf rust was scored following Roelfs’ (1985) standard after inoculation of 12~14 days, and pictures were photographed as well.
T0 plants were bagged and selfed seeds were harvested. Between 5 and 20 T1 plants for each T0 plant were grown, inoculated, and scored as previously described. Tissue sampling for RNA and DNA extraction, PCR procedures were the same as for the T0 plants. The fourth leaves were collected at 6, 12, 24, 48, 72, and 120 hpi for histological observation. The infected leaves were examined by a rapid staining method using a protein-specific dye (Coomassie Brilliant Blue R-250). Stained leaf segments were examined with an Olympus BX-51 microscope (Olympus Corp., Tokyo) for infection sites and lengths of infection hyphae. Percentage of germinating spores was calculated by counting about 500 spores measured by ProgResCapturePro2.6 software. Fifty infection sites were examined on each randomly selected leaf segment pretreatment. Procedures and methods including identification of successful infection sites, hyphae staining, and measurement of fungal structures followed the method described by Wang et al. (2007).
Preparations for TcLr19PR1 Recombinant Protein and Its Antibody
Isolation of total RNA and synthesis of the first-strand cDNA were performed above. The ORF of TcLr19PR1 gene without signal peptide was amplified by PCR with the special primers: TcLr19PR1-F: 5′-CGCGGA TCC AACTCGCCTCAGGACTAC-3′, and TcLr19PR1-R: 5′-CCC AAGCTTTTAGTATGGTTTCTGTCCAATGAT-3′. PCR products were cloned into plasmid expression vector pEASY-E1 according to the instructions of pEASY-E1 expression kit (TransGen). Recombinant plasmid pEASY-TcLr19PR1 were transferred to Escherichia coli-competent cell DH5α. The positive recombinant plasmid pEASY-TcLr19PR1 which had the correct connection direction was identified by PCR with the special primers TcLr19PR1-F and T7 terminator (5'-TAGTTATTGCTCAGCGGTGG-3'). After sequencing identification, positive recombinant plasmids were transferred into E. coli strain BL21 (DE3). Single colony was cultured in liquid Luria-Bertani (LB) medium (with 50 mg/mL Ampicillin) at 37 °C overnight and used to inoculate a new LB liquid medium until the culture reached an optical density of 0.5–0.8 at a wavelength of 600 nm in a spectrophotometer. Different concentrations of isopropyl b-d-thiogalactopyranoside (IPTG: 0, 0.01, 0.1, 0.3, 0.5, 0.8, and 1.0 mmol/L) and induction times (2, 4, 6, 8, 10, and overnight) were used to determine the optimum conditions for producing the target protein. The expression of recombinant protein was induced at the optimal condition and purred with the Ni-Agarose HIS trap column (TransGen). The targeted protein were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) with 15 % polyacrylamide gels and stained with Coomassie brilliant blue R-250. Preparation and purification of the antibodies were entrusted to the Hebei Academy of Sciences Institute of Biology.
Immunodetection of TcLr19PR1 Recombinant Protein
The wheat proteins were extracted from the leaves inoculated with leaf rust race 07-10-421-3, respectively, at 0, 24, 48, 72, 96, 120, 144, and 168 hpi. One constitutively present band in the Ponceau S staining was used to normalize the protein concentration in the samples. For Western blotting, fusion protein and wheat proteins were subjected to 15 % SDS-PAGE and transferred to a PVDF membrane. After blocking the membrane using 3 % BSA in Tris-buffered saline (TBS) buffer at room temperature for 1 h, the membrane was incubated with 1:10,000 diluted polyclonal antibodies of TcLr19PR1 and 1:3,000 diluted goat anti-rabbit IgG antibody conjugated with alkaline phosphatase for 1 h at room temperature. The membrane was washed with 1× TBS, 0.1 % Tween-20 for four times at room temperature. Bound antibodies were visualized by color development for NBT formazan. When the color developed to a desirable intensity, the reaction was stopped by washing the membrane in deionized water. Image J software was used to scan the signal intensity and calculated the average and standard deviation among three repeats of WB analysis.
Results
Expression Analysis of Wheat TcLr19PR1 in Wheat Tissues
Real-time qRT-PCR specific primers were first tested with regular PCR to amplify the TcLr19PR1 gene using cDNA as a template in order to determine gene-specific amplification. A single 165-bp PCR fragment was amplified and confirmed by sequencing to be the expected fragment of the TcLr19PR1 gene (data not shown). The primers were then used in qRT-PCR analyses to determine the gene expression profiles of TcLr19PR1 in different wheat tissues. Gene expression level of TcLr19PR1 was measured relative to GAPDH expression in different tissues of seedling, including roots, stems, and leaves. Gene expression level of TcLr19PR1 showed significant variations among different plant tissues. More transcripts of TcLr19PR1 gene accumulated in leaves and stems than that in roots, and the accumulation of TcLr19PR1 transcripts in leaves was 3.5 times higher than that in roots and 2.0 times than that in stems (Fig. 1).
Expression profiles of the TcLr19PR1 gene in different wheat tissues. The y-axis indicates the amount of wheat TcLr19PR1 transcript is normalized to the GAPDH gene and express relative to the transcripts of the noninoculated control. The x-axis indicates the different wheat tissues. Error bars represent variation among three biological replicates. Reference gene is GAPDH; **p < 0.01; *p < 0.05; n = 3
TcLr19PR1 is Significantly Induced Upon Leaf Rust Pathogen
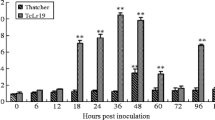
To investigate the expression profile of TcLr19PR1 in response to the infection by leaf rust pathogen, a more precise analysis of the expression using qPCR was performed. The transcripts quantity of TcLr19PR1 in incompatible interactions was detected at a low level at 0 hpi, then increased at 6 hpi, and peaked at 12 hpi (about 10-fold more expressed than the noninoculated control).The transcript levels then declined sharply to a relatively low expression from 18 to 24 hpi. At 60 hpi, there was a second expression peak that was lower than the first and 6-fold than the noninoculated control (Fig. 2). In compatible interactions, the TcLr19PR1 transcripts maintained at the lower levels were compared with noninoculated control at most time points. However, there were also expression peaks at 60 and 72 hpi, and then declined (Fig. 2). The accumulation of TcLr19PR1 transcripts in compatible interaction at 60 hpi was 1.9 times higher than that at 0 hpi. However, the expression levels in incompatible interactions were higher than that in compatible interactions except at 72 hpi, which was consistent with the result of semiquantitative PCR obtained by Wang (Wang et al. 2012).
Expression profiles of the TcLr19PR1 gene in Thatcher and TcLr19 leaves infected by the avirulent P. triticina isolate 07-10-421-3 at different postinoculation times. The y-axis indicates the amount of TcLr19PR1 transcript normalized to the GAPDH gene and express relative to that of noninoculated control, and the x-axis indicates sampling times. Error bars represent variation among three biological replicates. Reference gene is GAPDH. **p < 0.01; *p < 0.05; n = 3
Expression Analysis of TcLr19PR1 Induced by Signaling Molecules
The expression profile of TcLr19PR1 transcripts following the treatments of ABA and SA was investigated by qPCR respectively (Fig. 3). The TcLr19PR1 transcripts reached the peak level at 12 hpi rapidly after ABA treatment, then declined, and reached another peak level at 96 hpi. The accumulation of TcLr19PR1 transcripts at 12 hpi was 3.1 times higher than that at 0 hpi. For SA treatment, the TcLr19PR1 transcripts were detected at low level from 0 to 48 hpi, and suddenly peaked at 72 hpi (about 5.4 times higher than control), then declined. There were obvious differences in expression times between ABA and SA induction, but the accumulation of TcLr19PR1 transcripts were similar. The results showed that TcLr19PR1 gene could be induced by signaling molecules ABA and SA, thus we hypothesized that the expression of wheat TcLr19PR1 gene might be regulated by SA and ABA signaling pathway.
Expression profiles of the TcLr19PR1 gene in TcLr19 leaves treated by the SA and ABA at different postinoculation times. The y-axis indicates the amount of TcLr19PR1 transcript normalized to the GAPDH gene and express relative to that of noninoculated control, and the x-axis indicates sampling times. Error bars represent variation among three biological replicates. Reference gene is GAPDH; **p < 0.01; n = 3
Identification of Resistance to Leaf Rust in Transgenic Wheat
To further assess the role of TcLr19PR1 in responses to the leaf rust infection, transgenic plants were generated using particle bombardment-mediated transformation on susceptible wheat cultivar Zhengzhou 5389. Upon inoculation of T0 plants with leaf rust P. triticina race FHJT, all lines did not display an obvious resistant phenotype, but the severity of the infection was lessened compared with the control group (Table 1). To determine the presence of TcLr19PR1 gene in T0 transgenic wheat plants, 20 transgenic plants were subjected to PCR analysis using the forward primer of TcLr19PR1 gene and the reverse primer of the bar gene. A 3,000-bp DNA fragment as expected was amplified from 3 T0 transgenic plants and positive control (pAHC-TcLr19PR1), which was not amplified from the negative control (Zhengzhou 5389 and pAHC25) (Fig. 4a). The ultimate transformation efficiency was 0.58 % (the proportion of positive plants of callus from immature embryos). A total of 87 positive plants with TcLr19PR1 gene were obtained among 128 T1 transgenic plants derived from the 3 T0 transgenic plants (Table 1).
Transgenic plants showed segregation for resistance to leaf rust. a PCR amplification of genomic DNA using the forward primer of TcLr19PR1 gene and the reverse primer of the bar gene. TcLr19 T1-negative transgenic plants derived from T0 lines and susceptible control could not amplify the aim band and all T1-positive transgenic plants could amplify the aim band. b Leaf samples showing typical rust symptoms 14 days after inoculation with Puccinia triticina race (FHJT). 1, TcLr19 high resistant control; 2, Zhengzhou 5389 high susceptible control; 3, T1-negative transgenic plants derived from T0 lines; 4–7, T1-positive transgenic plants derived from T0 lines
To test the T1 plants resistance to leaf rust, the individual 87 positive plants, wheat-leaf rust resistance NILs TcLr19, and susceptible cultivar Zhengzhou 5389 were used for infectivity analysis after inoculation with P. triticina, and different degrees of infection (severity) were visualized on the surface of leaf blades on 10 dpi according to susceptible and resistant behavior of different plants. The results showed that TcLr19 did not show any infection whereas Zhengzhou 5389 started appearing infection courts within 4 dpi and scattered pustules surrounded by pale-halo region of chlorosis on the surface of leaf. Compared with the control group, the occurrence of disease symptoms of positive transgenic plants was postponed and the severity of the infection was lessened, which was reflected by less leaf rust uredinia (Fig. 4b). Furthermore, percentage of germinating spores was measured by microscopic observation and found that the germination of spores on transgenic plants was inhibited compared with those in Zhengzhou 5389 (Fig. 5), and the percentage of germinating spores in Zhengzhou 5389 was much higher than that in TcLr19 and transgenic plants especially at 24 hpi (Table 2). In addition, specific infection structures of leaf rust fungi were analyzed, including the length of hyphae, the number of hyphal branches, and the formation of haustorial mother cells. The fungal hyphal lengths were shorter (p < 0.01) compared with those in the control treatment (Zhengzhou 5389) from 12 to 120 hpi, and the numbers of hyphal branches were lower (p < 0.01) compared with those in the control treatment (Zhengzhou 5389) at 12 hpi (Table 2; Fig. 5). The delayed symptoms implicated that TcLr19PR1 gene has the role against wheat-leaf rust infection to some extent, reflected by reducing the severity of the infection.
Accumulation of TcLr19PR1 Protein Is Strongly Increased in the Transgenic Lines Upon Leave Rust Pathogen Infection
To detect the accumulation level of TcLr19PR1 protein in transgenic lines, polyclonal antibody was raised in rabbits against His-tagged TcLr19PR1 protein that was heterologously produced in E. coli. SDS-PAGE showed that the MW of the recombinant protein was about 16 kDa (Supplementary Fig. 1), which is consistent with the deduced amino acids composition of TcLr19PR1 and with the related sequence on pEASY plasmid. The ELISA result indicated that the antibody concentration of the antiserum reached 1:768,000, which was sufficient for further experimentation. The generated antibody was then used to detect the accumulation level of TcLr19PR1 in one of the transgenic lines that showed the significant increase of TcLr19PR1 upon rust pathogen challenge. As shown in Fig. 6, application of the TcLr19PR1 antibody resulted in a 16-kDa signal band detected in the positive at the expected height for TcLr19PR1, in the transgenic plant and its parental line Zhengzhou 5389 infected by leaf rust from 0 to 168 hpi. It was obvious that the signal intensity of TcLr19PR1 band in the transgenic line steadily increased from 0 to 120 hpi, then declined at 144 hpi. However, in parental line Zhengzhou 5389, a much weaker signal was detected compared with the transgenic line. Notably, the accumulation of TcLr19PR1 is consistent and maintained similar levels from 0 to 168 hpi in parental line as well. These results strongly implied that accumulation of TcLr19PR1 protein was correlated with resistance against leaf rust infection that has been demonstrated in Fig. 4.
Accumulation of TcLr19PR1 protein in transgenic plant upon leaf rust infection. One of transgenic lines and its parental line Zhengzhou 5389 were inoculated with leaf rust pathogen and the protein was sampled from 0 to 168 hpi. Western blot was probed with the TcLr19PR1 antibody. The purified TcLr19PR1 protein was used as a positive control (+). Plasmid pEASY was used as a negative control (−). Lower panel, one constitutively present band in the Ponceau S staining was used to normalize for loading variations. Bar graphs are summary data of normalize densimetric ratio
Discussion
Previous studies showed that PR1 expression is activated by various biotic and abiotic stresses, such as pathogen infection, salt, drought, and heavy metals (Mitsuhara et al. 2008; Thierry et al. 1995). For example, PsPR1 obtained from leaves of P. suffruticosa was significantly induced after C. canadense inoculation, and its expression peaked in 24 hpi, which implicated that PsPR1 might be involved in the disease defense (Yang et al. 2013). Similarly, VvPR1 isolated from leaves of Vitis vinifera was highly induced by Plasmopara viticola inoculation and expression level of PsPR1 reached the highest peak at 24 hpi, suggesting that VvPR1 might be related to biotic stresses (Hou et al. 2012). Our present studies showed that the expression of TcLr19PR1 was highly induced by leaf rust pathogen in incompatible interaction in contrast to compatible interaction at the onset stage of the infection (Fig. 2). However, the expression of TcLr19PR1 displayed certain variations. It was hardly detected at 24 hpi in both interactions, but accumulation of TcLr19PR1 transcripts peaked for the second time at 60 hpi. All in all, there was a consistent trend that significantly higher and rapid accumulation of TcLr19PR1transcripts in the incompatible combination compared with the compatible combination at most observed time points. The expression profiles of TcLr19PR1 were similar to the previous studies though the time points of gene expression were slightly different, which may be potentially caused by individual features of independent plant or pathogen that was investigated.
About the tissue-specific expression of PR1 gene, the real-time PCR analysis indicated that the expression level of PsPR1 obtained from leaves of P. suffruticosa was the highest in sepal but the lowest in leaves (Yang et al. 2013). In this study, the tissue organ-specific expression profiles of the TcLr19PR1 showed that TcLr19PR1 is expressed in leaves, stems, and roots, but its transcripts were detected abundantly in leaves and weakly stems in and roots. TcLr19PR1 was widely distributed in different tissues but differentially expressed in these tissues, which is in accordance with the research conducted by Tornero on PR1 gene from tomato (Tornero et al. 1997); TcLr19PR1 was constitutively expressed, which suggested that TcLr19PR1 directly interacted with pathogen and formed a barriers to tackle the invasion of pathogen together with other PRs.
Extensive studies have shown that phytohormones, such as SA and ABA, played important roles in adaptation to the biotic and abiotic stresses and can induce expression of PRl (Sabater et al. 2010; Le et al. 2009). SA has received particular attention to function as a signal molecule in acquired resistance to pathogens in several plant species, resulting in the synthesis of certain PRs and SAR (Yin and Hou 2007). ABA has durable roles not only being involved in abiotic stress responses but also plays an important role in adaptation to various biotic stresses. ABA is known to function in response to biotic stresses through cross-interaction with SA, JA, and ET in signal transduction pathway, thus conferring resistance of plants against adverse environment. Many elicitors enable the induction of PR1 in plant, but differential elicitors are required for the PR1 induction in individual plant species (Lorenzo et al. 2003; Nakano et al. 2006). VvPR1 from Vitis vinifera was induced by SA and ABA, and the response to ABA was earlier than SA (Hou et al. 2012). Our studies showed that TcLr19PR1 was significantly induced by signaling molecules such as SA and ABA, but differences of induction time were observed in expression of TcLr19PR1, which was consistent with the previous studies (Hou et al. 2012). Furthermore, the induction of TcLr19PR1 by ABA was earlier than SA, implying that ABA might be an upstream signaling molecule to activate the downstream signaling SA, which led to significant difference in expression pattern of TcLr19PR1.
Due to the longer time consumption of conventional wheat breeding and of map-based cloning of resistance genes being challenged by huge wheat genome, it is essential to identify and exploit new genetic resources such as resistance-related genes that exhibit effective capability to alleviate disease. Molecular and genetic studies of wheat-leaf rust have been carried out by wheat researchers worldwide. This study was focused on elucidating the behavior of the defense gene, TcLr19PR1, in susceptible and resistant plants during leaf rust infection, as well as the role of leaf rust-resistance gene Lr19 during wheat-leaf rust fungus interaction. In order to get insight into the role of PR1 in wheat growth and resistance to leaf rust pathogens attack, the expression of TcLr19PR1 and its function were analyzed. The results indicated that the expression level of TcLr19PR1 is much higher in the resistant plant than in the susceptible one after leaf rust pathogen challenging, SA treatment, and ABA treatment. Analysis of T1-positive transgenic plants showed challenged with leaf rust pathogen indicated that TcLr19PR1gene was involved in the wheat defense via attenuating the pathogen infection process, and Western blotting results correlated with the phenotype due to the interaction of the gene product. However, the segregation analysis of the putative transgenic plants could not be reflected by obvious phenotype due to TcLr19PR1 belonging to the defense gene and not to the resistance gene, so we have to obtain more generation of the transgenic plant used to confirm homozygous in order to further study TcLr19PR1 gene contributing to the defense against P. triticina infection. In future experiments, we should exploit more PR genes from wheat to broaden our understanding toward the role of PR genes in plant defense against pathogen attack.
Abbreviations
- PR:
-
Pathogenesis related
- HR:
-
Hypersensitive response
- ABA:
-
Abscisic acid
- SA:
-
Salicylic acid
- SAR:
-
Systemic acquired resistance
- ET:
-
Ethylene
- Hpi:
-
Hours postinoculation
- qRT-PCR:
-
Quantitative reverse transcription-polymerase chain reaction
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
References
Aglika E (2005) Pathogenesis-related proteins: research progress in the last 15 years. Gen Appl Plant Physiol 31:105–124
Antoniw JF, Ritter CE, Pierpoint WS, Van Loon LC (1980) Comparison of three pathogenesis-related proteins from plants of two cultivars of tobacco infected with TMV. J Gen Virol 47:79–87
Bolton MD, Kolmer JA, Garvin DF (2008) Wheat leaf rust caused by Puccinia triticina. Mol Plant Pathol 9:563–575
Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42:185–209
Hamamouch N, Li CY, Seo PJ, Park CM, Davis EL (2011) Expression of Arabidopsis pathogenesis-related genes during nematode infection. Mol Plant Pathol 12:355–364
Hammond-Kosack KE, Jones JD (1996) Resistance gene-dependent plant defense response. Plant Cell 8:1773–1791
Hou LX, Gao C, Che YM, Zhao FG, Liu X (2012) Gene cloning and expression analysis of pathogenesis-related protein 1 in Vitis vinifera L. Plant Physiol J 48:57–62
Komer JA (1996) Genetics of resistance to wheat leaf rust. Ann Rev Phytopathol 34:435–455
Le HG, Heitz TMP, Mutterer J, Walter BCJ (2009) Characterization of Vitis vinifera NPR1 homologs involved in the regulation of pathogenesis-related gene expression. BMC Plant Biol 9:54–64
Li ZJ, Dhekney S, Sadanand A, Gray DJ (2011) PR-1 gene family of grapevine: a uniquely duplicated PR-1 gene from a Vitis interspecific hybrid confers high level resistance to bacterial disease in transgenic tobacco. Plant Cell Rep 30:1–11
Liu JJ, Ekramoddoullah AKM (2006) The family 10 of plant pathogenesis-related proteins: their structure, regulation, and function in response to biotic and abiotic stresses. Physiol Mol Plant Pathol 68:3–13
Lorenzo O, Piqueras R, Sanchez SJ, Solano R (2003) Ethylene response factor 1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15:165–178
Mitsuhara I, Iwal T, Seo S, Yanagawa Y, Kawahigasi H, Hirose S, Ohkawa Y, Ohashi Y (2008) Characteristic expression of twelve rice PR-1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180). Mol Gen Genomics 279:415–427
Nakano T, Suzuki K, Ohtsuki N, Tsujimoto Y, Fujimura T, Hideaki S (2006) Identification of genes of the plant-specific transcription-factor families cooperatively regulated by ethylene and jasmonate in Arabidopsis thaliana. J Plant Res 119:407–413
Reddy PP (2013) Pathogenesis-related protein. In: Reddy PP (ed) Recent advances in crop protection. Springer, Heidelberg, pp 260–267
Roelfs AP (1985) Specificity and methods of study. In: Bushnell WR, Roelfs AP (eds) The cereal rusts: origins, specificity, structure and physiology. Academic, Orlando, pp 131–164
Ryals JA, Euenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8:1809–1819
Sabater JAB, Almagro L, Belchí NS, Ferrer MA, Barceló A, Pedreño M (2010) Induction of sesquiterpenes, phytoesterols and extracellular pathogenesis-related proteins in elicited cell cultures of Capsicum annuum. J Plant Physiol 167:1273–1281
Saghai-Maroof MA, Soliman KM, Jorgesen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley Mendelian inheritance, chromosomal location and population dynamics. Proc Natl Acad Sci U S A 81:8014–8018
Sels J, Mathys J, De Coninck BM, Cammue BP, De Bolle MF (2008) Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol Biochem 46:941–950
Singh R, Datta D, Priyamvada SS, Tiwari R (2004) Marker-assisted selection for leaf rust resistance genes Lr19 and Lr24 in wheat (Triticum aestivum L.). J Appl Genet 45(4):399–403
Sujon S, Young JK, Eui NK (2005) Overexpression of a pepper basic pathogenesis-related protein 1 gene in tobacco plants enhances resistance to heavy metal pathogen stresses. Plant Cell Reports 24:216–224
Thierry N, lsabelle G, Thierry B, Rene G, Annick S, Michel L, Bernard F, Egon M (1995) Pathogenesis-related PR-1 proteins are antifungal. Plant Physiol 108:17–27
Thomas DS, Kenneth JL (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108
Tornero P, Gadea J, Conejero V, Vera P (1997) Two pr-1 genes from tomato are differentially regulated and reveal a novel ode of expression for a pathogenesis-related gene during the hypersensitive response and development. Mol Plant Microbe Int 10:624–634
Uwe C (2006) Systemic acquired resistance. Plant Signal Behav 1:179–184
Van Loon LC, Van SEA (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55:85–97
Van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Ann Rev Phytopathol 44:135–162
Wang HZ, Xing LP, Li WL, Chen PD (2006) Disease- resistance function Analysis of wheat chitinase andβ-1,3-glucanase genes by a transient expression system. J Triticeae Crops 26:7–12
Wang C, Huang LL, Buchenauer H, Han QM, Zhang HC, Kang ZS (2007) Histochemical studies on the accumulation of reactive oxygen species (O2− and H2O2) in the incompatible and compatible interaction of wheat: Puccinia striiformis f. sp. tritici. Physiol Mol Plant Pathol 71:230–239
Wang S, Wang HY, Liu DQ (2012) Cloning and analysis of PR1 gene fromeTcLr19 wheat in the defense responses to Puccinia triticina. J Agric Univ Heibei 35:1–6
Yang DC, Zhang YX, Zheng GS (2013) Gene cloning and expression analysis of pathogenesis-related protein 1 of Paeonia suffruticosa. Acta Hortic Sin 40:1583–1590
Yin LL, Hou XJ (2007) The recent advances of salicylic acid as signal molecules of resistance in plant. Chin Agric Sci Bull 123:338–342
Acknowledgments
This study was supported financially by the Natural Science Foundation of Hebei Provice (no. C2012204005) and National key Basic Research Program of China (no. 2013CB127700).
Author information
Authors and Affiliations
Corresponding authors
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 237 kb)
Rights and permissions
About this article
Cite this article
Gao, L., Wang, S., Li, XY. et al. Expression and Functional Analysis of a Pathogenesis-Related Protein 1 Gene, TcLr19PR1, Involved in Wheat Resistance Against Leaf Rust Fungus. Plant Mol Biol Rep 33, 797–805 (2015). https://doi.org/10.1007/s11105-014-0790-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0790-5