Abstract
Sorbitol is the primary photosynthetic product and the translocatable and storage carbohydrate in apple (Malus domestica) and other fruit tree species within the Rosaceae family. Sorbitol-6-phosphate dehydrogenase (S6PDH, EC 1.1.1.200) is the key enzyme in the biosynthesis of sorbitol. In this study, we isolated two full-length genomic sequences for S6PDH from “Gala” apple. The two sequences have same six exons and first two introns, but the sizes of their last three introns are different. The two sequences were mapped to the same loci on chromosome 10. Immunogold electron microscopy analysis demonstrates that the S6PDH is localized mainly in leaf cytosol and chloroplasts. We also isolated and analyzed the promoter region of S6PDH and constructed a series of promoter deletion derivatives with β-glucuronidase (GUS) gene to identify the upstream region of the S6PDH gene required for promoter activity. The GUS activity in Agrobacterium-mediated transient transformation of tobacco leaves reveals that the -1719 region is more important for gene expression contrasting with other regions in the S6PDH promoter. The promoter region can be induced by cold, dark, and abscisic acid treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sorbitol, a sugar alcohol, is commonly found among the woody members of Rosaceae, including Malus, Prunus, Pyrus, and Eriobotrya (Bieleski 1982; Soria-Guerra et al. 2011). Besides being the preferred sugar for total photosynthesis products (Bieleski 1982; Loescher 1987), it is the primary form of translocatable and storage carbohydrates in those plants (Zhou et al. 2003). Sorbitol may also serve as an osmoprotectant and be involved in providing tolerances against abiotic and biotic stresses. It plays a key role in osmotic adjustment under drought (Lo Biancoa et al. 2000; Kobashi et al. 2000; Zhang et al. 2010; Li et al. 2011) and cold conditions and NaCl hardiness (Kanayama et al. 2006). Sorbitol is also involved in the tolerance to boron deficiency (Brown and Hu 1996; Brown et al. 1999) and the resistance of disease (Suleman and Steiner 1994).
Sorbitol synthesis shares a common hexose phosphate pool with sucrose production in cytosol. In source leaves, glucose-6-phosphate is converted to sorbitol-6-phosphate by sorbitol-6-phosphate dehydrogenase (S6PDH, EC 1.1.1.200, also named aldose-6-phosphate reductase) (Kanayama et al. 1992). Afterward, sorbitol-6-phosphate is used to form sorbitol via sorbitol-6-phosphatase (EC 3.1.3.50) by dephosphorylation (Zhou et al. 2003). The reaction catalyzed by S6PDH is the key regulatory step in sorbitol synthesis (Cheng et al. 2005). The majority of sorbitol degradation depends on the presence of NAD+-sorbitol dehydrogenase (EC 1.1.1.14), which catalyzes sorbitol by converting it to fructose in Rosaceae sink tissue (Oura et al. 2000).
Sorbitol-6-phosphate dehydrogenase is believed to play a critical role in the biosynthesis of sorbitol in the members of Rosaceae (Hirai 1981; Tao et al. 1995; Cheng et al. 2005). It has been purified and well characterized from loquat and apple (Yamaki 1980; Hirai 1981). The cDNA that encodes for this enzyme has been cloned from apple (Kanayama et al. 1992). In its leaves, S6PDH activity is closely related to the transition from sink to source (Loescher et al. 1982). The expression pattern for the gene encoding S6PDH is similar to that for the enzyme activity (Sakanishi et al. 1998). It is reported that the expression of S6PDH gene in apple and peach could be enhanced by abscisic acid, low temperature, high salinity, and drought (Kanayama et al. 2006; Lo Bianco et al. 2000; Cui et al. 2004). Apple S6PDH has been transformed into other plant species such as tobacco and persimmon (Tao et al. 1995; Sheveleva et al. 1998; Gao et al. 2001) to increase their sorbitol content and then their osmotic tolerance. This gene has also been transformed into apple in both sense and antisense orientations to improve our understanding of the relationship between sorbitol and sucrose (Kanamarua et al. 2004; Cheng et al. 2005; Teo et al. 2006; Zhou et al. 2006).
Although relatively abundant literatures on S6PDH are available, information is scarce on its molecular characterization, e.g., genomic structure, intercellular localization, and promoter. The research, reported herein, details the results of genomic structure, sub-cellular localization, and promoter of the S6PDH. Our objective is to gain some new information on the characterization of S6PDH and the regulating mechanism of sorbitol production in apple.
Materials and Methods
Plant Materials
Two-year-old apple plants (Malus domestica Borkh. cv. Gala), grafted onto rootstock M. sieversii (Ledeb.) Roem., were grown in pots at the experimental orchard of the Northwest A&F University in Yangling, China. Standard horticultural practices were followed for disease and pest control. The mature and healthy leaves were sampled and immediately frozen in liquid nitrogen and stored at −72°C.
Six-week-old Nicotiana tabacum cv. NC89 plants were grown in an environmental growth chamber under controlled conditions of 16-h light (12,000 lux)/8-h dark cycle, 65% relative humidity, and a 25°C (day)/21°C (night) temperature cycle.
RNA and DNA Extractions
Total RNA was extracted according to a modified cetyltrimethylammonium bromide (CTAB) method described by Chang et al. (1993). Prior to reverse-transcription, RNase-free DNAse I (Invitrogen, USA) was used to remove DNA contamination, according to the manufacturer's instructions. Genomic DNA was extracted from the leaves by a CTAB-based protocol (Dellaporta et al. 1983).
Cloning of MdS6PDH cDNA and Sequence Analysis
Based on the reported apple S6PDH sequences (D11080), we used Primers S6PDH1 and S6PDH2 (Table 1) to amplify the open reading frame (ORF) of the full-length cDNA. First strand cDNA was synthesized with a RevertAidTM First Strand cDNA Synthesis Kit (Fermentas). PCR was performed with rTaq DNA Polymerase (TaKaRa), applying the manufacturer's recommended reaction conditions. Products were gel-purified and cloned into the pMD18-T cloning vector. Afterward, positive clones were sequenced with an ABI 3730 sequencer.
Cloning and Analysis of the S6PDH Genomic Sequence
Primers S6PDH1 and S6PDH2 were also used to amplify the genomic sequence of DNA templates from M. domestica “Gala”. Fragments were amplified using LA Taq DNA Polymerase (TaKaRa) according to the manufacturer's recommendations. PCR products were ligated to pMD-18 (TaKaRa) and sequenced.
Expression of Apple S6PDH in Escherichia Coli and Antiserum Preparation
Full-length S6PDH cDNA was amplified with primers for S6PDH5 and S6PDH6 (Table 1) from the vector with MdS6PDH gene. BamHI and SalI sites were incorporated into the forward and reverse primers, respectively. After digestion with BamHI/SalI, the PCR product was cloned into vector pET 32a(+) (Novagen). The ligated products were used to transform BL21 (DE3)-competent cells. A recombinant plasmid, pET–S6PDH, was confirmed by restriction enzyme-digestion and sequencing. Competent cells harboring pET–S6PDH were cultured and induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside. Cells were pelleted and lysed in a 2× sample buffer of 0.1 M Tris-Cl (pH 6.8), 4% SDS, 0.2% bromophenol blue, 20% glycerol, and 0.1 M DTT. The cell extract was analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). The recombinant His-tagged proteins were purified by nickel affinity chromatography according to the manufacturer's protocol (Bio-Rad) and were analyzed by SDS-PAGE.
The purified fusion protein (10 mg) was used for standard immunization protocols in rabbits, from which the polyclonal antiserum was collected. Afterward, the IgG fraction (anti-GluGgt) of the antiserum was purified on a protein A-agarose column based on instructions from the supplier (GE Healthcare BioSciences, Uppsala, Sweden). Pre-immune serum was obtained from the rabbits before immunization.
Sub-Cellular Immunogold Labeling
We followed the method of sub-cellular immunogold labeling essentially as described by Zhang et al. (2004) and Fan et al. (2009). The sections were examined with a JEM-100S electron microscope. Specificity and reliability of the immunogold labeling were tested by two negative controls. In the first, rabbit pre-immune serum, rather than rabbit antiserum, was used to test the specificity of the antiserum. In the second control, the antiserum was omitted to test for possible nonspecific labeling of the goat anti-rabbit IgG antibody–gold conjugate. More than three repetitions of these experiments were conducted for each sample.
Promoter Isolation and Cis-Regulatory Element Prediction
We isolated the S6PDH promoter with a Genome Walking Kit (TaKaRa), according to the manufacturer's protocol, using Primers Spromoter1, Spromoter2, and Spromoter3 (Table 1). Products were gel-purified and cloned into the pMD18-T cloning vector. Afterward, positive clones were sequenced with an ABI 3730 sequencer. Their sequences were analyzed via plant cis-acting regulatory DNA elements (PLACE) and PlantCARE databases (Higo et al. 1999; Lescot et al. 2002).
Construction of Promoter::GUS Fusion Vectors
A series of nested 5′ deletions of S6PDH promoter fragments were generated using PCR amplification from pMD-Spromoter. Forward primers, SpromoterS1–S6 (Table 1) were designed to correspond to the -2396, -2122, -1719, -1066, -646, and -251 sequences of the S6PDH promoter, and the reverse primer SpromoterA (Table 1) was located in the 3′ end of the promoter. SmaI and SalI sites were incorporated into the forward and reverse primers, respectively. After digestion with SmaI/SalI, the PCR product was cloned into vector pC0390GUS (Xu et al. 2010) immediately upstream of the ATG start codon for β-glucuronidase. These new S6PDH promoter constructs, verified by sequencing, were introduced into Agrobacterium strain EHA105. We used CaMV35S::GUS (pC35SGUS) as the positive control and pC0390GUS as the negative (Xu et al. 2010).
Agrobacterium-Mediated Transient Assay and GUS Activity Analysis in Tobacco Plants
Agrobacterium-mediated transient assays of tobacco leaves were performed as described previously (Sparkes et al. 2006). The fully expanded leaves that were still attached to the intact tobacco plants were conducted by this protocol. Agro-infiltrated leaf samples from each treatment site were collected for each construct to determine β-glucuronidase (GUS) activity.
Quantitative GUS assays were performed as described previously (Jefferson 1987). They were carried out using 4-methyl umbelliferyl glucuronide (Sigma–Aldrich China) as substrate. Fluorescence of the methyl umbelliferone products was quantified using a Hitachi 850 fluorescence spectrophotometer (Hitachi, Tokyo, Japan). The total concentration of protein extracts from the tested samples was normalized through a protocol as described previously (Bradford 1976). GUS activity was expressed as nanomolar of 4-methylumbelliferone (4-MU, Sigma–Aldrich) generated per minute per milligram of soluble proteins.
Abiotic Treatments for Agro-Infiltrated Tobacco Plants
Cold, dark, and abscisic acid (ABA) treatment were conducted on tobacco 48 h after agro-infiltration. Cold treatment of agro-infiltrated plants was conducted at 4°C in an incubator for 5 h and of infiltrated plants at 28°C which were used as controls. Dark treatment of agro-infiltrated plants was conducted at 28°C under dark for 5 h and of control plants at 28°C under light (200 μmol m−2 s−1). For ABA treatment, agro-infiltrated plants were sprayed with 1 mM ABA for 48 h, and controls were sprayed with sterile water.
Results
Isolation and Analysis of Full-Length Apple S6PDH cDNA
Full-length S6PDH cDNA was isolated from mature apple leaves. The amplified cDNA fragment, named MdS6PDH, was about 1,080 bp long. This gene has been submitted to the GenBank database with accession number AY786539. The cDNA contains a 79-bp 5′-untranslated region (UTR), a 933-bp ORF, and a 68-bp 3′-UTR. The ORF of MdS6PDH encodes 310 amino acids. MdS6PDH showed 98% and 100% identity in their nucleotide sequence and amino acid sequence levels, respectively, with the reported apple S6PDH (GenBank entry D11080). MdS6PDH also showed 80–100% identity with 16 S6PDH or S6PDH-like genes in the apple genome (Velasco et al. 2010).
Genomic Structure and Chromosomal Localization of the Apple S6PDH Gene
PCR amplification of the full-length S6PDH gDNA produced two different-sized fragments — SgDNA1 (3595 bp, JF764598) and SgDNA2 (3402 bp, JF764599). Alignment of these two gDNA sequences with the cDNA sequence of MdS6PDH (AY786539) revealed that each comprises six exons and five introns (Fig. 1). The consensus sequences GT and AG were found at the 5′ and 3′ borders of all introns, which strictly follow the convention for “GT–AG” splicing sites of eukaryotic introns, as proposed by Breathnach and Chambon (1981).
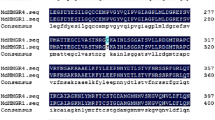
Sequence alignment analysis revealed that the six exons and the first two introns of SgDNA1 and SgDNA2 were 100% identical; therefore, the deduced amino acid sequences of cDNAs for SgDNA1 and SgDNA2 were the same. However, the two sequences differed in the size of their last three introns (Fig. 1). Blast analysis indicated that SgDNA2 was 99% identical with the reported apple S6PDH gDNA (AF057134). To further ascertain the differences in the two gDNAs, the sequence of the last three introns were compared (Fig. 2). Intron 3,intron 4, and intron 5 of SgDNA1 are 32.8%, 74.34%, and 33.73% homologous to those of SgDNA2, respectively. The results indicated that the variation in the last three introns was rooted in base deletion and insertion.
Homology searches of the genome sequence of M. domestic cv. Golden Delicious (http://genomics.research.iasma.it/index.html) were conducted with the two cloned S6PDH nucleotide sequences. The genome analysis revealed that SgDNA1 and SgDNA2 had high sequence similarity (97.1% and 98.65%) from 11,175,634 bp to 11,179,027 bp in chromosome 10. The present results suggested that SgDNA1 and SgDNA2 were mapped in the same loci, and both existed as alleles.
Antibody Preparation and Sub-Cellular Immunogold-Labeling of MdS6PDH
To study the distribution and sub-cellular localization of S6PDH proteins in apple leaves, we selected the full-length 310 amino acids of MdS6PDH for antibody preparation. Only one signal was detected by immunoblotting crude protein preparations from the leaves with the MdS6PDH antiserum (data not shown), which indicates that the anti-MdS6PDH serum was recognized S6PDH in apple.
Tissue sections from mature leaves were analyzed with the anti-MdS6PDH serum and an immunogold-labeling secondary antibody. Using transmission electron microscopy of cells within mature tissues, we observed gold particles in both the chloroplast (Fig. 3a, b) and cytoplasm (Fig. 3c, d). No label was seen in other organelles. In experiments to assess the reliability of this immuno-labeling procedure, the control sections displayed no labeling either (Fig. 3e, f).
Transmission electron microscopy result of apple leaf mesophyll cells with anti-MdS6PDH serum and goat anti-rabbit IgG antibody conjugated with 10 nm gold. Particles are distributed mainly in chloroplast (a and b) and cytosol (c and d). No immunogold particles were found in pre-immune serum control (e) or antiserum omission control (f). Ch chloroplast; Cy cytosol
Isolation of Apple S6PDH Promoter Sequences and Characterization Analysis
After isolating the S6PDH promoter by PCR and a Genome Walking Kit, we detected a 2.4-kb fragment of product. This promoter, JF827272, was 2,396 bp long (Fig. 4). Its sequence was analyzed for the presence of cis-acting elements involved in regulating S6PDH expression. PLACE and PlantCARE databases were used to identify several potential regulatory elements that corresponded to known cis-acting elements of eukaryotic genes. Detailed analysis of the cis-regulatory elements occurring in S6PDH promoters revealed that most elements were classified into three groups according to their functions: hormone-responsive elements, light-responsive elements, and stress-responsive elements (Table 2).
To test which part of the promoter sequence has high transcriptional activity, a series of deletions (-2396, -2122, -1719, -1066, -646, and -251) were transcriptionally fused to the promoterless reporter gene β-glucuronidase (GUS) (Fig.5). Each construct was introduced into tobacco leaves and tested for GUS activity. Here, the pC35SGUS construct was used as the positive control while the promoterless pC0390GUS served as the negative. The highest levels of GUS activity were detected in tobacco leaves harboring the construct containing the -1719 region of the S6PDH promoter. The -2122 and -1066 regions had a relatively low level of GUS activity. The lowest levels of GUS activity were detected in -2396, -646, and -251 regions (Fig.6). These results indicated that in the S6PDH promoter sequence, the -1719 region is more important for promoter activity than other regions.
Analysis of cis-regulatory elements of S6PDH promoters revealed that light-responsiveness motifs and stress-regulated motifs were present in the S6PDH promoter (Table.2). The effects of these elements were determined. We fused the 2396-bp fragment to the promoterless GUS reporter gene. This construct was introduced into tobacco leaves and tested for GUS activity. GUS assays displayed that S6PDH promoter was evidently induced by cold treatment, dark treatment, and ABA treatment. The inducible GUS activity after a 5-h cold treatment was approximately 2.21-fold, compared to the controls at 28°C (Fig.7a). S6PDH promoter-driven GUS activity in dark treatment was 6.61-fold lower than in control leaves (Fig.7b). With regard to ABA treatment, the S6PDH promoter was activated with a 2.76-fold higher inducible GUS activity compared with controls (Fig.7c).
Discussion
Although S6PDH has been studied in several plant species, little is known about its molecular characteristics. We systematically investigated its genomic structure, sub-cellular localization, and promoter.
In analyzing the molecular basis for S6PDH expression, we obtained two different genomic sequences — SgDNA1 and SgDNA2. The latter shares high sequence identity with apple S6PDH gDNA (AF057134). SgDNA1 and SgDNA2 differ in the sizes of their last three introns; however, they distribute in the same location on chromosome 10. It was reported that there are 16 S6PDH or S6PDH-like genes in the apple genome. They are located in chromosome 10 (Velasco et al. 2010). Therefore, SgDNA1 and SgDNA2 may be alleles. More work is needed to investigate whether there are different genomic sequence types of S6PDH in other Malus germplasm resource.
Little information was previously available about how S6PDH is distributed within the sub-cellular compartments of leaves. Our new discovery of its localization is essential for furthering our understanding on the mechanisms for sorbitol metabolism. Here, we have confirmed that S6PDH is distributed not only in the cytosol but also in the chloroplasts of leaf cells. Yamaki (1981) has also determined that S6PDH is found predominantly in the chloroplasts of apple cotyledons. Sorbitol and other sugars have also been detected in the vacuoles, protoplasts, and extracellular free spaces (Yamaki 1982). In mature peach leaves, sorbitol is present in the chloroplasts and cytosol, with the concentration being 1.5-fold higher in the former (Nadwodnik and Lohaus 2008). Wang et al. (2009) found that sorbitol dehydrogenase, the key enzyme for sorbitol metabolism, is distributed predominantly not only in the cytosol but also in the chloroplasts of the leaf. All of these results suggest that sorbitol can be synthesized and decomposed in the chloroplast and cytosol, which might explain our results for such localization on both sites in apple. In the cytosol, sorbitol that was synthesized by S6PDH will be used for phloem loading and export from source organs to sink organs. In the chloroplast, one possible explanation for S6PDH function is that S6PDH might be involved in regulating starch synthesis by Calvin cycle. On the other hand, sorbitol acts as an osmoregulatory substance to help maintain the osmotic balance. Therefore, the S6PDH present in the chloroplast and cytosol may take part in the regulation of the sorbitol concentration, maintaining the osmotic balance between chloroplast, cytosol, and apoplast.
To characterize the functional organization of the S6PDH promoter involved in expression, we made 5′-serially deleted promoter constructs fused to the GUS reporter gene and tested their expression by Agrobacterium-mediated transient assay system in tobacco leaves. The results demonstrated that the -1719 region of the S6PDH promoter had the highest levels of GUS activity. One possible explanation is that both activation and repression regulatory elements co-exist in S6PDH promoter sequence. Positive regulatory elements, maybe, are present at positions from -1719 to -251, and negative regulatory elements may be located from -2396 to -1719. Examining the S6PDH promoter sequence, we found several stress-regulated and light-responsive elements, such as ABRE, CGTCA-motif, LTR, MBS, TC-rich repeats, TGACG-motif, ACE, ATC-motif, ATCT-motif, G-box, and MRE. These are homologous to cis-acting elements previously identified in plants. This paper displayed that S6PDH promoter was evidently induced by stress and light treatment.
Many physiological processes in plants are affected by different stresses (Gao et al. 2010; Stolf-Moreira et al. 2011; Zhang et al. 2011). Sorbitol is induced to accumulate in abiotic stress and has been proposed to act as an osmoregulatory substance to help maintain the osmotic balance under adverse growing conditions. As the key enzyme in sorbitol biosynthesis, expression of S6PDH can also be enhanced by ABA treatment or the exposure to low temperatures and high salinity in apple, as well as sorbitol content (Kanayama et al. 2006). A linear increase in S6PDH activity with drought stress correlates with a significant accumulation of sorbitol in phloem of peach (Escobar-Gutiérrez et al. 1998). These results showed that S6PDH gene and S6PDH promoter had similar expression pattern against stress conditions. It means that the transcriptional level regulation may be a key step for S6PDH gene expression at adverse situation.
In summary, two different genomic sequences for S6PDH from apple, designated as SgDNA1 and SgDNA2, were isolated and may be alleles on chromosome 10. Our immuno-electron microscopy revealed that S6PDH is localized on the cytosol and chloroplasts of apple leaves. Analysis of its gene promoter region leads us to believe that this promoter responds to light and various environmental stresses. Therefore, we can conclude that S6PDH is multifunctional for sorbitol biosynthesis under various stresses and photosynthate demands.
References
An C, Ichinose Y, Yamada T, Tanaka Y, Shiraishi T, Oku H (1993) Organization of the genes encoding chalcone synthase in Pisum sativum. Plant Mol Biol 21(5):789–803
Bieleski RL (1982) Sugar alcohols. In: Loewus F, Tanner W (eds) Encyclopedia of plant physiology new series, Vol. 13A. Springer, Berlin, pp 158–192
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Breathnach R, Chambon P (1981) Organization and expression of eukaryotic split genes coding for proteins. Annu Rev Biochem 50:349–383
Brown PH, Hu H (1996) Phloem mobility of boron is species dependent: evidence for phloem mobility in sorbitol-rich species. Ann Bot 77:497–505
Brown PH, Bellaloui N, Hu H, Dandekar AM (1999) Transgenically enhanced sorbitol synthesis facilitates phloem boron transport and increases tolerance of tobacco to boron deficiency. Plant Physiol 19:17–20
Chang S, Puryear J, Cairney J (1993) Simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Report 11:113–116
Cheng LL, Zhou R, Reidel EJ, Sharkey TD, Dandekar AM (2005) Antisense inhibition of sorbitol synthesis leads to up-regulation of starch synthesis without altering CO2 assimilation in apple leaves. Planta 220:767–776
Cui SM, Sadayoshi K, Ogawa Y, Nii N (2004) Effects of water stress on sorbitol content in leaves and roots, anatomical changes in cell nuclei, and starch accumulation in leaves of young peach trees. J Japan Soc Hortic Sci 73(1):25–30
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA mini-preparation, version II. Plant Mol Biol Report 1:19–21
Diaz de Leon F, Klotz KL, Lagrimini M (1993) Nucleotide sequence of the tobacco (Nicotiana tabacum) anionic peroxidase gene. Plant Physiol 101(3):1117–1118
Escobar-Gutiérrez AJ, Zipperlin B, Carbonne F, Moing A, Gaudillère JP (1998) Photosynthesis, carbon partitioning and metabolite content during drought stress in peach seedlings. Aust J Plant Physiol 25:197–205
Fan RC, Peng CC, Xu YH, Wang XF, Li Y, Shang Y, Du SY, Zhao R, Zhang XY, Zhang LY, Zhang DP (2009) Apple sucrose transporter SUT1 and sorbitol transporter SOT6 interact with cytochrome b5 to regulate their affinity for substrate sugars. Plant Physiol 150:1880–1901
Feldbrugge M, Sprenger M, Dinkelbach M, Yazaki K, Harter K, Weisshaar B (1994) Functional analysis of a light-responsive plant bZIP transcriptional regulator. Plant Cell 6(11):1607–1621
Feldbrugge M, Sprenger M, Hahlbrock K, Weisshaar B (1997) PcMYB1, a novel plant protein containing a DNA-binding domain with one MYB repeat, interacts in vivo with a light-regulatory promoter unit. Plant J 11(5):1079–1093
Gao M, Tao R, Miura K, Dandekar AM, Sugiura A (2001) Transformation of Japanese persimmon (Diospyros kaki Thunb.) with apple cDNA encoding NADP-dependent sorbitol-6-phosphate dehydrogenase. Plant Sci 160(5):837–845
Gao Y, Zhao Y, Li TT, Ren CX, Liu Y, Wang ML (2010) Cloning and characterization of a G protein β subunit gene responsive to plant hormones and abiotic stresses in Brassica napus. Plant Mol Biol Report 28:450–459
Hennig J, Dewey RE, Cutt JR, Klessig DF (1993) Pathogen, salicylic acid and developmental dependent expression of a RT beta-1,3-glucanase/GUS gene fusion in transgenic tobacco plants. Plant J 4(3):481–493
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucl Acid Res 27:297–300
Hirai M (1981) Purification and characteristics of sorbitol-6-phosphate dehydrogenase from loquat leaves. Plant Physiol 67:221–224
Jefferson R (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Report 5:387–405
Kanamarua N, Itob Y, Komorib S, Saitob M, Katob H, Takahashib S, Omurac M, Soejimab J, Shiratakea K, Yamadaa K, Yamaki S (2004) Transgenic apple transformed by sorbitol-6-phosphate dehydrogenase cDNA: switch between sorbitol and sucrose supply due to its gene expression. Plant Sci 167(1):55–61
Kanayama Y, Mori H, Imaseki H, Yamaki S (1992) Nucleotide sequence of a cDNA encoding sorbitol-6-phosphate dehydrogenase from apple. Plant Physiol 100:1607–1608
Kanayama Y, Watanabe M, Moriguchi R, Deguchi M, Kanahama K, Yamaki S (2006) Effects of low temperature and abscisic acid on the expression of the sorbitol-6-phosphate dehydrogenase gene in apple leaves. J Jpn Soc Hortic Sci 75(1):20–25
Kim SY, Wu R (1990) Multiple protein factors bind to a rice glutelin promoter region. Nucl Acid Res 18(23):6845–6852
Kim JK, Wu R (1992) Nucleotide sequence of a high-pI rice (Oryza sativa)-amylase gene. Plant Mol Biol 18(2):399–402
Kobashi K, Gemma H, Iwahori S (2000) Abscisic acid content and sugar metabolism of peaches grown under water stress. J Am Soc Hortic Sci 125:425–428
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucl Acid Res 30:325–327
Li F, Lei HJ, Zhao XJ, Tian RR, Li TH (2011) Characterization of three sorbitol transporter genes in micropropagated apple plants grown under drought stress. Plant Mol Biol Report. doi:10.1007/s11105-011-0323-4
Liaud MF, Zhang DX, Cerff R (1990) Differential intron loss and endosymbiotic transfer of chloroplast glyceraldehyde-3-phosphate dehydrogenase genes to the nucleus. Proc Natl Acad Sci 87(22):8918–8922
Lo Biancoa R, Riegera M, Sung SS (2000) Effect of drought on sorbitol and sucrose metabolism in sinks and sources of peach. Physiol Plant 108:71–78
Loescher WH (1987) Physiology and metabolism of sugar alcohols in higher plants. Physiol Plant 70(3):553–557
Loescher WH, Marlow GC, Kennedy RA (1982) Sorbitol metabolism and sink–source interconversions in developing apple leaves. Plant Physiol 70:335–339
Nadwodnik J, Lohaus G (2008) Subcellular concentrations of sugar alcohols and sugars in relation to phloem translocation in Plantago major, Plantago maritima, Prunus persica, and Apium graveolens. Planta 227:1079–1089
Oura Y, Yamada K, Shiratake K, Yamaki S (2000) Purification and characterization of a NAD+-dependent sorbitol dehydrogenase from Japanese pear fruit. Phytochemistry 54(6):567–572
Pastuglia M, Roby D, Dumas C, Cock JM (1997) Rapid induction by wounding and bacterial infection of an S gene family receptor-like kinase in Brassica oleracea. Plant Cell 9:1–13
Pichersky E, Bernatzky R, Tanksley SD, Breidenbach RB, Kausch AP, Cashmore AR (1985) Molecular characterization and genetic mapping of two clusters of genes encoding chlorophyll a/b-binding proteins in Lycopersicon esculentum (tomato). Gene 40(2–3):247–258
Rouster J, Leah R, Mundy J, Cameron-Mills V (1997) Identification of a methyl jasmonate-responsive region in the promoter of a lipoxygenase 1 gene expressed in barley grain. Plant J 11(3):513–523
Sakanishi K, Kanayama Y, Mori H, Yamada K, Yamaki S (1998) Expression of the gene for NADH-dependent sorbitol-6-phosphate dehydrogenase in peach leaves of various developmental stages. Plant Cell Physiol 39:1372–1374
Sheveleva EV, Marquez S, Chmara W, Zegeer A, Jensen RG, Bohnert HJ (1998) Sorbitol-6-phosphate dehydrogenase expression in transgenic tobacco. High amounts of sorbitol lead to necrotic lesions. Plant Physiol 117:831–839
Soria-Guerra RE, Rosales-Mendoza S, Gasic K, Wisniewski ME, Band M, Korban SS (2011) Gene expression is highly regulated in early developing fruit of apple. Plant Mol Biol Report. doi:10.1007/s11105-011-0300-y
Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1(4):2019–2025
Stolf-Moreira R, Lemos EGM, Carareto-Alves L (2011) Transcriptional profiles of roots of different soybean genotypes subjected to drought stress. Plant Mol Biol Report 29:19–34
Suleman P, Steiner PW (1994) Relationship between sorbitol and solute potential in apple shoots relative to fire blight symptom development after infection by Erwinia amylovora. Phytopathology 84(10):1244–1250
Takaiwa F, Oono K, Wing D, Kato A (1991) Sequence of three members and expression of a new major subfamily of glutelin genes from rice. Plant Mol Biol 17(4):875–885
Tao R, Uratsu SL, Dandekar AM (1995) Sorbitol synthesis in transgenic tobacco with apple cDNA encoding NADP-dependent sorbitol-6-phosphate dehydrogenase. Plant Cell Physiol 36(3):525–532
Teo G, Suzuki Y, Uratsu SL, Lampinen B, Ormonde N, Hu WK, DeJong TM, Dandekar AM (2006) Silencing leaf sorbitol synthesis alters long-distance partitioning and apple fruit quality. Proc Natl Acad Sci 103(49):18842–18847
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, Salvi S, Pindo M, Baldi P, Castelletti S, Cavaiuolo M, Coppola G, Costa F, Cova V, Dal Ri A, Goremykin V, Komjanc M, Longhi S, Magnago P, Malacarne G, Malnoy M, Micheletti D, Moretto M, Perazzolli M, Si-Ammour A, Vezzulli S, Zini E, Eldredge G, Fitzgerald LM, Gutin N, Lanchbury J, Macalma T, Mitchell JT, Reid J, Wardell B, Kodira C, Chen Z, Desany B, Niazi F, Palmer M, Koepke T, Jiwan D, Schaeffer S, Krishnan V, Wu C, Chu VT, King ST, Vick J, Tao Q, Mraz A, Stormo A, Stormo K, Bogden R, Ederle D, Stella A, Vecchietti A, Kater MM, Masiero S, Lasserre P, Lespinasse Y, Allan AC, Bus V, Chagne D, Crowhurst RN, Gleave AP, Lavezzo E, Fawcett JA, Proost S, Rouze P, Sterck L, Toppo S, Lazzari B, Hellens RP, Durel CE, Gutin A, Bumgarner RE, Gardiner SE, Skolnick M, Egholm M, Van de Peer Y, Salamini F, Viola R (2010) The genome of the domesticated apple (Malus x domestica Borkh.). Nat Genet 42:833–839
Wang XL, Xu YH, Peng CC, Fan RC, Gao XQ (2009) Ubiquitous distribution and different subcellular localization of sorbitol dehydrogenase in fruit and leaf of apple. J Exp Bot 60(3):1025–1034
White AJ, Dunn MA, Brown K, Hughes MA (1994) Comparative analysis of genomic sequence and expression of a lipid transfer protein gene family in winter barley. J Exp Bot 45:1885–1892
Xu WR, Yu YH, Ding JH, Hua ZY, Wang YJ (2010) Characterization of a novel stilbene synthase promoter involved in pathogen- and stress-inducible expression from Chinese wild Vitis pseudoreticulata. Planta 231:475–487
Yamaguchi SK, Shinozaki K (1993) Arabidopsis DNA encoding two desiccation-responsive rd29 genes. Plant Physiol 101(3):1119–1120
Yamaki S (1980) Property of sorbitol-6-phosphate dehydrogenase and its connection with sorbitol accumulation in apple. Hortic Sci 15:268–270
Yamaki S (1981) Subcellular localization of NADP-dependent sorbitol-6-phosphate dehydrogenase in protoplast from apple cotyledons. Plant Cell Physiol 22:359–367
Yamaki S (1982) Distribution of sorbitol, neutral sugars, free amino acids, malic acid and some hydrolytic enzymes in vacuoles of apple cotyledons. Plant Cell Physiol 23:881–889
Zhang LY, Peng YB, Pelleschi ST, Fan Y, Lu YF, Lu YM, Gao XP, Shen YY, Delrot S, Zhang DP (2004) Evidence for apoplasmic phloem unloading in developing apple fruit. Plant Physiol 135:574–586
Zhang J, Yao Y, Streeter JG, Ferree DC (2010) Influence of soil drought stress on photosynthesis, carbohydrates and the nitrogen and phosphorus absorb in different section of leaves and stem of Fugi/M.9EML, a young apple seedling. Afric. J Biotechnol 9(33):5320–5325
Zhang X, Zhen JB, Li ZH, Kang DM, Yang YM, Kong J, Hua JP (2011) Expression profile of early responsive genes under salt stress in upland cotton (Gossypium hirsutum L.). Plant Mol Biol Report. doi:10.1007/s11105-010-0269-y
Zhou R, Cheng LL, Wayne R (2003) Purification and characterization of sorbitol-6- phosphate phosphatase from apple leaves. Plant Sci 165:227–232
Zhou R, Cheng LL, Dandekar AM (2006) Down-regulation of sorbitol dehydrogenase and up-regulation of sucrose synthase in shoot tips of the transgenic apple trees with decreased sorbitol synthesis. J Exp Bot 57(14):3647–3657
Acknowledgments
This work was supported by the earmarked fund for China Agriculture Research System. The authors are grateful to Priscilla Licht for her help in revising our English composition.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liang, D., Cui, M., Wu, S. et al. Genomic Structure, Sub-Cellular Localization, and Promoter Analysis of the Gene Encoding Sorbitol-6-Phosphate Dehydrogenase from Apple. Plant Mol Biol Rep 30, 904–914 (2012). https://doi.org/10.1007/s11105-011-0409-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-011-0409-z