Abstract
Drought is one of the most severe environmental factors that impair plant growth and agricultural production. To investigate how Malus prunifolia, an excellent apple rootstock with strong drought tolerance, adapts to stress and to identify genes for improving this important trait, we constructed a suppression subtractive hybridization cDNA library from seedling leaves under water stress. The cDNA for a drought-inducible glycine-rich RNA-binding protein (GR-RBP) was isolated, and the gene was characterized for its role in the response of Malus seedlings to drought stress. cDNA clone MpGR-RBP1 has 781 nucleotides, with an open reading frame of 516 nucleotides. The deduced 171 amino acids contain an amino-terminal RNA recognition motif and a carboxyl-terminal glycine-rich domain, with structural similarity to a class of stress-induced GR-RBP proteins found in other plants. Phylogenetic analysis confirmed that the MpGR-RBP1 protein belongs to the plant GR-RBP family, members of which play important roles in posttranscriptional regulation of gene expression under various stress conditions. The expression profile of MpGR-RBP1 transcripts was detected by quantitative real-time polymerase chain reaction (PCR) and semiquantitative reverse transcriptase PCR. MpGR-RBP1 was expressed in both roots and leaves, with expression being higher in the latter. This is the first report of this class of protein in Malus plants, and the putative role of MpGR-RBP1 is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant glycine-rich RNA-binding proteins (GR-RBPs) (Burd and Dreyfuss 1994; Fu et al. 2007) are characterized by at least one RNA recognition motif (RRM) present at the N-terminus (also known as an RNA-binding domain or ribonucleoprotein [RNP] domain) and a glycine-rich domain at the C-terminal (Bandziulis et al. 1989; Albà and Pagès 1998). Similarly, a group of structural proteins in the cell walls, i.e., glycine-rich proteins (GRPs), contain glycine-rich domains (Sachetto-Martins et al. 2000). Whereas GRPs are well-characterized as cell-wall proteins, GR-RBPs are involved in posttranscriptional RNA processing in the cytoplasm as well as in the nucleus (Ziemienowicz et al. 2003; Fusaro et al. 2007). To distinguish between these two groups, Lorković and Barta (2002) have proposed that RRM-containing GRPs be renamed as GR-RBPs. An RRM has two short, highly conserved sequences—an octamer designated RNP1 and a hexamer designated RNP2—plus several other, mostly hydrophobic, conserved amino acids interspersed throughout the motif (Query et al. 1989). A different type of RNA-binding motif in the N-terminus of GR-RBPs is the cold shock domain (CSD), where only the RNP1 sequence is conserved (Sachetto-Martins et al. 2000). The CSD is proposed to be an ancient molecule that was present before the origin of single-cell life and is the most evolutionary conserved nucleic acid-binding domain within prokaryotes and eukaryotes (Nakaminami et al. 2006). Nevertheless, the function of GR-RBPs in eukaryotes, including plants, remains largely unknown (Kim et al. 2007a; Lorković 2009).
GR-RBPs have now been isolated from cyanobacteria and the cells of plants and mammals (Shinozuka et al. 2006). Some cDNAs that encode plant GR-RBPs have been identified from both angiosperms and gymnosperms and are highly conserved (Stephen et al. 2003). Bocca et al. (2005) have utilized ForEST database and the phylogenetic tree method to cluster plant GR-RBPs into groups based upon their combination of structural domains. Those in group 1 have an RRM conserved motif at the N-terminal end, followed by a glycine-rich region with GGYGG repeats (Sachetto-Martins et al. 2000). Group 2 members, also known as RZ proteins, contain an RRM at the N-terminus and a glycine-rich region interspersed with a (CCHC)-type zinc finger at the C-terminus. This group appears to be plant-specific (Lorković and Barta 2002; Kim et al. 2007c). In group 3, GR-RBPs are organized with a CSD at the N-terminus plus one to seven CCHC zinc fingers in their glycine-rich region, which is also termed the cold shock proteins. Finally, group 4 presents two copies of the RRM motif followed by a C-terminal glycine-rich region. It is therefore interesting to determine whether these different structural features have a functional importance in plants. Although some GR-RBPs have been shown to play roles in posttranscriptional regulation of gene expression in Arabidopsis plants under cold, high-salinity, or dehydration stress, other functions must still be verified (Kim et al. 2005; Kim and Kang 2006; Kim et al. 2007a; 2008; Kim et al. 2007b).
Plant GR-RBPs are induced by various stresses, especially low temperatures. Since the first gene encoding GR-RBP was identified in immature maize embryos (Gόmez et al. 1988), other such cDNAs have been isolated from monocots, dicots, and gymnosperms, including WhGRP-1 in wheat (Guiltinan and Niu 1996), ngRBP in tobacco (Naqvi et al. 1998), SCRGP-1 in wild potato (Baudo et al. 1999), PgRNP in white spruce (Richard et al. 1999), sbGR-RNP in broomcorn (Aneeta et al. 2002), LpGRP1 in perennial ryegrass (Shinozuka et al. 2006), and NtGRP1 in tobacco (Lee et al. 2009). All of the proteins encoded by these genes share similar structural features, including an RRM at the N-terminal half and glycine-rich domains at the C-terminal half. Their expression has been linked to both developmental (Bandziulis et al. 1989; Staiger et al. 2003) and physical influences, including cold, wounding, ABA, jasmonic acid, high salinity, water stress, viral infection, and pathogens (Sachetto-Martins et al. 2000). Although their occurrence has been reported in several species, only a few plant GR-RBPs have been studied for their functions (Lorković and Barta 2002; Fu et al. 2007). Some may act as key posttranscriptional regulators through the trafficking, stabilization, or processing of specific mRNAs in response to pathogens (Fu et al. 2007). In addition, several GR-RBPs circadian oscillations of transcript levels have been demonstrated to be regulated by the circadian clock. Notably, the clock-regulated AtGRP7 (AtGR-RBP7) influences the amplitude of its transcript oscillation by negative feedback at the posttranscriptional level. This autoregulation relies on AtGRP7 binding to its own pre-mRNA and cause alternative splicing (Staiger et al. 2003; Schöning et al. 2007; 2008). Several Arabidopsis GR-RBPs also serve as RNA chaperones for cold adaptation (Kim and Kang 2006; Kim et al. 2007a; b). Arabidopsis GR-GRP7 is strongly expressed in the guard cells and regulates the opening and closing of stomata in plants under dehydration or salt stress (JS Kim et al. 2008). However, the mechanisms by which GR-RBPs contribute at the molecular level are still largely unknown (Lorković 2009).
Drought is one of the most serious problems for sustainable agriculture worldwide. Woody plants utilize mechanisms for resistance and adaptation to a limited water supply. Drought tolerance is a complex trait that is influenced by the coordinated expression of a network of genes. It is affected by numerous anatomical, physiological, biochemical, developmental, and environmental factors (Ramanjulu and Bartels 2002; Zhu 2002). For promoting agricultural and environmental sustainability, it is important to breed or genetically engineer crops with improved stress tolerance. To understand the molecular mechanism for such tolerance in fruit crops, we focused on Malus prunifolia (Willd.) Borkh., a wild species that is naturalized to the desert mountain regions of northwest China. This rootstock is important for the propagation of apples in China and displays strong disease resistance and stress tolerance. In this study, we described the isolation and identification of a new full-length cDNA, MpGR-RBP1 (M. prunifolia glycine-rich RNA-binding protein1), using in silico cloning and reverse transcriptase polymerase chain reaction (RT-PCR). To get an insight into its behavior and regulation by drought stress, we have characterized the expression profile of mRNA from these apple leaves and roots during drought treatment.
Materials and Methods
Plant Materials and Drought Treatment
Two-year-old seedlings of M. prunifolia were grown in pots in a greenhouse at the College of Horticulture, Northwest A&F University, Yangling (34°20′N, 108°24′E), Shaanxi Province, China. Drought treatments were previously described by Ma et al. (2008). Field experiments were conducted between 20 March and 10 August 2009. After 3 months of development under well-watered conditions, control seedlings continued to receive this standard care while others were progressively stressed by withholding normal irrigation. Leaves and roots from both control and drought-stressed plants were sampled at day 0, 2, 4, 6, 8, 10, and 12 of treatment. All samples were rapidly frozen in liquid nitrogen and stored at −80°C.
Total RNA Extraction and mRNA Isolation
Total RNA was extracted according to the method described by Chang et al. (1993). Poly(A)+ RNA was purified with a poly(A)+ Ttract® mRNA Isolation Systems III kit (Promega, USA) according to the manufacturer's instructions. Three independent biological replications were performed for each experiment.
Suppression Subtractive Hybridization cDNA Library Construction and EST Analysis
Total RNA was extracted from Malus leaves after 6, 8, and 10 days of drought treatment for suppression subtractive hybridization (SSH) and at all sampling time points for gene expression analysis. Total extracted RNA from stressed seedlings was mixed equally as the tester, whereas total RNA from control plants, collected at each corresponding time point, was used as the driver. Our SSH library was constructed according to the method of Diatchenko et al. (1996), using a PCR-select™ cDNA Subtraction kit (Clontech, USA) and 2 μg of Poly(A)+ mRNA isolated from leaf tissues. Briefly, the tester and driver cDNAs were digested with RsaI, a restriction enzyme that yields blunt-ended cDNA fragments. The tester cDNA pool was then divided into two portions, each of which was ligated with a different cDNA adapter sequence. Two sequential hybridizations were performed. First, an excess of driver was added to each tester subpool, and the samples were heat-denatured and allowed to anneal to each other. This resulted in the generation of several different hybrid sequences of cDNA. Second, the two tester subpools were mixed together in the presence of excess driver, but without denaturing, to form new hybrids. The ends of the differentially expressed cDNA sequences were then filled in by DNA polymerase, and two rounds of PCR were performed to enrich these cDNA clones. During PCR, the nondifferentially regulated hybrid clones were either not amplified or did not exhibit exponential amplification. Thus, only differentially regulated genes were enriched by this process. After two rounds of subtractive hybridization and two rounds of suppressive PCR, the products were ligated into the pMD19-T cloning vector (TaKaRa, China), transformed into DH5α cells (Tiangen, China), and plated onto Luria–Bertani agar plates containing ampicillin (100 μg mL−1). Based on their blue/white coloring, recombinant colonies were then selected from those plates. To identify which M. prunifolia genes are differentially expressed under drought treatment, all positive clones obtained from the subtraction libraries were subjected to sequencing, clustering, BLAST alignment, functional annotation, and classification into different categories (unpublished data).
cDNA Synthesis and In Silico Cloning of MpGR-RBP1
A 371-bp cDNA fragment of SSH clone D1403, showing high similarity to GR-RBP, was used as a query probe to search the EST database in GenBank. Homologous EST clones were retrieved and assembled. To verify this assembly, we used forward primer Mpf (5′-GTCCCATTTGTCACTAGGGTTACTG-3′) and reverse primer Mpr (5′-ATCTCAAAAGTCCCAAACCACCTAA-3′) to amplify the open reading frame (ORF) of the full-length cDNA. Complementary DNA was synthesized with 2 μg of total RNA, using Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase (Promega) and the random hexamer primer. PCR was performed with an initial denaturation step at 95°C for 10 min; followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 90 s; then a final extension at 72°C for 10 min. Products were gel-purified and cloned into the pMD19-T cloning vector. Afterward, both strands of the DNA fragments from positive clones were sequenced with an ABI 3730 sequencer.
Analyses of Sequences and Phylogenetics
DNA sequence similarities were analyzed using the programs provide by NCBI BLAST (http://www.ncbi.nlm.nih.gov/blast/). The protein theoretical molecular weight and isoelectric point were predicted using compute pI/Mw (http://au.expasy.org/tools/). A hydropathic index was calculated according to the Kyte–Doolittle scale (http://www.expasy.org/tools/protscale.html). The signal peptide of the putative protein was forecasted using SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/), and prediction of protein localization sites was analyzed using PSORT (http://psort.ims.u-tokyo.ac.jp/form.html). The protein conserved domain was predicted using SMART (http://smart.embl-heidelberg.de/smart/change_mode.pl) network services. The protein structure homology modeling prediction and analysis were performed using 3D-JIGSAW (http://bmm.cancerresearchuk.org/3djigsaw/) and SWISS-MODEL (http://swissmodel.expasy.org/) (Arnold et al. 2006). The deduced amino acid sequence was aligned and the phylogenetic tree was constructed by the ClustalW method using MEGA4.0 software (http://www.megasoftware.net/).
Quantitative Real-Time PCR
To remove any contaminating genomic DNA prior to cDNA synthesis, we treated the RNA with RNase-free DNAse I (Invitrogen, USA) according to the manufacturer's instructions. Total RNA was quantified on a NanoDrop™ 1000 spectrophotometer before and after this DNAse I treatment, and its quality and integrity were checked by electrophoresis through agarose gels stained with ethidium bromide. For real-time RT-PCR, first-strand cDNA was synthesized with 2 μg of total RNA in a volume of 20 μL, using a SYBR®PrimeScript™ RT-PCR Kit II (TaKaRa) plus random hexamers and oligo(dT) primers. After reverse transcription, the reaction product was diluted tenfold with sterile water. Real-time PCR was performed on an iQ5.0 instrument (Bio-Rad, USA) using SYBR Green qPCR kits (TaKaRa) according to the manufacturer's instructions. Primer sequences for MpGR-RBP1 and reference genes were as follows: MpGR-RBP1, forward primer 5′-CGTCGTGAGGGTGGCTATG-3′ and reverse primer 5′-AAAGTCCCAAACCACCTAACAC-3′ (119-bp product); apple EF-1α, forward 5′-ATTCAAGTATGCCTGGGTGC-3′ and reverse 5′-CAGTCAGCCTGTGATGTTCC-3′ (174-bp product); and apple actin, forward 5′-CCAAAGGCTAATCGGGAGAA-3′ and reverse 5′-ACCACTGGCGTAGAGGGAAAG-3′ (105-bp product). Real-time PCR reactions were done in 20-μL volumes containing a 10 μM concentration of each primer, 40 ng of cDNA, and 10 μL of SYBR Premix Ex Taq™ II. Thermal cycling conditions included an initial heat-denaturing step at 95°C for 3 min, and then 40 cycles of 95°C for 20 s, 58°C for 20 s, and 72°C for 20 s. Fluorescence was measured at the end of each cycle. A melting curve analysis was performed by heating the PCR product from 58 to 95°C. Expression data for MpGR-RBP1 were presented as relative units after normalization to the EF-1α control, using the 2− ΔΔCT method. Values for mean expression and SD were calculated from the results of three independent experiments.
Semiquantitative RT-PCR
The relative amount of gene transcription by cloned cDNAs was quantified via RT-PCR. Primer sequences and first-strand cDNA were the same as those used for quantitative real-time PCR. A PCR Master Mix Kit (TaKaRa) was used with 2 μL (10-dilution) of first-strand cDNA as template plus 0.2 μmol L−1 of each primer (final concentration) in a total volume of 25 μL. The linear range of detection for each transcript was monitored after 35 cycles for MpGR-RBP1 in the leaves or roots, or 30 cycles each for EF-1α or actin. Except for cycle numbers, all other reaction conditions were identical. A 15-μL sample of each PCR product was detected on a 1.2% (w/v) agarose gel, then visualized and photographed under UV light after staining with ethidium bromide. Three repetitions were compared for each sample.
Results
Isolation of Full-Length M. prunifolia GR-RBP cDNA
Screening of our SSH cDNA library for drought-stressed seedling leaves from M. prunifolia revealed a 371-bp fragment containing a short polyA-tail, D1403, which shared high sequence identity with genes that encode GR-RBPs. Based on this finding, we used in silico cloning and RT-PCR to isolate a novel full-length cDNA sequence (781 bp) with a complete ORF (Fig. 1).
Nucleotide sequence of MpGR-RBP1 cDNA and deduced amino acid sequence. Predicted protein sequence is shown beneath nucleotide sequence. RRM at N-terminal is shaded, and glycine-rich domain within C-terminal is underlined. RNP1 and RNP2 motifs are underlined and marked below sequence. Repeats of Tyr–(Gly)n–Arg–Arg–Glu–(Gly)2 and Tyr–Ser–Arg–(Gly)4 motif are underlined with arrows. Numbers at left indicate positions in recovered nucleotide sequence, those at right show amino-acid positioning from N-terminus. Initiation codon (ATG) and stop signals (TAG) are underlined in bold. Stop codons are represented by asterisk
The gene has been submitted to the GenBank database with accession number HM042682. The sequence surrounding the first ATG codon at nucleotide position 32 was in the context of GCCATGG, thus agreeing with that of the Kozak (1987) consensus initiator A/GXXATGG. The cDNA contained a 31-bp 5′-untranslated region (UTR), a 516-bp ORF, and a 234-bp 3′-UTR. The ORF of the MpGR-RBP1 gene encodes 171 amino acids with an estimated molecular mass of 16.69 kDa and an isoelectric point of 6.36. The deduced protein, MpGR-RBP1, has 38.01% glycine (G), which is due to the glycine-rich region. Transmembrane and signal peptide prediction show that MpGR-RBP1 might belong to nonsecretory protein.
High Conservation of MpGR-RBP1
We compared the predicted amino acid sequence of MpGR-RBP1 with known GenBank sequences through BLASTP and PSI-BLAST searches. These revealed that our overall amino acid sequence was very similar to stress-responsive plant GR-RBPs such as Pa-RRM-GRP1 from sweet cherry (Stephen et al. 2003). This apple protein sequence showed high identity to sweet cherry (93.0%, AAL13082), soybean (86.0%, AAD48471), geranium (84.9%, AAB63581), tobacco (84.3%, ACD03270), potato (82.9%, ABB87126), citrus (82.6%, BAA92156), Arabidopsis (80.2%, AAM62447), and alfalfa (80.7%, AAF06329), while it exhibited lower identities to maize (74.4%, AAM16020), broomcorn (73.3%, AAG23220), and wheat (72%, BAF30986). In addition, the putative MpGR-RBP1 protein was almost the same size as GR-RBP protein from broomcorn (AAG23220) and barley (CAA88558) (Table 1). Because of these apparent structural characteristics, we designated the gene as MpGR-RBP1 (M. prunifolia glycine-rich RNA-binding protein1).
The predicted amino acid sequence had two main features, an amino-terminal RRM and a glycine-rich carboxy-terminal domain. The RRM was characterized by 77 amino-acid residues, with a hydrophobic segment of six residues (RNP2) and an octapeptide motif of eight residues (RNP1) (Fig. 2a) (Stephen et al. 2003). The glycine-rich domain ranged from 88 to 168 amino-acid residues interspersed with two short, highly conserved sequences—Gly/Gly/Tyr/Gly/Gly(GGYGG) and Arg/Arg/Glu(RRE)—both of which are also abundant in an ABA-inducible GR-RBP from maize embryos (Gόmez et al. 1988). Furthermore, the C-terminal glycine-rich domain was mainly interposed with arginine and tyrosine (the Arg/Gly/Tyr(RGY)-rich domain) (Kumaki et al. 2004). The classic RGY-rich domain contained two similar peptides (YGGGGRREGG and YGGGGGRREGG) and three consensus repeats (YSRGGGG) (Fig. 1). The predicted secondary structure provided long stretches of extended conformation. Such a structural pattern for this RRM (Fig. 2b) fit well with a model of four antiparallel β-strands and two α-helices arranged in a β1–α1–β2–β3–α2–β4 fold with side chains (Birney et al. 1993). The three-dimensional (3-D) model of the protein was built using 3D-JIGSAW program (Fig. 3), and the protein structure homology modeling alignment and analysis were performed using SWISS-MODEL (Arnold et al. 2006). Depending on the SWISS-MODEL Web server, we built and evaluated the MpGR-RBP1 protein homology model based on homologous template: 3bs9B.pdb. Accordingly, the 3-D model of MpGR-RBP1 showed that the N-terminal RRM folds (βαββαβ topology) of MpGR-RBP1 structure most closely matched the RRM2 of the RNA binding protein TIA-1 (T-cell-restricted intracellular antigen-1) (Kumar et al. 2008; PDB-code = 3bs9B; RMSD = 1.95 Å and 38.5% sequence identity for residues 7–84). Therefore, these results suggested that MpGR-RBP1 is a member of the GR-RBPs family and is structurally conserved.
Putative conserved domain structure and schematic model for predicted amino acid sequence. a Model shows amino-terminal RRM and carboxyl-terminal glycine-rich domain. Within RRM are two conserved regions, hydrophobic six-residue RNP2 motif, and eight-residue RNP1. Numbers at top refer to amino-acid positioning from N-terminal methionine. b Putative conserved domain structure of RRM. Secondary-structure prediction indicates that RRM structure comprises four antiparallel β-strands and 2 α-helices arranged in β–α–β–β–α–β fold with side chains
Structural mode of MpGR-RBP1, with RRM folds (βαββαβ topology). α-Helices are colored red, β-sheet are colored yellow, and strands are colored blue–green, respectively. Three-dimensional representations are done with RasTop (http://www.geneinfinity.org/rastop/)
Phylogenetic Analysis of the Predicted MpGR-RBP1 Protein
In order to investigate the evolutionary relationships of MpGR-RBP1 protein among plant GR-RBPs, GRPs, and human hnRNPs, we constructed a phylogenetic tree (Fig. 4) that enabled us to group all of them into three distinct clusters. MpGR-RBP1 protein belonged to a cluster that does not include GRPs or human hnRNPs. MpGR-RBP1 was closely related to Pa-RRM-GRP1 with 80.6% nucleotide identity, which was consistent with our observations described above. Phylogenetic analysis demonstrated that plant GR-RBPs within the same species are more closely related to each other than to GR-RBPs from different species, e.g., NgRBP and NtGRP1 from tobacco or AtGR-GBP7 and AtGR-GBP8 from Arabidopsis, with Pa-RRM-GRP1 (Prunus) and MpGR-RBP1 (Malus) being subordinate to the Rosaceae family. ATRZ-1A is phylogenetic closeness to NSRZ-1 in that they have one zinc fingers in their glycine-rich domain, while the Arabidopsis cold induced proteins AtGR-GBP2 (namely AtGRP2) has two zinc fingers as described above. A recent research shows that AtGR-RBP2 is localized into mitochondria of Arabidopsis and may play a role in the multiple posttranscriptional mechanisms involved in mitochondrial gene expression (Vermel et al. 2002).
Phylogenetic tree of MpGR-RBP1 with other plant GR-RBPs, GRPs, and human hnRNPs was reconstructed by the neighbor-joining method. The tree is based on an alignment corresponding to full-length amino acid sequences, using ClustalW and MEGA 4.0. Branches are labeled with gene names, and MpGR-RBP1 is indicated by asterisk. The number shown at the branches denote the bootstrap majority consensus values of 1,000 replicates. The GenBank accession numbers for the sequence designations are as follows: A. thaliana (AtGRP3, S47409), A. thaliana (AtGRP5, S47414), Boea hygrometrica (BhGRP1, EU003996), Nicotiana glutinosa (ngRBP, AF005359), N. tabacum (NtGRP1, EU569289), A. thaliana (AtGR-RBP7, AAD23639), M. sativa (MsGRP1, AF191305), P. avium (Pa-RRM-GRP1, AY050483), A. thaliana (AtGR-RBP8, NP_195637), Z. mays (ZmGRP2, NP_00115156), A. thaliana (ATRZ-1A, NM_113549), Nicotiana sylvestris (NSRZ-1, BAA06012), A. thaliana (AtGR-RBP2, CAB36849), Homo sapiens (CIRP, NP_001271), H. sapiens (RBM3, NP_006734), and H. sapiens (hnRNPA0, NP_006796)
Another phylogenetic cluster contains CIRP (cold-inducible RNA-binding protein; A18 hnRNP), RBM3 (also known as IS1-RNPL or RNPL), and hnRNPA0 (also known as hnRPA0). The first two hnRNPAs have a single RRM domain. In contrast, the third one has two repeats of quasi-RRM domains that bind RNAs and a glycine-rich C-terminus (Rousseau et al. 2002). Independent clusters are used to distinguish among GRPs, BhGRP1, AtGRP5, and AtGRP3. Our results indicated that the current clustering of MpGR-RBP1 is based on its complete primary sequences and species.
Stress-Responsive Expression Pattern of MpGR-RBP1
To investigate the effect of drought stress on MpGR-RBP1 expression, we performed quantitative real-time RT-PCR. This gene was induced in both roots and leaves during treatment. Transcripts within the latter were initially increased up to 26-fold over that of the control (as measured on day 0) in the first 8 days of drought stress before decreasing by approximately sevenfold at day 12. In contrast to this strong up-regulation in the leaves, expression in the roots increased by only about sevenfold during the first 8 days of treatment, followed by a slight decrease by day 10 (Fig. 5). Real-time PCR results showed that transcription levels were much higher in the leaves than in root tissues.
Quantitative real-time RT-PCR analysis of the MpGR-RBP1 expression level in leaves and roots from Malus prunifolia under drought stress. Two-year-old potted seedlings were grown in 14-h light/10-h dark cycles during water treatment period. Leaves and roots of plants were harvested at midday, respectively. Transcript levels in seedlings stressed for 0, 2, 4, 6, 8, 10, or 12 days were plotted as expression (n-fold) relative to nonstressed control (day 0). Amount of transcript was normalized to apple EF-1α expression level. Mean values and standard errors (bars) were obtained from three independent experiments per time point
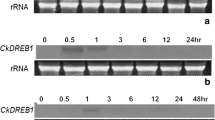
Expression was analyzed at the mRNA level at different time points during the stress period. Semiquantitative RT-PCR showed that transcripts of MpGR-RBP1 were elevated in both leaves and roots between days 4 and 8 (Fig. 6). After peaking, this expression began to decline from day 10 to 12. By comparison, no significant changes were observed in the transcript levels of EF-1α and actin. These RT-PCR results were in agreement with data from our quantitative real-time PCR analysis.
Semiquantitative RT-PCR analysis of MpGR-RBP1 expression under well-watered conditions (CK) and in response to drought (4, 6, 8, 10, or 12 days). Leaves and roots were harvested at indicated times after stress period. Accumulation of MpGR-RBP1 was monitored by semiquantitative RT-PCR. EF-1α or actin was used as constitutive controls
Discussion
Plant GR-RBPs play an important role in normal developmental processes and also participate in adaptations to various environmental conditions. Although a few such GR-RBPs involved in abiotic stress responses have been identified in Arabidopsis, rice, and tobacco (Sahi et al. 2007; JS Kim et al. 2008; Lee et al. 2009), little information has been reported for GR-RBPs in Malus plants against water stress. Therefore, it is necessary to investigate this relationship because the development of drought-resistant rootstocks through the modification of stress-related genes can lead to enhanced fruit crop productivity.
Here, we reported the isolation and characterization of a novel GR-RBP gene, MpGR-RBP1. Like the orthologous genes in human and other plants, MpGR-RBP1 protein displays considerable sequence conservation in the RRM motif and the glycine-rich region, suggesting conservation of functional properties. The RRM conserved motif at the N-terminal end contains two short sequences, RNP1 and RNP2. Evidence indicates that RRMs are present in a variety of RBPs and are also found in a few single-stranded DNA binding proteins (Birney et al. 1993; Lorković and Barta 2002). Proteins with RRM are involved in the posttranscriptional regulation of gene expression and pre-rRNA processing, as well as in RNA processing at several levels (Burd and Dreyfuss 1994; Guiltinan and Niu 1996). The 3-D model of MpGR-RBP1 shows significant homology with human RNA binding protein TIA-1. Human TIA-1 regulates alternative pre-mRNA splicing in the nucleus, and mRNA translation in the cytoplasm, by recognizing uridine-rich sequences of RNAs (Kumar et al. 2008). This suggests that MpGR-RBP1 might have similar functions to the human protein.
Database searches of our sequence demonstrated that MpGR-RBP1 has significant homology to GR-GRP protein family genes that are responsive to abiotic stress. Phylogenetic analysis further demonstrated that MpGR-RBP1 can be highly homologous to some other plant GR-RBPs, in particular, the one encoded by Pa-RRM-GRP1 (AY050483 from Prunus avium), rather than to Arabidopsis GR-RBP7 and GR-RBP8. These GR-RBP proteins are distinct from members of the group of GRPs, which lack a conventional RRM, such as a drought-induced cell-wall GRP protein AtGRP3 and a vacuole-located GRP AtGRP5 (Mangeon et al. 2009). The current data indicate that the Arabidopsis genome encodes eight GR-RBPs (AtGR-RBP1–AtGR-RBP8), all characterized by the presence of an N-terminal RRM domain and a C-terminal tail of varying lengths and all rich in glycine residues. Among them, AtGR-RBP7 and AtGR-RBP8 are homologous to each other (76.9% identity) and probably perform related functions (Fu et al. 2007; Lorković 2009). However, AtGR-GRP7 differs from AtGR-GRP2 and AtGR-GRP4 in that it has a much shorter N-terminal region and a much longer glycine-rich C-terminal region (JS Kim et al. 2008).
Plants produce a large number of kingdom-specific RNA-binding proteins. RBPs contain one or more RRMs at the N-terminus and a variety of auxiliary motifs at the C-terminus, such as glycine-rich, arginine-rich, SR-repeat, RD-repeat, and acidic domains (JS Kim et al. 2008; Lee et al. 2009). Plant GR-RBPs belong to the family of RNA-binding proteins that has in common a protein domain that is necessary and sufficient for RNA binding. Over time, only those proteins required for basic mechanisms in posttranscriptional regulation of gene expression have been preserved in all eukaryotic lineages (Lorković and Barta 2002; Fu et al. 2007). Our results further support that MpGR-RBP1 protein is in that plant GR-GRP family. However, because GR-RBPs are encoded in the genomes of different species, certain variations do exist, and so researchers must still determine whether functioning of a particular protein is similar or equivalent to other plant GR-RBPs.
Abiotic stresses, such as drought, modulate expression at the posttranscriptional level as well as at the transcriptional level (Sunkar and Zhu 2004). Posttranscriptional mechanisms for the regulation of gene expression involve a control of pre-mRNA processing, transportation to cytoplasm, and mRNA stability. These regulatory mechanisms are mediated by specific interaction between RNA-binding proteins and RNA molecules (Shinozuka et al. 2006). Plant GR-RBPs are involved in posttranscriptional RNA processing in response to various stress conditions. For example, transcripts of rice Osgr-rbp4 are constitutively expressed as well as regulated by high temperatures (Sahi et al. 2007). A recent report shows that AtGR-GRP7 and AtGR-GRP8 are rapidly up-regulated in response to oxidative stress (Schmidt et al. 2010). Here, we show that MpGR-RBP1 gene expression is strongly up-regulated upon drought stress. This observation is consistent with the results shown by Lee et al. (2009), where tobacco NtGRP1 is also up-regulated by drought, albeit to a much lower degree compared to flooding stress. On the contrary, the transcript level of GR-RBP4 in Arabidopsis decreases slightly with dehydration stress (Kwak et al. 2005). Recent studies have shown that Arabidopsis GR-RBP2, -4, and -7, as well as atRZ-1a, are differentially involved in plant responses to abiotic stresses also, but may be either ABA independent (AtGR-RBP2 and AtGR-RBP7) or dependent (atRZ-1a). Thus, both transcriptional reprogramming and posttranscriptional mechanisms are utilized for coping with stress (Lorković 2009).
Furthermore, to avoid bias while investigating MpGR-RBP1 transcription, we selected two internal control genes, actin and EF-1α, both of which are consistently expressed in apple and grapevine during biotic or abiotic stress (Nicot et al. 2005; Paris et al. 2009). Our results show that EF-1α gene used as a standard is more validated than actin, which is consistent with the results that EF gene proved to be the most stable among all the housekeeping genes tested (Nicot et al. 2005). Expression of MpGR-RBP1 was increased in the roots and even more so in the leaves. Similarly, in Arabidopsis, real-time RT-PCR and GUS expression analyses have shown that AtGR-RBP4 is abundantly expressed in young plants, root tips, and flowers, but only weakly in mature leaves and stems (Kwak et al. 2005). Overexpression of AtGR-GRP2 and AtGR-GRP7 enhances cold and freezing tolerances, and AtGR-RBPs exert RNA chaperone activity during the process of cold adaptation (Kim et al. 2007a). Therefore, we can speculate that the elevated expression of MpGR-RBP1 also plays a critical role for the drought adaptation of Malus plant.
In conclusion, the results of the current study provide clear evidence that MpGR-RBP1 is structurally conserved and may be involved in posttranscriptional RNA processing in response to water stress. Although we have not yet fully described its functioning, our results suggest that this protein is active in the adaptation process because its mRNA level increases following exposure to drought treatment. Therefore, it will be important to further evaluate the regulatory function of MpGR-RBP1 in Malus plants and the molecular mechanisms that control its expression under different environmental conditions.
Abbreviations
- GR-RBP:
-
Glycine-rich RNA-binding protein
- GRP:
-
Glycine-rich protein
- SSH:
-
Suppression subtractive hybridization
- RT-PCR:
-
Reverse transcriptase polymerase chain reaction
- ORF:
-
Open reading frame
- UTR:
-
Untranslated region
References
Albà MM, Pagès M (1998) Plant proteins containing the RNA-recognition motif. Trends Plant Sci 3:15–21
Aneeta Sanan-Mishra N, Tuteja N, Sopory SK (2002) Salinity- and ABA-induced up-regulation and light-mediated modulation of mRNA encoding glycine-rich RNA-binding protein from Sorghum bicolor. Biochem Biophys Res Commun 296(5):1063–1068
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL Workspace: a Web-based environment for protein structure homology modelling. Bioinformatics 22(2):195–201
Bandziulis RJ, Swanson MS, Dreyfuss G (1989) RNA-binding proteins as developmental regulators. Genes Dev 3:431–437
Baudo MM, Meza-Zepeda LA, Palva ET, Heino P (1999) Isolation of a cDNA corresponding to a low temperature- and ABA-responsive gene encoding a putative glycine-rich RNA-binding protein in Solanum commersonii. J Exp Bot 50(341):1867–1868
Birney E, Kumar S, Krainer AR (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res 21(25):5803–5816
Bocca SN, Magioli C, Mangeon A, Junqueira RM, Cardeal V (2005) Survey of glycine-rich proteins (GRPs) in the Eucalyptus expressed sequence tag database (ForEST). Gen Mol Biol 28(3):608–624
Burd CG, Dreyfuss G (1994) Conserved structures and diversity of functions of RNA binding proteins. Science 265:615–621
Chang S, Puryear J, Cairney J (1993) Simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11:113–116
Diatchenko L, Lau YFC, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD (1996) Suppression subtractive hybridization: a method for generating differentially regulated tissue-specific cDNA probes and libraries. Proc Natl Acad Sci 93:6025–6030
Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR (2007) A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447:284–288
Fusaro AF, Bocca SN, Ramos RL, Barrôco RM, Magioli C, Jorge VC, Coutinho TC, Rangel-Lima CM, De Rycke R, Inzé D, Engler G, Sachetto-Martins G (2007) AtGRP2, a cold-induced nucleo-cytoplasmic RNA-binding protein, has a role in flower and seed development. Planta 225(6):1339–1351
Gόmez J, Sánchez MD, Stiefel V, Rigau J, Puigdomènech P, Pagès M (1988) A gene induced by the plant hormone abscisic acid in response to water stress encodes a glycine-rich protein. Nature 334:262–264
Guiltinan MJ, Niu XP (1996) cDNA encoding a wheat (Triticum aestivum cv Chinese Spring) glycine-rich RNA-binding protein. Plant Mol Biol 30(6):1301–1306
Kim YO, Kang H (2006) The role of a zinc finger-containing glycine-rich RNA-binding protein during the cold adaptation process in Arabidopsis thaliana. Plant Cell Physiol 47(6):793–798
Kim YO, Kim JS, Kang H (2005) Cold-inducible zinc finger-containing glycine-rich RNA-binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. Plant J 42(6):890–900
Kim JS, Park SJ, Kwak KJ, Kim YK, Kim JY, Song J, Jang B, Jung CH, Kang H (2007a) Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Res 35(2):506–516
Kim JY, Park SJ, Jang BS, Jung CH, Ahn SJ, Goh CH, Cho K, Han O, Kang H (2007b) Functional characterization of a glycine-rich RNA-binding protein 2 in Arabidopsis thaliana under abiotic stress conditions. Plant J 50(3):439–451
Kim YO, Pan S, Jung CH, Kang H (2007c) A zinc finger-containing glycine-rich RNA-binding protein, atRZ-1a, has a negative impact on seed germination and seedling growth of Arabidopsis thaliana under salt or drought stress conditions. Plant Cell Physiol 48(8):1170–1181
Kim JS, Jung HJ, Kim KA, Goh CH, Woo Y, Oh SH, Han YS, Kang H (2008) Glycine-rich RNA-binding protein 7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J 55:455–466
Kozak M (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15(20):8125–8132
Kumaki Y, Nitta K, Hikichi K, Matsumoto T, Matsushima N (2004) Side chain–side chain interactions of arginine with tyrosine and aspartic acid in Arg/Gly/Tyr-rich domains within plant glycine-rich RNA binding proteins. J Biochem 136(1):29–37
Kumar AO, Swenson MC, Benning MM, Kielkopf CL (2008) Structure of the central RNA recognition motif of human TIA-1 at 1.95 Å resolution. Biochem Biophys Res Commun 367:813–819
Kwak KJ, Kim YO, Kang H (2005) Characterization of transgenic Arabidopsis plants overexpressing GR-RBP4 under high salinity, dehydration, or cold stress. J Exp Bot 421(56):3007–3016
Lee MO, Kim KP, Kim BG, Hahn JS (2009) Flooding stress-induced glycine-rich RNA-binding protein from Nicotiana tabacum. Mol Cells 27(1):47–54
Lorković ZJ (2009) Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci 14(4):229–236
Lorković ZJ, Barta A (2002) Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res 30(3):623–635
Ma X, Ma F, Mi Y, Ma Y, Shu H (2008) Morphological and physiological responses of two contrasting Malus species to exogenous abscisic acid application. Plant Growth Regul 56:77–87
Mangeon A, Magioli C, Menezes-Salgueiro AD, Cardeal V, de Oliveira C, Galvão VC, Margis R, Engler G, Sachetto-Martins G (2009) AtGRP5, a vacuole-located glycine-rich protein involved in cell elongation. Planta 230(2):253–265
Nakaminami K, Karlson DT, Imai R (2006) Functional conservation of cold shock domains in bacteria and higher plants. Proc Natl Acad Sci 103(26):10122–10127
Naqvi SMS, Park KS, Yi SY, Lee HW, Bok SH, Choi D (1998) A glycine-rich RNA-binding protein gene is differentially expressed during acute hypersensitive response following tobacco mosaic virus infection in tobacco. Plant Mol Biol 37(3):571–576
Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56(421):2907–2914
Query CC, Bentley RC, Keene JD (1989) A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70 K U1 snRNP protein. Cell 57(1):89–101
Ramanjulu S, Bartels D (2002) Drought- and desiccation-induced modulation of gene expression in plants. Plant Cell Environ 25:141–151
Richard S, Drevet C, Jouanin L, Seguin A (1999) Isolation and characterization of a cDNA clone encoding a putative white spruce glycine-rich RNA binding protein. Gene 240(2):379–388
Rousseau S, Morrice N, Peggie M, Campbell DG, Gaestel M, Cohen P (2002) Inhibition of SAPK2a/p38 prevents hnRNPA0 phosphorylation by MAPKAP-K2 and its interaction with cytokine mRNAs. EMBO J 21(23):6505–6514
Sachetto-Martins G, Franco LO, de Oliveira DE (2000) Plant glycine-rich proteins: a family or just proteins with a common motif? Biochim Biophys Acta 1492(1):1–14
Sahi C, Agarwal M, Singha A, Grover A (2007) Molecular characterization of a novel isoform of rice (Oryza sativa L.) glycine rich-RNA binding protein and evidence for its involvement in high temperature stress response. Plant Sci 173(2):144–155
Schmidt F, Marnef A, Cheung MK, Wilson I, Hancock J, Staiger D, Ladomery M (2010) A proteomic analysis of oligo(dT)-bound mRNP containing oxidative stress-induced Arabidopsis thaliana RNA-binding proteins ATGRP7 and ATGRP8. Mol Biol Rep 37(2):839–845
Schöning JC, Streitner C, Page DR, Hennig S, Uchida K, Wolf E, Furuya M, Staiger D (2007) Autoregulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J 52(6):1119–1130
Schöning JC, Streitner C, Meyer IM, Gao Y, Staiger D (2008) Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res 36(22):6977–6987
Shinozuka H, Hisano H, Yoneyama S, Shimamoto Y, Jones ES, Forster JW, Yamada T, Kanazawa A (2006) Gene expression and genetic mapping analyses of a perennial ryegrass glycine-rich RNA-binding protein gene suggest a role in cold adaptation. Mol Genet Genom 275(4):399–408
Staiger D, Zecca L, Wieczorek Kirk DA, Apel K, Eckstein L (2003) The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J 33:361–371
Stephen JR, Dent KC, Finch-Savage WE (2003) A cDNA encoding a cold-induced glycine-rich RNA binding protein from Prunus avium expressed in embryonic axes. Gene 320:177–183
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Paris R, Cova V, Pagliarani G, Tartarini S, Komjanc M, Sansavini S (2009) Expression profiling in HcrVf2-transformed apple plants in response to Venturia inaequalis. Tree Genetics & Genomes 5(1):81–91
Vermel M, Guermann B, Delage L, Grienenberger JM, Maréchal Drouard L, Gualberto JM (2002) A family of RRM-type RNA-binding proteins specific to plant mitochondria. Proc Natl Acad Sci 99(9):5866–5871
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Ziemienowicz A, Haasen D, Staiger D, Merkle T (2003) Arabidopsis transportin1 is the nuclear import receptor for the circadian clock-regulated RNA-binding protein AtGRP7. Plant Mol Biol 53(1):201–212
Acknowledgments
This work was supported by the Hi-Tech Research and Development Program of China (2008AA10Z157) and by the Modern Agricultural Industry Technology System in China. The authors are grateful to Priscilla Licht for help in revising our English composition.
Author information
Authors and Affiliations
Corresponding author
Additional information
Shuncai Wang and Dong Liang contributed equally to this work
Rights and permissions
About this article
Cite this article
Wang, S., Liang, D., Shi, S. et al. Isolation and Characterization of a Novel Drought Responsive Gene Encoding a Glycine-rich RNA-binding Protein in Malus prunifolia (Willd.) Borkh.. Plant Mol Biol Rep 29, 125–134 (2011). https://doi.org/10.1007/s11105-010-0221-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-010-0221-1