Abstract
Loquat (Eriobotrya japonica) is an underutilized fruit crop that originated in China and for which only a small number of molecular markers are available. This number can be increased by identifying apple SSRs that are transferable to loquat cultivars/accessions to provide new insight into the level of genetic diversity within loquat and synteny with apple. We evaluated 71 apple SSR markers distributed across 17 linkage groups, and identified 39 SSRs transferable to loquat. Testing 54 loquat accessions, from Japan, Spain, four provinces in China, and two wild species gave a total of 155 different alleles with a mean value of 3.38 per locus. The mean effective number of alleles was 2.21, and the mean observed heterozygosity was 0.47. These values indicate a high degree of genetic diversity in the set of Chinese loquat accessions analyzed. Unweighted pair-group method analysis based on simple matching coefficent clustered the accessions into two groups, cultivated and wild loquat. The cultivated loquat can be subdivided into three subgroups which generally reflect their geographic origin in China. The Spanish cultivars clustered with those of the Jiangsu and Zhejiang provinces. A core set of five SSR markers could distinguish most accessions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Loquat [Eriobotrya japonica (Thunb) Lindl.], a subtropical evergreen fruit tree of the Rosaceae subfamily Maloideae, originated in southwest China and has been cultivated for over 2,000 years (Qiu and Zhang 1996). It has been introduced to more than 30 countries including Japan, Mediterranean countries in Europe, India, Australia, New Zealand, Madagascar, and South Africa, while commercial cultivation is limited to a few countries. In China, loquat blooms in the fall and early winter, and the fruit ripens between May and June. Loquat leaves and fruits are traditionally used for treating coughs and as expectorant, and the flower is an excellent source of honey. In 2005, around 131,000 ha were planted worldwide (120,000 ha in China), with a production of more than 549,220 t (400,000 t in China). In China, the Sichuan, Fujian, Jiangsu, and Zhejiang provinces are the main production areas (Lin 2007). The loquat cultivars from these areas are the most well-known and commonly planted in China, mainly ‘Dawuxing’, ‘Longquan No.1’, ‘Zaozhong No.6’, ‘Jiefangzhong’, ‘Dahongpao’, ‘Luoyangqing’, ‘Ruantiaobaisha’, ‘Baiyu’, ‘Qingzhong’ and ‘Guanyu’.
The pedigrees of the majority of loquat cultivars are unknown as historically. The current system used for cultivar classification gives little information on genetic identity and variability as it is mainly based on morphological traits linked to ecotype, flesh color, fruit shape, usage, and ripening time (Martínez-Calvo et al. 2008). Genetic diversity and the relationships among different cultivars of loquat are of great importance for the conservation of genetic resources, breeding initiatives, and national and international exchange of materials.
Using molecular markers has significantly contributed to our understanding of genetic diversity and relatedness in various crops. Microsatellites or simple sequence repeats (SSRs) are markers generally used to detect polymorphism (Weber and May 1989). The applicability of SSR markers for apple, loquat, and peach has been reported (Soriano et al. 2005; Qiao 2008; Watanabe et al. 2008; Gisbert et al. 2009a/b). Soriano et al. (2005) used 30 SSRs from apple to assay the genetic relationship in loquat, 13 of which amplified polymorphic products and distinguished 34 of the 40 loquat accessions originating from different European countries. SSR markers have also been used to identify the polyploidy level of loquat (Watanabe et al. 2008). Recently, Gisbert et al. (2009a) constructed the first loquat linkage maps with 83 SSRs and 94 AFLPs, which will also be very useful in genetic analysis and comparative genome research with other Rosaceae fruits. However, an in-depth analysis of the genetic diversity of loquat species, using large numbers of SSR markers and accessions from different geographical areas of the world, is still lacking.
In this paper, we tested the transferability of 71 apple SSRs (Silfverberg-Dilworth et al. 2006; Gisbert et al. 2009a) to loquat to study the genetic diversity of, and fingerprint, 51 loquat accessions of local cultivars from the Jiangsu and Zhejiang and other Chinese provinces, and three cultivars from Spain and Japan.
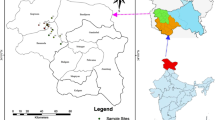
Materials and Methods
Plant Materials
Fifty two loquat cultivars/accessions were examined (Table 1), of which 27 (including the Japanese cultivar ‘Moriowase’) were from the Taihu Extension Center for Evergreen Fruit (Jiangsu province), 14 from the Yuhang District Loquat Institute (Zhejiang province), six from Fujian Putian College, and five from an orchard at Southwest University. In addition, two DNA sample of Spanish cultivars ‘Marc’ and ‘Peluches’ were provided by Professor Shunquan Lin at the South China Agricultural University. The two wild species, ‘Liye’ (Eriobotrya prinoides var. prinoides) and ‘Daduhe’ (E. prinoides var. daduheensis, a natural hybrid of E. prinoides var. prinoides and E. japonica) accessions of the native wild species E. prinoides were included as an out-group reference. The geographical distribution of Chinese accessions is shown in Fig. 1, and some of the cultivars with known genetically relationship shown in Fig. 2
Illustration of known pedigrees of some accessions. Information is from Table 1, X indicates crossing or seeding progeny, thundering symbol indicates mutant
DNA Extraction and Examination
DNA extraction was based on the method of Roche et al. (1997), modified by adding 0.1 g PVP powder to grind the leaf material. After extraction, 2-5 μl DNA solution was loaded on 1.0% agarose gel to check the quality.
SSR Primers
We selected a total of 71 SSR markers, including six from Soriano et al. (2005), covering the 17 linkage groups in apple (Silfverberg-Dilworth et al. 2006; Patocchi et al. 2008), with at least two well-separated loci from each linkage group. The primer sequences and polymerase chain reaction (PCR) conditions were obtained from the apple SSR database (http://www.hidras.unimi.it) (Gianfranceschi and Soglio 2004). Primers were synthesized by Shanghai InvitrogenTM Life Technologies.
PCRs and Electrophoresis
PCR amplification was in a total volume of 20 μl, containing: 10 ng genomic DNA, 2 μl of 10× PCR buffer (with MgCl2), 0.1 μl of 10 mM dNTP mixture, 0.5 μl each of forward and reverse primer (10 pmol/μl), and 0.5 U Taq polymerase (Takara Biotechnology Company, Dalian). The reactions were performed using an Eppendorf Mastercycler (Gradient, No. 5331-41264, Germany) with the following conditions: 94°C for 2 min and 30 s, then 35 cycles of 94°C for 30 s, 50-62°C for 30 s, and 72°C for 30 s, and a final step at 72°C for 10 min. The annealing temperature for each primer pair was optimized using a PCR gradient (T = 57°C, R = 3°Cs, G = 8°C). Twenty microliter PCR products were mixed with 5 μl formamide loading buffer (98% formamide, 10 mM EDTA, 0.25% bromophenol blue, 0.25% xylene cyanol, pH 8.0), heat-denatured at 95°C for 5 min, and 5 μl of each mixture and a molecular size marker of pBR322 DNA-MspI Digest (New England BioLabs) loaded onto a 6% denaturing polyacrylamide gel (7 M urea) in 1× TBE buffer (89 mM Tris-borate, 2 mM EDTA, pH 8.0). Gels were run at 60 W, 45°C for 1 to 1.5 h in a sequencing gel electrophoresis apparatus (DYCZ-20C, Beijing Liuyi Instrument Factory, China) and silver stained (Brant 1991).
Allele Data and Statistic Analysis
Allele sizes were estimated by comparison to pBR322 DNA-MspI Digest, and named in alphabetical order (A, the largest). Where only one band was present, this allele was assumed to be homozygous. Cultivars with known pedigree (Fig. 2) were used to evaluate the scores and the validity of the parentages. All accessions, except the wild species, were analyzed as a single population for number of alleles per locus (Na), effective number of alleles (Ne), Shannon index (I), Nei’s expected heterozygosity (He), observed heterozygosity (Ho), and Wright's fixation index (F). The chi-square test for Hardy–Weinberg equilibrium (P hw) of primers was using the POPGENE program (version 1.32), with 0.01 significance and 10,000 times simulation. A dendrogram with all 54 accessions, based on the matrix of the genetic identities, was constructed using unweighted pair-group method using arithmetic average (UPGMA) with the NTSYSpc 2.10e software (Rohlf 2000). The data were transformed to a matrix of similarity coefficients using simple matching (SM) coefficient. The Eigen procedure of NTSYSpc 2.10e software was used for principal component analysis.
Results
Screening Suitable SSR Markers and Their Variation
Of the 71 apple SSR primer pairs tested, more than half were found to be transferable to loquat: 39 gave polymorphic alleles in loquat (Table 2), 16 get a single or two alleles in all accessions (Table 3), and 16 primers produced smear bands. Four markers (AT000400, CH01d03, CH01h02, and CH03c02) were judged to be multi-locus as they amplified more than three alleles in most accessions. The size range of the alleles in loquat differed from that of apple. The 39 polymorphic SSR markers gave a total of 155 alleles, two to seven alleles per marker with an average of 3.38. The effective number of alleles varied from 1.04 for CH01f02 to 4.00 for CH02d12, with an average of 2.21. The Shannon index, as a measure of gene diversity, ranged from 0.10 for CH01f02 to 1.49 for CH03a09, with an average of 0.84. The Ho ranged from approximately zero for CH01f02 to 0.83 for Hi08a04, with a mean of 0.47. Similar values were calculated for He. The fixation index (F) ranged from −0.70 for Hi08a04 to 1.00 for CH01f02, with an average of 0.07. There was significant deviation from the HW equilibrium (P < 0.05) for 20 loci (Table 2).
In the dendogram constructed from UPGMA cluster analysis of the similarity matrix, with 155 SSR alleles, the accessions are clustered in two groups, the commonly cultivated loquats and the two wild species (Fig. 3) at a 0.48 threshold of SM coefficient. The cultivated loquat accessions were further subdivided into three subgroups (A-C), at a threshold of 0.723, and generally reflected their geographic origin. Subgroup A included all local accessions from the geographically close Zhejiang and Jiangsu provinces, being temperate zone accessions, plus the ‘Longquan No.1’ from Sichuan and ‘Changhong’ from Fujian, and the two Spanish cultivars. Subgroup B included the subtropical Fujian cultivars (‘Zaozhong No.6’, ‘Jiefangzhong’, ‘Xiangzhong’, ‘Taicheng No.4’ and ‘Baili’), but also the Sichuan cultivar ‘Jinfeng’ and the Japanese cultivar ‘Moriowase’. The third subgroup included only one cultivar: ‘Dawuxing’ of Sichuan. The clusters thus generally reflected the geographic origin of their members.
Dendrogram of the 54 loquat accessions with 39 primer pairs generated by UPGMA cluster analysis from the similarity matrix obtained using simple matching (SM) coefficient; filled square indicates Japanese and Spanish accessions, other symbols as Fig. 1
PCA analyses, carried out using the similarity matrices for the 39 SSR markers (Fig. 4), confirmed the UPGMA cluster analysis. The two wild species (group V) were separated from the cultivated accessions, and the cultivated accessions were classified in four groups, mainly according to their geographical distribution. All the Zhejiang accessions, ten Jiangsu accessions, the Japanese cultivar ‘Moriowase’, and two Spanish cultivars, ‘Marc’ and ‘Peluches’, were in group I. The 15 Jiangsu accessions clustered in group II, all the Fujian cultivars in group III, and ‘Dawuxing’ and ‘Longquan No.1’, the two most commonly cultivated varieties in Sichuan province, in group IV. The first two principal components explained 9.86% and 8.63% of the total variation, respectively.
Compared to the cultivated varieties, half the markers gave distinct alleles with the wild species ‘Liye’, while ’Daduhe’ had more common alleles, in agreement with its origin as a hybrid between a wild, E. prinoides var. prinoide, and cultivated loquat (Table 1).
Genetic Identity of Different Accessions
Thirty nine SSR markers distinguished all accessions except four cultivars: the two Spanish cultivars ‘Marc’ and ‘Peluches’ could not be distinguished from each other, and ‘Meiyu’ could not be distinguished from ‘Changlv No.2’. The scores of these markers confirmed pedigrees (Fig. 2), such as that of ‘Zaozhong No.6’ (a ‘Jiefangzhong’ × ‘Moriowase’) and ‘Changlv No.5’ (a ‘Baiyu’ × ‘Tianzhong’), and ‘Xiangzhong’ was confirmed as a descendant of ‘Jiefangzhong’, since for each SSR marker one of the alleles of ‘Jiefangzhong’ was present in ‘Xiangzhong’. ‘Moriowase’ was also confirmed as a sport of ‘Mogi’ by comparing published marker scores for Mogi (Watanabe et al. 2008) with our scores for Moriowase (our study did not include Mogi). Identical alleles were found for the three common SSR: 140/140 for CH03a09, 185/185 for CH05a04, and 212/216 for CH02d10a.
For genetic identity comparison, it is important to discriminate the greatest number of accessions with the least number of markers. Based on the number of effective alleles, we selected a set of five SSR markers (CH03a09, CH02c06, CH04g12, CH02d12, and CH05h05) able to distinguish all the accessions except bud sports.
Discussion
We used comparative genomics to transfer apple SSR markers for genetic diversity and identity studies in Chinese loquat. Cultivar grouping based on SSR markers reflected their geographic origin, and a core set of five polymorphic SSR markers allowed efficient and easy identity assessments of the cultivars. The relationship between the cultivated loquat and closely related wild species was also addressed. This research will help to construct better reference linkage mapping populations.
Transferability and Polymorphism of Apple SSR to Loquat
The transferability of microsatellites among Rosaceae has been described (Dirlewanger et al. 2002; Wünsch and Hormaza 2007, Sargent et al. 2009) and molecular marker linkage maps of apple and Prunus have been shown to have a high level of macro-synteny (Dirlewanger et al. 2004). Eriobotrya has the same chromosome number (2n = 34) as apple. Microsatellite markers used in loquat linkage mapping have been mainly derived from Malus, Prunus, and Pyrus (Soriano et al. 2005; Watanabe et al. 2008; Gisbert et al. 2009a) with only 21 polymorphic simple sequence repeats derived from an enriched loquat genomic library (Gisbert et al. 2008). The ratio of apple SSR that are polymorphic in loquat is not high, varying between 31% (14/45, Watanabe et al. 2008) and 55% (39/71, this research), with intermediate values (43%, 13/30) reported by Soriano et al. (2005).
The level of polymorphism of an SSR marker seems to greatly depend on the germplasm on which it is tested. Gisbert et al. (2009b) amplified 12 different alleles using Hi03a03 and distinguished most of their cultivars, whereas we only amplified three alleles with this SSR, as shown in Tables 4 and 5, the observed Ho we found was lower than that found by other authors. This is most likely due to the accessions used in this study being mostly seedling progenies or bud mutations of ancient cultivars with a narrow genetic background. More Chinese and European accessions are needed, including wild species and semi-cultivated varieties, to reveal the total genetic diversity of loquat.
The size of the alleles and the number of loci has also been found to vary when using SSR markers. For example, with locus CH01f02 in loquat, Soriano et al. (2005) found three alleles between 170 and 180 bp long, and Gisbert et al. (2009b) found four alleles between 160 and 185 bp, whereas we found only two alleles of 160 and 162 bp. Another example is pear, where nine alleles between 155 and 180 bp long have been found in Tunisian pears (Brini et al. 2008), while eight alleles from 156 to 178 bp were found in European pears (Wünsch and Hormaza 2007). This may be caused by artificial deviation as a result of comparing the alleles with DNA markers.
Quality of Marker Scoring
In most diversity studies of loquat, the consistency of the marker scores cannot be checked. Errors in scoring remain unnoticed and lead to false levels of high diversity between accessions. As a control in our marker scoring system, we included some cultivars with known genetic relationships, such as ‘Zaozhong No.6’ and ‘Changlv No.5’, where, for each SSR locus, one allele of a progeny was present in each parent to give 100% scoring accuracy. We also compared our data with the literature to confirm the consistency of our scores: our scores on Moriowase were consistent with those of Watanabe et al. (2008) on Mogi, from which Moriowase is derived.
Grouping and Relationships of Accessions
Most of the new loquat cultivars are derived from natural hybridization (seeds) or bud sports, as E. japonica is both self-compatible and cross-compatible. Seedlings are usually quite heterogeneous and can easily be distinguished from each other, while accessions derived from mutation (and selected out by growers) cannot be distinguished. This is true for ‘Meiyu’ and ‘Changlv No.2’, with identical scores for each of our 39 SSR markers, and for the two Spanish cultivars ‘Peluches’ and ‘Marc’. Our results are in agreement with those of Vilanova et al. (2001), who observed no difference with 23 polymorphic RAPD primers, and Soriano et al. (2005) who found a genetic distance so low (0.02) that it could be based on a single erroneous score of one of the 30 tested SSR markers. Peluches is thought to be a mutant of Algerie (Gisbert et al. 2009b), and it is probably also the case that Marc is derived from Algerie or one of its mutants.
Loquat was introduced to Japan from China in ancient times, and spread to Europe and other continents, so Japanese loquat accessions are expected to have close relationship to Chinese accessions. The Spanish loquat accessions, ‘Peluches’ and ‘Marc’, clustered with Zhejiang and Jiangsu, which indicates that Spanish loquat was directly introduced from the Zhejiang or Jiangsu provinces. This is consistent with the results of Cai et al. (2003), using allozyme analysis, and Qiu and Zhang (1996). This is not surprising, as the Zhejiang province has been a major loquat producing area and famous for cultivars with good flavor quality since the Tang Dynasty (618-907 AD). The cluster analysis of the estimated genetic distance grouped the accessions according to their pedigrees and geographic origin. This result is similar to that found by Soriano et al. (2005).
Core set of Reference Accessions and Markers
A core set of highly divergent reference cultivars would be useful for evaluating the allelic diversity of new markers. For Malus sieversii, such a core collection was constructed based on 128 SSR alleles and 109 M. sieversii accessions. Accessions were selected by a preferred-allele sampling strategy combined with SM, Jaccard, and Nei & Li genetic distances, using a stepwise clustering approach (Zhang et al. 2009). Following the same procedure (with a threshold of 0.85 for SM coefficient), we obtained a core reference set of 20 loquat accessions based on our germplasm: ‘Jidanhong’, ‘Luoyangqing’, ‘Dayeyangdun’, ‘Jiajiao’, ‘Bahong’, ‘Hongmao’, ‘Dahongsha’, ‘Taicheng No.4’, ‘Jiefangzhong’, ‘Changhong’, ‘Tongpi’, ‘Gaoliangjiang’, ‘Ruantiaobaisha’, ‘Biqizhong’, ‘Dazhong’, ‘Bingtangzhong’, ‘Guanyu’, ‘Dawuxing’, ‘Longquan No.1’, and ‘Marc’. Considering the literature (Yang et al. 2007; Tang 1997), it seems useful to extend this set with the two additional wild accessions: Daduhe and Liye.
To identify the maximum number of accessions with the minimum number of markers, we drew up a set of five core reference markers (CH03a09, CH02c06, CH04g12, CH02d12, and CH05h05) able to distinguish all our accessions except the sports (Figs. 5 and 6).
Linkage Mapping of Loquat
The first loquat linkage map was constructed with ‘Algerie’ and ‘Zaozhong No. 6’, which contained only 75% of the tested SSR markers, 25% of the markers being homozygous in at least one of the parents, and only five of their 111 SSRs (MS06g03, AU223657, CH03d07, CH04g12, and CH05h05) suitable for the ‘Marc’ x ‘Zaozhong No. 6’ cross (Gisbert et al. 2009b). Heterozygosity and similarity analyses of the tested cultivars could support the generation of a mapping population, with ‘Daduhe’ (E. prinoides var. Daduheensis) and ‘Ruantiaobaisha’ two candidate parents for crossing.
References
Brant J (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83
Brini W, Mars M, Hormaza JI (2008) Genetic diversity in local Tunisian pears (Pyrus communis L.) studied with SSR markers. Sci Hortic 115:337–341
Cai LH, Huang HW, Deng XX, Zhang WC (2003) Allozyme analysis of genetic diversity, genetic structure and interspecific relationship in genus Eriobotrya. First International Symposium on Loquat. Zaragoza: CIHEAM-IAMZ. 31-40
Dirlewanger E, Cosson P, Tavaud M, Aranzana MJ, Poizat C, Zanetto A, Arús P, Laigret F (2002) Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.). Theor Appl Genet 105:127–138
Dirlewanger E, Graziano E, Joobeur T, Garriga-Calderé F, Cosson P, Howad W, Arús P (2004) Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proc Natl Acad Sci 101:9891–9896
Gianfranceschi L, Soglio V (2004) The European project HiDRAS: innovative multidisciplinary approaches to breeding high quality disease resistant apples. Acta Hort 663:327–330
Gisbert AD, López-Capuz I, Soriano JM, Llácer G, Romero C, Badenes ML (2008) Development of microsatellite markers from loquat, Eriobotrya japonica (Thunb.) Lindl. Mol Ecol Resour 9(3):803–805
Gisbert AD, Martínez-Calvo J, Llácer G, Badenes ML, Romero C (2009a) Development of two loquat [Eriobotrya japonica (Thunb.) Lindl.] linkage maps based on AFLPs and SSR markers from different Rosaceae species. Mol Breeding 23:523–538
Gisbert AD, Romero C, Martínez-Calvo JM, Leida C, Llácer G, Badenes ML (2009b) Genetic diversity evaluation of a loquat (Eriobotrya japonica (Thunb) Lindl) germplasm collection by SSRs and S-allele fragments. Euphytica 168:121–134
Guilford P, Prakash S, Zhu JM, Rikkerink E, Gardiner S, Bassett H, Forster R (1997) Microsatellites in Malus × domestica (apple): abundance, polymorphism and cultivar identification. Theor Appl Genet 94:249–254
Liebhard R, Gianfranceschi L, Koller B, Ryder CD, Tarchini R, Van de Weg WE, Gessler C (2002) Development and characterization of 140 new microsatellites in apple (Malus × domestica Borkh.). Mol Breeding 10:217–241
Lin SQ (2007) World loquat production and research with special reference to China. Acta Hort 750:37–43
Martínez-Calvo JM, Gisbert AD, Alamar MC, Hernandorena R, Romero C, Llácer G, Badenes ML (2008) Study of a germplasm collection of loquat (Eriobotrya japonica Lindl.) by multivariate analysis. Genet Resour Crop Evol 55:695–703
Patocchi A, Fernández-Fernández F, Evans K, Gobbin D, Rezzonico F, Boudichevskaia A, Dunemann F, Stankiewicz-Kosy M, Mathis F, Durel CE, Gianfranceschi L, Costa F, Toller C, Cova C, Mott D, Komjanc M, Barbaro E, Kodde L, Rikkerink E, Gessler C, Van de Weg WE (2008) Development and test of 21 multiplex PCRs composed of SSRs spanning 80% of the apple genome. Tree Gen Genomes 5:211–223
Qiao YC (2008) Construction of molecular genetic linkage map and genetic diversity of genus Eriobotrya. Dissertation, South-China Agr Univ
Qiu WL, Zhang HZ (1996) Fruit flora of China (longan and loquat). China Forestry Press, Beijing
Roche P, Alston FH, Maliepaard C, Evans KM, Vrielink R, Dunemann F, Markussen T, Tartarini S, Brown LM, Ryder C, King GJ (1997) RFLP and RAPD markers linked to the rosy leaf curling aphid resistance gene (Sd 1) in apple. Theor Appl Genet 94:528–533
Rohlf FJ (2000) NTSYS-pc: Numerical taxonomy and multivariate analysis system, version 2.1. Exeter Publications, New York
Sargent DJ, Fernandéz-Fernandéz F, Ruiz-Roja JJ, Sutherland BG, Passey A, Whitehouse AB, Simpson DW (2009) A genetic linkage map of the cultivated strawberry (Fragaria × ananassa) and its comparison to the diploid Fragaria reference map. Mol Breeding 24:293–303
Silfverberg-Dilworth E, Matasci CL, Van de Weg WE, Van Kaauwen MPW, Walser M, Kodde LP, Soglio V, Gianfranceschi L, Durel CE, Costa F, Yamamoto T, Koller B, Gessler C, Patocchi A (2006) Microsatellite markers spanning the apple (Malus × domestica Borkh) genome. Tree Genet Genomes 2:202–224
Soriano JM, Romero C, Vilanova S, Llácer G, Badenes ML (2005) Genetic diversity of loquat germplasm (Eriobotrya japonica (Thunb) Lindl) assessed by SSR markers. Genome 48:108–114
Tang B (1997) A study of the sibship among Eriobotrya japonica Lindl, E. prinoides Rehd. & Wils. var. daduheensis H.Z. Zhang and E. prinoides Rehd.&wils. J Chongqing Teachers College (Natural Science Edition). 14:18-25
Vilanova S, Badenes ML, Martínez-Calvo J, Llácer G (2001) Analysis of loquat germplasm (Eriobotrya japonica Lindl) by RAPD molecular markers. Euphytica 121:25–29
Vinatzer BA, Patocchi A, Tartarini S, Gianfranceschi L, Sansavini S, Gessler C (2004) Isolation of two microsatellite markers from BAC clones of the Vf scab resistance. Plant Breed 123:321–326
Watanabe M, Yamamoto T, Ohara M, Nishitani C, Yahata S (2008) Cultivar differentiation identified by SSR markers and the application for polyploidy loquat plants. J Jpn Soc Hortic Sci 77:388–394
Weber JL, May PE (1989) Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet 44:388–396
Wünsch A, Hormaza JI (2007) Characterization of variability and genetic similarity of European pear using microsatellite loci developed in apple. Sci Hortic 113:37–43
Yang XH, Liu CM, Lin SQ (2007) The Genetic relationship among common loquat, Daduhe loquat and Oakleaf loquat, according to RAPD and AFLP analysis. Subtropical Plant Sci 36:9–12
Zhang CY, Chen XS, Zhang YM, Yuan ZH, Liu ZC, Wang YL, LIN Q (2009) Method of constructing core collection for Malus sieversii in Xinjiang, China using molecular markers. Agric Sci China 8:276–284
Acknowledgments
This work was supported by the Special Research Fund for Public Welfare in Chinese Agriculture (contract no. 200903041) and the Ministry of Education of the PR China Visiting PhD Students, also by Southwest University Postgraduate Science and Technology Innovation Foundation Projects (contract no. 2006013). We gratefully acknowledge Lin S.Q. of South China Agricultural University and Wu J.C. of Putian College for kindly providing the Spanish and Fujian Province accessions, respectively. We thank Dr. Luud Gilissen for his critical comments.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix 1
XLS 106 kb
Rights and permissions
About this article
Cite this article
He, Q., Li, X.W., Liang, G.L. et al. Genetic Diversity and Identity of Chinese Loquat Cultivars/Accessions (Eriobotrya japonica) Using Apple SSR Markers. Plant Mol Biol Rep 29, 197–208 (2011). https://doi.org/10.1007/s11105-010-0218-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-010-0218-9