Abstract
To conduct RNAi interference of Lyc-β and Lyc-ε genes, two plant expression vectors were constructed by inserting the intron fragments of the gusA gene into the two target gene fragments, which were designed in anti-sense directions. After the Agrobacterium tumefaciens-mediated transformation, 13 transgenic tomato plants (seven and six for Lyc-β and Lyc-ε, respectively) were obtained, which was further validated by PCR. Real-time PCR revealed that the messenger RNA abundance of Lyc-β gene and Lyc-ε gene in transgenic tomato plants was significantly reduced to 8.95% and 13.16%, respectively, of the level of the wild-type plant. Subsequent high-performance liquid chromatography analysis found that transgenic tomato plant had significantly increased lycopene content, with the highest value of 13.8 μg/g leaf dry weight, which was about 4.2-fold that of wild-type plant. Moreover, Lyc-β and Lyc-ε interference gene effects were observed on downstream products as well. β-Carotene and lutein contents decreased in Lyc-β RNAi lines, ranging from 40.7 to 117.3 μg/g and 4.9 to 23.5 μg/g leaf dry weight, respectively. In Lyc-ε RNAi lines, β-carotene content increased, ranging from 195.8 to 290.2 μg/g, while lutein content decreased, ranging from 3.7 to 11.3 μg/g. For total carotenoids, Lyc-β RNAi lines resulted in 2.9-fold decrease, while Lyc-ε RNAi lines yielded 1.7-fold increase in contents when compared to wild-type control. This study demonstrated that RNAi gene technology is an effective method for enhancing lycopene content in plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
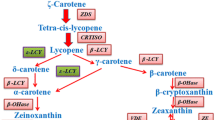
Carotenoids are groups of isoprenoid-derived molecules widely produced by photosynthetic organisms, bacteria, and fungi. The key step of carotenoid biosynthesis is a head-to-head coupling of two molecules of GGPP to yield phytoene (15-cis-phytoene, a colorless C40 compound) by phytoene synthase (PSY), with prephytoene diphosphate as an intermediate (Galpaz et al. 2006). Phytoene is converted to all-translycopene (a red pigment) by four desaturation reactions [mediated by phytoene desaturase (PDS) and ζ-carotene desaturase (ZDS)] and by an isomerization reaction (mediated by CRTISO), thus compound lycopene (Howitt and Pogson 2006). Finally, lycopene is metabolized by lycopene β-cyclases (LYC-β) and lycopene ε-cyclases (LYC-ε) producing α-carotene or β-carotene that are precursors of xanthophylls or abscisic acid (Baroli and Niyogi 2000; Howitt and Pogson 2006; Cunningham and Gantt 2001; Aluru et al. 2008) (Fig. 1).
Carotenoid biosynthetic pathway in plant. The pathway shows the primary steps found in many plant species. Solid arrows represent react steps. The following intermediate compounds and enzyme are represented. IPP isopentenyl pyrophosphate, DMAPP dimethylallyl pyrophosphate, GGPP geranylgeranyl pyrophosphate, ABA abscissic acid, PSY phytoene synthase, PDS phytoene desaturase, ZDS ζ-carotene desaturase, CRTISO carotenoid isomerase, LYC-β, lycopene β-cyclase, LYC-ε lycopene ε-cyclase, YHD carotene hydroxylases
Many genes of the carotenoid biosynthetic pathway have been cloned and manipulated in a large group of plants. In maize, overexpression of the bacterial genes crtB (for phytoene synthase) and crtI (function as PDS and ZDS in plants), significantly increased total carotenoids (34-fold) with the preferential accumulation of β-carotene in the maize endosperm (Aluru et al. 2008). In tomato, overexpression of the LYC-β and β-carotene hydroxylase remarkably increased β-carotene, β-cryptoxanthin, and zeaxanthin (Dharmapuri et al. (2002). Also in tomato, suppression of Def1 gene (an endogenous photo morphogenesis regulatory gene) increased notably both carotenoid and flavonoid contents (Davuluri et al. 2005), while suppression of the zeaxanthin epoxidase (ZEP) gene (which converts zeaxanthin to violaxanthin) resulted in the reduction of abscisic acid content in transgenic plants (Galpaz et al. 2008).
Tomato is a good source of carotenoids with especially high lycopene content. Therefore, regulating the biosynthetic enzyme in the carotenoid biosynthetic pathway through genetic engineering is a convenient and feasible manner to increase the content of lycopene in tomato fruits. The increase in lycopene content in tomato has been attempted by different strategies. Most of the studies carried so far mainly through overexpression methods to achieve an increase in plant carotenoids (Fraser et al. 2002; Diretto et al. 2007a). On the other hand, some studies started to manipulate the contents of carotenoids in plants using gene silencing techniques (Rosati et al. 2000; Wan et al. 2007). Carotenoid biosynthesis pathway is one of important biosynthetic pathway in plant. It is known that a certain number of secondary metabolites can accumulate in plants through the manipulation of related enzymes in carotenoid biosynthesis pathway. In the carotenoid biosynthesis pathway, lycopene can be cyclized by LYC-β and LYC-ε enzymes to produce β- and α-carotene. Current research mainly focused on the overexpression and silencing of the upstream genes, such as Psy and Pds. Ronen et al. (2000) reported that increasing the Lyc-β gene expression may effectively increaseβ-carotene content in the tomato fruit and the lycopene content drops at the same time. By overexpressing an additional bacterial crtB in tomato, Fraser et al. (2002) increased phytoene, lycopene, carotenoids, and lutein levels in transgenic plant. Overexpressing of tobacco Psy gene increased the content of lycopene (Busch et al. 2002), but the transgenic plants showed abnormal morphology and growth, and lethal phenotypes. This problem was solved in potato by overexpressing them under tissue-specific promoters (Diretto et al. 2007a). Recently, RNA interference technology has been used to increase carotenoids in plants. By silencing Lyc-β gene expression in tomato, Wan et al. (2007) found that the transgenic plants had a maximum 3.2-fold lycopene as that of wild-type plants as measured by spectrophotometer. In potato, silencing Lyc-ε gene increased β-carotene and total carotenoids 14- and 2.5-fold, respectively (Diretto et al. 2006, 2007b). Interestingly, silencing β-carotene hydrogenase (HYD) gene in the background of Lyc-ε RNAi plants a significant increased β-carotene and total carotenoids content (38- and 4.5-fold, respectively).
Presently, most studies undertaken to increase carotenoid content in plants focused on overexpression of the upstream genes, such as Psy and Pds, in the carotenoid biosynthetic pathway. In this study, two RNAi expression vectors were constructed to silence Lyc-β and Lyc-ε genes, respectively, and then separately transformed into tomato. The relative messenger RNA (mRNA) abundance of Lyc-β/ε genes and the contents of lycopene, β-carotene, lutein, and total carotenoids were analyzed in different transgenic plants.
Materials and Methods
Construction of Vectors
The total RNA was isolated from tomato leaves with UNIQ-10 Column RNA Isolation kit (Sangon, SK1322). Reverse transcript assay was performed using the MMLV Reverse Transcript Kit (Sangon, K1622). PCR primers used for the amplification of Lyc-β (X-1S and X-1AS), Lyc-ε (Y-1S and Y-1AS) genes and gusA (GusA-1S, GusA-1AS, GusA-2S and GusA-2AS) gene intron fragments were listed in Table 1. The Pfu PCR assays, for the amplification of the Lyc-β and Lyc-ε gene, were carried out in 50 μL reactions containing a Super High PCR master [3.0 mM MgCl2, 0.2 mM dNTP, and 2.5 U Super Pfu DNA polymerase (Sangon SK2084)], 0.4 μM of each primer, and 2.5 μL reverse transcript products. The PCRs were carried out in a MyCycle Thermal cycler System (Bio-Rad). The PCRs condition were as follows: one cycle denaturalization step at 94°C for 3 min, 30 cycles of denaturalization step at 94°C for 45 s, primers annealing at 59°C (X-1S and X-1AS)/52°C (Y-1S and Y-1AS) for 45 s and extension at 72°C for 30 s, with the final extension step of 72°C for 7 min. Amplification products were run in a 1% (w/v) TAE agarose gel and visualized by staining with 0.5 μg ethidium bromide per milliliter. The amplification products were cloned into pUCm-T vector (Sangon), and its identification was verified by DNA sequencing (Sangon Company). The plasmids were referred to as pPUCM-X-1 and pPUCM-Y-1 separately. For the amplification of the fragment of gusA gene intron, the PCR primers GusA-1S/GusA-1AS and GusA-2S/GusA-2AS were used with the annealing temperature at 57°C and 50°C separately. The PCR products were amplified using the same PCR conditions. The plasmids of pGusA-1 and pGusA-2 were then obtained after the cloning and sequencing verification.
The pPUCM-X-1 was digested with XhoI and BamHI, and then the small XhoI–BamHI fragments were inserted into pPUCM-X-1 at the SalI–BamHI sites. Following the digestion with BamHI and its dephospharylation, this constructor was inserted into a BamHI–BamHI intron fragment of pGusA-1 to generate pPUCM-X1-RNAi. To that end, a series containing a gusA gene intron flanked by Lyc-β fragments were obtained. Final assembly of plant expressive vector was performed as follows. The pPUCM-X1-RNAi was digested with HindIII following the blunting with Klenow enzyme, and then it was digested with SacI. The small fragments were inserted into the p2300-121 expressive vector (Ma et al. 2009) at SacI–SmaI sites. The resulting plasmid containing the expression cassettes of the P35s-Lycβ-Tnos in series was designated as p2300-121-X1-RNAi (Fig. 2a). Following the comprehensive digestion analyze, the vector p2300-121-X1-RNAi was verified by sequencing and used for the interference of Lyc-β gene. For construction of the Lyc-ε gene interference vector, pPUCM-Y-1 and pGusA-2 were used to construct p2300-121-Y1-RNAi (Fig. 2b) using the same strategy as described above.
The diagram of Lyc-β (a) and Lyc-ε (b) genes’ RNAi vectors. P35s-NPTII-Tnos represent the antibiotic resistance gene used as selectable marker. The Lycβ/ε S-gusA1-Lyc β/ε AS represents the Lyc-β/ε genes RNAi sequences; it is promoted by P35s and terminated by Tnos. LB and RB represents left and right borders respectively
Genetic Transformation
Agrobacterium tumefaciens-mediated transformation of tomato (Solanum lycopersicum Mill. cv. Leeger 87-5) was performed following the protocol described by Zhang et al. (2006). Kanamycin-resistant plantlets were grown under controlled greenhouse conditions at 25°C and 50% relative humidity. Plants were grown under 16/8 h (light/dark) photoperiod with supplementary light provided by 400 W fluorescent tubes (200 µmol m−2 s−1).
Screening of Transgenic Plants
The genomic DNA was extracted from the transgenic and wild-type tomato leaves with UNIQ-10 Column DNA Isolation kit (Sangon, SK1204). The fragments of P35s promoter, used for screening of transgenic plants, was amplified with the primers 5′-AGGCTTACGCAGCAGGTC-3′ and 5′-GCAGAGGCATCTTCAACG-3′. The PCR amplifications were carried out in 20 μL reactions containing a PCR buffer (500 mM KCL, 100 mM Tris/HCL, pH 9), 1.5 mM MgCl2, 0.4 mM dNTP, 0.2 mM each of primer, 1 U of Taq DNA polymerase, and 100 ng DNA template. The condition of PCR reactions were as follows: one cycle denaturalization step at 94°C for 5 min, 30 cycles of denaturalization step at 94°C for 45 s, primers annealing at 55°C for 45 s, and extension at 72°C for 1 min, with the final extension step of 72°C for 7 min.
Real-Time PCR Analysis
Total RNA was extracted from transgenic and wild-type tomato leaves, and its concentration and purity were calculated by measuring A260 and A280 value with Nano Drop Meter (Gene Cop). Two micrograms of total RNA was reverse transcribed into complementary DNAs (cDNAs). Real-time PCRs were performed in an optical 96-well plate with the Mx3000sp QPCR system (Stratagene). The primer X-1S/X-1AS and Y-1S/Y-1AS were used to determine the abundance of Lyc-β and Lyc-ε genes. The ubiquitin (ubi) gene, amplified by primer UBI-s/UBI-as, was used as the reference to normalize data. Twenty-microlitre reactions contained 10 μL SYBR® Green Real time PCR Master Mix, 0.4 μL of each primer (20 mM), and 1 μL previously synthesized cDNA. The following standard thermal profile was used for PCRs: one cycle denaturalization step at 94°C for 30 s, 40 cycles of denaturalization step at 94°C for 5 s, primers annealing at 59°C (X-1S and X-1AS)/52°C (Y-1S and Y-1AS)/55°C (UBI-S and UBI-AS) for 5 s and extension at 72 for 20 s. A dissociation step was performed after amplification to confirm the presence of a single amplification. Three technical replicates were carried out at the same time. The standard curve was made by the 10-fold dilutions of pPUCM-X-1, pPUCM-Y-1, and pUBI vectors ranging from 10 to 106 copy number, respectively. The standard curves were as follows: pPUCM-X-1, Y = −3.464 × log(X) + 47.70 (Eff. = 94.4%; RSq, 0.996); pPUCM-Y-1, Y = −2.273 × log(X) + 42.38 (eff. = 175.4%, RSq, 0.994); pUBI, Y = −3.348 × log(X) + 45.28 (Eff. = 98.9%; RSq, 0.999). The gene copies were calculated through the standard curves, and then converted into gene concentration by the following formulae: (gene concentration) = (gene copies) × (plasmid length) × 660 × 6.02 × 1023. At last, the mRNA relative abundance of target gene equaled the ratio of target gene concentration and reference gene concentration (Schmidt and Parrott 2001).
Carotenoids Extraction and HPLC Analysis
Carotenoids were extracted from 0.5 g leaves of transgenic and wild-type tomato separately. A solvent extraction system assisted by ultrasonication was run at room temperature. For the extraction of lycopene, the methanol was firstly added into samples and filtered until all the color was removed from the plant materials. The toluene of 5 mL was used as the solvent, and the extraction was performed for 30 min at 20°C under a 200-W ultrasonic power (Kunshan). The insoluble materials were removed by centrifugation (4,000×g for 4 min). The final extracts were filtered through nylon filters, 0.45 μm pore size, and stored at −70°C. All of the above steps were done in dark. Chromatography was carried out with an Agilent liquid chromatography system (Agilent 1100). Lycopene in tomato leaves was determined by reversed phase high-performance liquid chromatography (HPLC) using an Agilent C18 column (150 × 4.6 mm and 5 μm particle diameter) at 20°C, at with methanol/acetonitrile/dichloromethane (7:7:2 v/v) at 1 ml/min. The eluate was monitored at 472 nm. Peak was identified by it retention time, and absorption spectra was compared to the known standards (Sigma-Aldrich, L9879). Amounts were quantified using the standards curve in concentrations of 20, 10, 2, 1, and 0.2 μg/mL. Three replicates were preformed to decrease the experiment error.
For the extraction of β-carotene and lutein, the petroleum was used as the solvent. The ultrasonic power was exchanged at 140 W, and the acetonitrile/dichloromethane/ isopropanol (8:1:1 v/v) was used as mobile phase in differently. The peak assignment of β-carotene and lutein, monitored at 450 nm, was based on the retention time of their commercial standards (Sigma-Aldrich, C9750, X6250).
Results
RNAi Vectors
According to the sequences of LYC-β (X86452) and LYC-ε (Y14387) genes, two segments of 302 bp were amplified using PCR from tomato leaf total RNA. At the same time, the partial fragments (168 bp) and the full length fragments (238 bp) of the gusA first intron were obtained from the plasmid pCAMBIA-2301 by PCR. All the cloned fragments were subcloned and sequenced, with the 100% homology to the corresponding gene fragments.
Using pCAMBIA-2300, two RNAi expressive vectors were constructed to silence LYC-β and LYC-ε genes separately. The recombine vector constructs is shown in Fig. 2. After the confirmation by sequencing and enzyme digestion analysis, the resultant plant expressive vectors were then used for the transformation into tomato plants.
PCR Detection on Transgenic Tomato
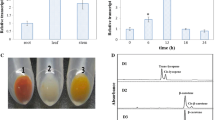
The transformed plants were selected firstly based on their resistances to the kanamycin. Total 15 resistance plants were obtained to perform PCRs detection by the amplification of 35s promoter. The predicted 35s fragments of 550 bp were amplified in plasmid pCAMBIA-2300 (positive control) and transgenic tomatoes but not in the wild-type tomatoes (negative control). Finally, 13 transgenic plants were obtained (Fig. 3). These transgenict tomatoes were planted in the glasshouse; they did not show any visible abnormal phenotype, and all exhibited the same growth as wild-type plants, indicating that the RNAi of the Lyc-β or Lyc-ε gene does not affect the plant growth.
Photographs of ethidium bromide-stained 1% (w/v) gels carrying PCR products obtained from 35s promoter. The lane labeled M contain the 1 kb DNA ladder (GeneRulerTM DNA Ladder Mix) as size marker; the lane labeled 1 shows the result from plasmid pCAMBIA-2300; the lane labeled 2 shows the result from wild-type tomato plant; the lanes labeled 3–10 show the results from Lyc-β gene RNAi plants; the lanes labeled 11–17 show the results from Lyc-ε gene RNAi plants
Lyc-β and Lyc-ε Genes mRNA Abundance
To examine the effect of RNAi on the expression of the Lyc-β and Lyc-ε in transgenic plants, real-time PCR was performed on six transgenic plants for each gene. The target gene and the reference gene were detected at the same time. The value of ∆Ct [(target gene)−(reference gene)] of the transgenic lines were higher than that of the wild type (Fig. 4c). The dissolution curves showed that the reaction was independent, and all reactions had only one PCR production (Fig. 4d). The endogenous Lyc-β or Lyc-ε genes relative abundance was analyzed separately, while the ubi gene was used as reference gene to eliminate the error of reverse transcription reactions. As shown in Fig. 5, reverse transcription PCRs revealed that mRNA abundance of Lyc-β and Lyc-ε were significantly reduced in all tested 12 RNAi plants. Transgenic plant line X1-2 exhibited the lowest mRNA abundance of Lyc-β gene, which was only 1.44% of that of the wild-type plants. Silencing Lyc-β gene did not change the mRNA abundance of Lyc-ε gene. Transgenic plant line Y1-3 exhibited the lowest mRNA abundance of Lyc-ε gene, which was 2.98% of that of the wild-type plants. Interestingly, silencing Lyc-ε gene increased slightly the mRNA abundance of Lyc-β gene, with the line Y1-3 rising to 116% of that of the wild-type plants. All in all, the average of the reduction of the mRNA abundance of Lyc-β gene by the transformation of p2300-121-X1-RNAi was 91.05%, while the average of the reduction of the mRNA abundance of Lyc-ε gene by the transformation of p2300-121-Y1-RNAi was 86.84%. Compared with the p2300-121-Y1-RNAi, the p2300-121-X1-RNAi vector showed a better silencing effect.
The real time PCR results. a Photographs of ethidium bromide-stained 1% (w/v) gels carrying PCR products obtained from the target gene real time PCR products; b photographs of ethidium bromide-stained 1% (w/v) gels carrying PCR products obtained from the reference gene real time PCR products; c the real-time PCR amplification curves. The X-axes showed cycle number, and the Y-axes show the fluorescence in the amplification curves. The reference gene abundance is higher than the target gene, so the Ct value is lower than the target gene; d the real-time PCR dissolution curves. The X-axes showed temperature, and the Y-axes shows the fluorescence in the dissolution curves. Both of the reference gene and target gene had a single peak in dissolution curves. It is means that the product of the amplification was unique; no other products were amplified
Lyc-β/ε genes mRNA abundance analysis in transgenic tomatoes. The lane labeled CK shows the Lyc-β/ε genes relative abundance in wild-type plant; the lanes labeled X1-1 to X1-6 showed the Lyc-β/ε genes relative abundance in p2300-121-X1-RNAi vecort transgenic tomatoes; the lanes labeled Y1-1 to Y1-6 showed the Lyc-β/ε gene mRNA relative abundance in p2300-121-Y1-RNAi vector transgenic tomatoes
Lycopene, β-carotene, Lutein, and Total Carotenoids Content
The lycopene, β-carotene, and lutein of tomato leaves from individual transgenic lines were analyzed through HPLC. Lycopene contents in the all six Lyc-β RNAi transgenic plants were increased significantly, with the transgenic line X1-2 showing the highest (13.8 μg/g). The average content of lycopene of Lyc-β RNAi transgenic plants was 7.1 μg/g, which was 2.2-fold of that of the wild-type plants (3.2 μg/g). The silencing of Lyc-β gene also impacted the production of downstream substances in carotenoids biosynthesis pathway. The contents of β-carotene and lutein in transgenic plants were both decreased, ranging from 40.7 to 117.3 μg/g and from 4.9 to 23.5 μg/g, respectively. The transgenic line X1-2 had the lowest β-carotene content of 40.7 μg/g (29% of the wild-type plant), while the transgenic line X1-1 had the lowest lutein content of 4.9 μg/g (11.8% of the wild-type plant). Accompanying with the reduction of β-carotene content, the total carotenoids contents of Lyc-β RNAi transgenic plants decreased 29.7–70.3%; The transgenic line X1-2 has the lowest total carotenoids content of 64.4 μg/g. This means that the total carotenoids content decreased 2.9-fold compared with the 185.1 μg/g from the control plant (Fig. 6a).
The alterations in pigment contents between the transgenic plants and the wild-type plants: a HPLC analyses of the lycopene, β-carotene, and lutein contents in the p2300-121-X1-RNAi vector transgenic plants; b HPLC analyses of the lycopene, β-carotene, and lutein contents in the p2300-121-Y1-RNAi vector transgenic plant. The CK showed the content of three pigments and total carotenoids in wild-type tomato leaves. X1-1 to X1-6 and Y1-1 to Y1-6 showed the content of three pigments and total carotenoids in different individual transgenic tomato plant lines. The lycopene and lutein contents were measured with the left y-axes; the β-carotene and the total carotenoids contents were measured with the right y-axes. DW, Dry weight
The lycopene contents were also increased in Lyc-ε transgenic plants. The transgenic line Y1-3 had the highest lycopene content of 8.5 μg/g (2.66-fold of that of the wild-type plants). The average content of lycopene of Lyc-ε transgenic plants was 6.2 μg/g (1.94-fold of that of the wild-type plants). It is noteworthy that the silencing of Lyc-ε gene exerted different impacts on the downstream substances. The content of lutein decreased 72.3–90.9%. The transgenic line Y1-3 has the lowest lutein content of 3.7 μg/g (9.1% of that of the wild-type plants). Conversely, the β-carotene and total carotenoids increased 38.9–105.8% and 13.2%–65.0%, respectively. The transgenic line Y1-6 had the highest β-carotene contents of 290.2 μg/g (2.06-fold of that of the wild-type plants) and the highest total carotenoids contents of 305.4 μg/g (1.7-fold of that of the wild-type plants; Fig. 6b).
Discussion
RNAi technology has become a powerful tool for the study of the functions of individual genes in a range of organisms, including plants, such as Arabidopsis, soybean, rice, maize, tomato, potato, and so on(Padmanaban et al. 2004; Subramanian et al. 2005; Miki et al. 2005; McGinnis et al. 2007; Liu et al. 2004; Kim et al. 2008). It becomes a common method for gene silence. In the siRNAi pathway, any of the 21–23 nucleotides’ change may disturb the work of the siRNA-targeted mRNA sequence. For vector-mediated RNAi, it has been shown that the RNAi fragment should include complete sequence of intron, and the intron boundary sequences play an important role in the formation of hairpin RNA structures (Li et al. 2006). The RNAi fragments designed in this experiment was 302 bp, which obtained from tomato through Pfu PCR with a 100% homological rates compared with the target mRNA. Two intron sequences used in the RNAi vectors for silencing Lyc-β and Lyc-ε gene were different; the one used for Lyc-ε RNAi was whole length of the intron sequence, and the one used for Lyc-β RNAi did not include its boundary sequence. The results showed that both of them had obvious silencing effects on their corresponding target mRNAs. Overexpressing the RNAi vector with the partial intron sequence resulted in an average of 91.05% deregulation of the expression of the target mRNA, while overexpressing the RNAi vector with the full intron sequence caused an average of 86.84% downregulation of the corresponding target genes. Therefore, the results suggested that the intron boundary sequence may be not the necessary component of the RNAi silence machinery.
Earlier studies revealed that overexpression of genes in the carotenoid biosynthetic pathway enhanced carotenoid content in plants. Moreover, increased downstream components were also observed, but only to limited levels. In this study, using reverse genetics to control Lyc-β or Lyc-ε gene expression enhanced lycopene content in tomato. Earlier, conserved sequences of interfering segments of Lyc-β and Lyc-ε genes were used in tobacco first, and subsequently, these were used in tomato plants. In tomato, both Lyc-β gene (X86452) and the chromoplast Lyc-β gene (AF254793) regulated lycopene degradation. The chromoplasts Lyc-β gene was expressed during flower and fruit color development, but the Lyc-β gene was expressed at all stages of plant growth. When the tomato Lyc-β gene and chromoplast Lyc-β gene sequence were aligned, before amplification of interfering segments, it was revealed that these two genes had 57% homology and without a 21 nucleotide sequence identity. Based on the RNAi protocol, the hairpine structure would be cut by Dicer 4, yielding 21–22nt double strands of small RNA in the cytoplasm. These would bind to Ago1, resulting in degradation of target mRNA in the cytoplasm. Therefore, the Lyc-β gene silencing would not have an effect on the function of the chromoplast Lyc-β gene. In this study, the Lyc-β and Lyc-ε genes were separately blocked through RNA interference and resulting in high accumulation of lycopene. Both vectors for Lyc-β and Lyc-ε genes used in this study had high degenerating effects on mRNA of target genes, average of 8.95% and 13.16% compared to those of control plants, respectively. Moreover, lycopene content in these transgenic tomato plants, average of 7.07 and 6.2 μg/g, respectively, did not increase significantly. This might be due to the presence of two branches downstream of the lycopene metabolic pathway. While one branch was blocked in this study, the other proceeded with production of downstream substances. Another likely reason might be attributed to the fact that the chromoplast Lyc-β and chromoplast Lyc-ε genes (that has not yet been reported) could regulate lycopene biosynthesis in tomato, and the Lyc-β/ε gene interference did not influence the chromoplast Lyc-β/ε gene, thus resulting in no significant change in lycopene content.
In the carotenoid biosynthesis pathway, the lycopene can be cyclized by lycopene β-cyclase and lycopene ε-cyclase. The lycopene β-cyclase exists in both of the two branches to yield the downstream substance β-carotene and lutein, but the lycopene ε-cyclase only exists in the lutein branch. Thus, when the Lyc-β gene was silenced, both of the two branch downstream substance was decreased, which is in agreement with the changes in the expression of the corresponding genes (Figs. 5 and 6a). In Lyc-ε RNAi plants, the changes in the mRNA abundance were consistent with the changes in the downstream substance contents. The changes in total carotenoids depend mainly on the changes in β-carotene in transgenic plants. Silencing Lyc-ε gene (blocking one way of lycopene catabolism) induced the accumulation of β-carotene, which contributed to the increase in the total carotenoids. On the other hand, silenced Lyc-β gene (blocking both ways of lycopene catalism) induced the increase of lycopene, but decreased total carotenoids. Our results supported the efficiency of RNAi in silencing the endogenous gene and confirmed that the silencing either Lyc-β or Lyc-ε gene impacted not only the lycopene content but also the downstream substances. Further study will focus on the transform of both Lyc-β RNAi and Lyc-ε RNAi into tomato plants and will examine their effects on the regulation of carotenoids.
Conclusion
The manipulation of tomato carotenoids can be done through the silencing of either Lyc-β or Lyc-ε gene using vector mediated RNAi technology. The strongest silencing of the target genes was obtained by use intron fragment without the boundary sequence, and the efficiency of the downregulation of the corresponding target genes by RNAi in transgenic plants was significant, which caused obvious changes in the contents of β-carotene , lycopene, total carotenoids, and downstream lutein. All those metabolic changes are consistent with the changes of corresponding genes resultings from the transformation of the RNAi vectors. Therefore, RNAi is an effective method for enhancing lycopene content in tomato plants.
References
Aluru M, Xu Y, Guo R, Wang Z, Li SS, White W, Wang K, Rodermel S (2008) Generation of transgenic maize with enhanced provitamin A content. J Exp Bot 59:3551–3562
Baroli I, Niyogi KK (2000) Molecular genetics of xanthophyll-dependent photoprotection in green algae and plants. Philos Trans R Soc Lond B Biol Sci 355:1385–1394
Busch M, Seuter A, Hain R (2002) Functional analysis of the early steps of carotenoid biosynthesis in tobacco. Plant Physiol 128:439–453
Cunningham FX Jr, Gantt E (2001) One ring or two? Determination of ring number in carotenoids by lycopene e-cyclases. PNAS 98:2905–2910
Davuluri GR, Van Tuinen A, Fraser PD, Manfredonia A, Newman R, Burgess D, Brummell DA, King SR, Palys J, Uhlig J, Bramley PM, Pennings HM, Bowler C (2005) Fruit-specific RNAi-mediate suppression of DETl enhances carotenoid andflavonoid content in tomatoes. Nat Biotechnol 23:825–826
Dharmapuri S, Rosati C, Pallara P, Aquilani R, Bouvier F, Camara B, Giuliano G (2002) Metabolic engineering of xanthophyll content in tomato fruits. FEBS Lett 519:30–34
Diretto G, Tavazza R, Welsch R, Pizzichini D, Mourques F, Papacchioil V, Bever P, Giuliano G (2006) Metabolic engineering of potato tuber carotenoids through tuber-specific silencing of lycopene epsilon cyclase. BME Plant Biol 6:13
Diretto G, Al-Babill S, Tavazza R, Papacchioli V, Beyer P, Giuliano G (2007a) Metabolic engineering of potato carotenoid content through tuber-specific overexpression of a bacterial mini-pathway. PLoS ONE 2:e350
Diretto G, Welsch R, Tavazza R, Mourques F, Pizzichini D, Bever P, Giuliano G (2007b) Silencing of beta-carotene hydroxylase increases total carotenoid and beta-carotene levels in potato tubers. BME Plant Biol 7:11
Fraser PD, Romer S, Shipton CA, Mills PB, Kiano JW, Misawa N, Drake RG, Schuch W, Bramley PM (2002) Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. PNAS 99:1092–1097
Galpaz N, Ronen G, Khalfa Z, Zamir D, Hirschberg J (2006) A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell 18:1947–1960
Galpaz N, Wang Q, Menda N, Zamir D, Hirschberg J (2008) Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J 53:717–730
Howitt CA, Pogson BJ (2006) Carotenoid accumulation and functions in seeds and non-green tissues. Plant Cell Environ 29:435–445
Kim YS, Lee YH, Kim HS, Kim MS, Hahn KW, Ko JH, Joung H, Jeon JH (2008) Development of patatin knockdown potato tubers using RNA interference (RNAi) technology, for the production of human-therapeutic glycoproteins. BMC Biotechnol 8:36
Li XP, Deng N, Ma YY, Li PL, Wang Y, Zheng R, Wang NN (2006) Construction of RNAi binary vector of soybean receptor-like kinase gene(rlpk2) and its soybean transformation. Fen Zi Xi Bao Sheng Wu Xue Bao 39:1–8
Liu YS, Roof S, Ye ZB, Barry C, Van Tuinen A, Vrebalov J, Bowler C, Giovannoni J (2004) Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. PNAS 101:9897–9902
Ma BG, Duan XY, Niu JX, Ma C, Hao QN, Zhang LX, Zhang LX (2009) Expression of stilbene sythase gene in transgenic tomato using salicylic acid-inducible Cre/loxP recombination system with self-excision of selectable marker. Biotechnol Lett 31:163–169
McGinnis K, Murphy N, Carlson AR, Akula A, Akula C, Basinger H, Carlson M, Hermanson P, Kovacevic N, McGill MA, Seshadri V, Yoyokie J, Cone K, Kaeppler HF, Kaeppler SM, Springer NM (2007) Assessing the efficiency of RNA interference for maize functional genomics. Plant Physiol 143:1441–1451
Miki D, Itoh R, Shimamoto K (2005) RNA silencing of single and multiple members in a gene family of rice. Plant Physiol 138:1903–1913
Padmanaban S, Lin X, Perera I, Kawamura Y, Sze H (2004) Differential expression of vacuolar H+-ATPase subunit c genes in tissues active in membrane trafficking and their roles in plant growth as revealed by RNAi. Plant Physiol 134:1514–1526
Ronen G, Carmel-Goren L, Zamir D, Hirschberq J (2000) An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of beta and old-gold color mutations in tomato. PNAS 97:11102–11107
Rosati C, Aquilani R, Dharmapuri S, Pallara P, Marusic C, Tavazza R, Bouvier F, Camara B, Giuliano G (2000) Metabolic engineering of beta-carotene and lycopene content in tomato fruit. Plant J 24:413–419
Schmidt MA, Parrott WA (2001) Quantitative detection of transgenes in soybean [Glycine max (L.) Merrill] and peanut (Arachis hypogaea L.) by real-time polymerase chain reaction. Plant Cell Rep 20:422–428
Subramanian S, Graham MY, Yu O, Graham TL (2005) RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and enhanced susceptibility to Phytophthora sojae. Plant Physiol 137:1345–1353
Wan Q, Zhang XG, Song M (2007) Fruit-specific RNAi-mediated restraining expression of Lcy gene to enhance lycopene content in tomatoes. Chin J Biotech 23:429–433
Zhang YY, Li HX, Ouyang B, Lu YE, Ye ZB (2006) Chemical-induced autoexcision of selectable markers in elite tomato plants transformed with a gene conferring resistance to lepidopteran insects. Biotechnol Lett 28:1247–1253
Acknowledgements
This research was supported by National Natural Science Foundation of China (30460081) and Scientific Research Fund of Xinjiang BingTuan, China (ZD2007JC06).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ma, C., Ma, B., He, J. et al. Regulation of Carotenoid Content in Tomato by Silencing of Lycopene β/ε-Cyclase Genes. Plant Mol Biol Rep 29, 117–124 (2011). https://doi.org/10.1007/s11105-010-0211-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-010-0211-3