Abstract
Cytokinin oxidase/dehydrogenase plays an important role in regulating plant growth and development. A novel gene of TaCKX3 was cloned from wheat by specific primers designed according to the Triticeae Full-Length CDS Database and was proved to be located on chromosome 7B by analyzing the nulli-tetrasomic lines of “Chinese Spring.” The genomic coding sequence was interrupted by three introns, which were 103, 421, and 458 bp, successively. The complementary DNA sequence of TaCKX3 is 60% identical to that of TaCKX1, and the homology of their deduced proteins is even low. The putative TaCKX3 protein shows highly homology with ZmCKX10 but not with other known cytokinin oxidase/dehydrogenase. However, it contains conserved motifs as other cytokinin oxidase/dehydrogenase, such as flavin adenosine dinucleotide binding domain and cytokinin binding domain. Consistent with ZmCKX10 and AtCKX7, nor the putative TaCKX3 has signal peptide at N terminus, which means that TaCKX3 functions in cytoplasm. Quantitive polymerase chain reaction analysis indicated that the expression of TaCKX3 gene is significantly up-regulated in germinating embryos treated by 6-BA and slightly up-regulated by NaCl or PEG-6000.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cytokinin oxidase/dehydrogenase (CKX; EC.1.5.99.12) enzyme is a flavoenzyme, containing flavin adenosine dinucleotide (FAD) as a cofactor. It is thought to be the only known plant enzyme that irreversibly degrade cytokinins (CKs) into inactive products by cleaving the N 6-unsaturated side chain of the active CKs, such as isopentenyladenine, zeatin, and their ribosides (Hare and Van Staden 1994; Jones and Schreiber 1997; Galuszka et al. 2001). As important plant hormones, CKs are considered to play a key role in plant growth and development, including cell division and differentiation, apical dominance, shoot and root balance, transduction of nutritional signals, fruit development, leaf senescence, and other processes (Mok and Mok 2001; Ashikari et al. 2005; Hitoshi 2006). The CKXs, which commonly exist in plants, are involved in the above processes by controlling the levels of CKs.
The CKX activity was first discovered in crude tobacco culture by Pačes et al. (1971) and was then extensively reported in various tissues and species (Galuszka et al. 2004). Since the first CKX gene was cloned from Zea mays by two independent teams simultaneously (Houba-Hérin et al. 1999; Morris et al. 1999), the amount of CKX genes have been identified from maize, Arabidopsis thaliana, rice, Dendrobium, barley, and wheat (Morris et al. 1999; Bilyeu et al. 2001; Werner et al. 2001; Schmülling et al. 2003; Yang et al. 2003a; Galuszka et al. 2004; Feng et al. 2008; Šmehilová et al. 2009). Phylogenetic analysis indicated that although the similarity of the entire protein sequences from different plant species is not very high, these CKX proteins contain two obviously conserved domains, one is for FAD binding located at N terminus and the other is for CK binding located at C terminus. These two conserved motifs are considered to be related with CKX catalytic activity.
The relationship between CKXs and CKs has been extensively investigated in order to figure out their functions in plant development. CKX activity in cultured tobacco cells was increased transitorily by applying external CKs (Terrine and Laloue 1980; Chatfield and Armstrong 1986; Kaminek and Armstrong 1990). In maize kernels, CKX activity is much higher after pollination, coincident with the increased CKs levels (Whitty and Hall 1974; Dietrich et al. 1995). These results showed that CKX activity may be induced by the levels of CKs. CKX overexpression in tobacco led to stunted shoots and enlarged root meristems with more branched roots (Werner et al. 2001). ZmCKX1-transformed maize plants displayed male-sterile phenotype with the accumulation of ZmCKX1 in reproductive tissues (Huang et al. 2003). Transgenic A. thaliana with DSCKX1 gene exhibited CK-deficient developmental processes due to decreased CKs levels (Yang et al. 2003b). Reduced expression of OsCKX2 caused CK accumulation in inflorescence meristems and increased the number of reproductive organs, resulting in enhanced grain yield (Ashikari et al. 2005).
Based on previously work, as an important gene family, several CKX genes have been identified from various plant species and proved to be functionally critical for the plant growth and development by decreasing CK levels. Hexaploid wheat also contain several CKX genes in its large genome, but there is little systematic identification of full-length CKX genes from this important economic species available in literature (Galuszka, et al. 2004; Feng et al. 2008). In this work, we identified a novel CKX gene, TaCKX3 from wheat, and initially analyzed its characteristics.
Materials and Methods
Plant Materials and Growth Conditions
In this study, wheat (Triticum aestivum L. cv. Yanyou 361) was used to clone TaCKX3 and nulli-tetrasomic (NT) lines of “Chinese Spring” (CS) was used to locate the cloned gene onto the chromosome.
Stress Treatments
To investigate the expression of TaCKX3 gene in germinating embryos under stress condition, the seeds of Y361 imbibed for 15 h were transferred to Petri dishes fitted with filter papers that were soaked in 250 mM NaCl, 10 μM 6-BA, and 20% PEG-6000 and then cultured for 12 h at growth chamber. The embryos were separated from endosperms after the processing. Distilled water treatment was used as the mock control.
DNA and RNA Isolation
Genomic DNA was extracted from wheat according to cetyltrimethylammonium bromide method described by Saghai-Maroof et al. (1984). Total RNA was isolated from treated germinating embryos by RNAprep pure Plant kit (Tiangen, Beijing, China) according to the manufacturer’s protocol and quantified by GeneQuant (Amersham, Sweden). Total RNA was separated on 1.5% agarose gel to assess the quality of the isolated RNA.
Cloning and Sequencing of the TaCKX3 Genomic DNA
Specific primer sets—forward, 5-ACCGCAGTGGGAAGAGAAGC-3, and reverse, 5-CAGCTATGATCGGTCGGTTCG-3—were used for gene amplification that were designed according to the sequence information deposited in the Triticeae Full-Length CDS Database (Mochida et al. 2009), which includes putative full-length complementary DNAs (cDNAs) for barley and wheat. The polymerase chain reaction (PCR) was performed in a 50-μL mixture containing approximately 200 ng of genomic DNA, 1× GC reaction buffer, 300 μM dNTPs, 250 ng of each primer, and 0.5 U LA GC Taq DNA polymerase (TaKaRa, Dalian, China). The thermal cycle profile was 95°C for 3 min, 36 cycles of 94°C for 40 s, and 68°C for 3.5 min, with a final extension of 68°C for 10 min. The PCR products were separated by 1% agarose gel electrophoresis. DNA from the target band was excised from the gels and purified, then ligated into the pMD 18-T vector (TaKaRa, Dalian, China) and sequenced by Sangon Company (Shanghai, China).
Sequence Analysis of TaCKX3
Similarity searches for nucleotides and deduced amino acids were conducted using the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST) network server (http://www.ncbi.nlm.gov/BLAST). The exon/intron constitution of TaCKX3 was determined by comparison between corresponding cDNA and genomic DNA sequence. The alignment of the putative amino acid sequences of TaCKX3 with other known CKXs from wheat, maize, rice, and A. thaliana was carried out using DNAMAN software. The phylogenetic tree at amino acid level was also drawn using DNAMAN based on the sequence alignment. Signal peptides were predicted with the program SignalP (http://www.cbs.dtu.dk/services/SignalP/). Its isoelectric point (pI) and molecular weight (MW) were analyzed by pI/MW program (available on the ExPaSy Web site at http://www.expasy.org/).
Chromosomal Localization of TaCKX3
To determine the chromosomal localization of the novel gene, specific primer sets—forward, 5-GAGGATGCGGGAGATGATGC-3, and reverse, 5-TATAAGTGATCCCGGTCAGG-3—were designed according to the sequence of the obtained TaCKX3 gene to amplify wheat genomic DNA. A series of wheat NT lines of CS were used in this work. The PCR products were separated with 6% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels, which stained by silver.
Quantitative Real-Time PCR Analysis
The first-strand cDNA was synthesized from 2 μg total RNA using the RevertAid First Strand cDNA Synthesis Kit (Fermentas, Canada) and oligo (dT) primer. Real-time PCR experiments were performed using SYBR Green RealMasterMix (Tiangen) and CFX96TM Real-Time System (Bio-Rad, Hercules, CA) according to the manufacturer’s protocols. β-Actin expression was used as an internal standard. Three biological replications were analyzed for each point. PCR was performed using the following primer sets: TaCKX3, forward, 5-CGTGGCTCAACCTCTTCGTC-3, reverse, 5-GTTCGGGTCCCACTTGCTC-3; actin, forward, 5-GCAATGTATGTCGCAATCCAG-3, reverse, 5-CTTCATTAGATTA TCCGTGAGGT -3.
Results
Cloning and Sequence Analysis of TaCKX3
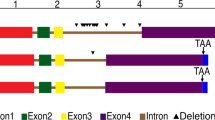
A 2,555-bp full-length DNA of TaCKX3 was obtained from wheat by using primer TaCKX3F and TaCKX3R. Comparison between cDNA and genomic sequence was performed to determine the introns' positions and sequence. Sequence analysis showed that the coding region of TaCKX3 has three introns, which are 103, 421, and 458 bp, successively (Fig. 1).
We performed a BLAST search of wheat EST GenBank database and found that the cDNA of TaCKX3 was more highly homologous with known TaCKX3 EST (BE404516.1) than any other TaCKX ESTs from wheat. According to the previous work (Galuszka et al. 2004), we named the novel gene as TaCKX3. The full-length nucleotide sequence was submitted to the GenBank Data Libraries under the accession number GQ925404.
Bioinformation Analysis of Amino Acid Sequence Deduced from TaCKX3
The TaCKX3 cDNA with a TAG stop codon encodes a 516-polypeptide. To analyze the homologous identity scores of the predicted protein with other known CKXs, sequence alignment of the deduced amino acid sequence with other known CKXs was performed and a phylogenetic tree based on evolutionary distances was constructed using DNAMAN program. As shown in the alignment, two functional domains with characteristics of TaCKX3 were identified by sequence comparison: the FAD binding domain (from positions 70 to 192 of amino acid sequences) and the CK binding domain (from positions 224 to 504 of amino acid sequences; Fig. 2). The alignment also displayed many conserved motifs, like GHS motif in FAD binding domain around position 80 and PHPWLN motif around 370 (Fig. 2). A PGQXIF signature (X stands for K or D amino acid) was present at the C-terminal ends of the proteins (Fig. 2). Some of the conserved regions may play important roles in catalytic action of CKX.
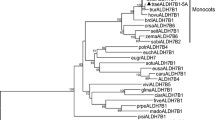
The phylogenetic tree revealed that the putative polypeptide of TaCKX3 has significant homology with ZmCKX10, up to 82% identity, but less than 35% identity scores to other known CKX used for comparisons (Fig. 3). The result showed that TaCKX3 is closely related to ZmCKX10 and has low homologies with other CKXs to some extent.
Phylogenetic tree of protein sequences of TaCKX1 and other known CKX from maize, rice, Arabidopsis, and barley. The numbers on the branches presented correspond to the degree of shared sequence similarity. TaCKX1 (DQ784573), TaCKX3 (GQ925404), ZmCKX1 (Y18377), ZmCKX2 (AJ606942), ZmCKX3 (AJ606943), ZmCKX10(ACJ06785), OsCKX1 (EAY72836), OsCKX2 (AP003244), OsCKX3 (AC051632), OsCKX5 (AP003344),HvCKX2 (AF540382), HvCKX3 (AY209184), AtCKX1 (NM219714), AtCKX2 (AF303978), AtCKX3 (AF303979), AtCKX4 (AF303980), AtCKX5 (AF303982), AtCKX6 (NM116209), and AtCKX7(AF303981)
The theoretical molecular mass of the encoded protein is 55.7 kDa with a calculated pI of 5.93 (http://expasy.org/tools/). TaCKX3 gene may be a nonsecreted CKX gene in wheat. The protein of TaCKX3 differs from that of TaCKX1, which carries typical N-terminal sequences of secretion pathway protein (Feng et al. 2008). As AtCKX7 and ZmCKX10 (Schmülling et al. 2003; Šmehilová et al. 2009), the deduced sequence of TaCKX3 has no signal peptide, and therefore, TaCKX3 may carry out functions in cytoplasm.
Chromosomal Localization
PCR using DNA of CS NT lines as templates showed that Y361 and other 20 NTs produced the target band except N7BT7A (Fig. 4). Therefore, the novel gene of TaCKX3 was located on wheat chromosome 7B.
Expression Levels of TaCKX3 Gene in Germinating Embryos
The expression of the TaCKX3 gene in germinating embryos after different stress treatments was measured by real-time quantitative reverse transcription PCR analysis. The water-treated germinating embryos have little expression of TaCKX3 (Fig. 5). Compared with the water mock treatment, all of NaCl, 6-BA, and PEG-6000 treatments increase TaCKX3 mRNA expression in germinating embryos (Fig. 5). The expression of TaCKX3 in germinating embryos treated with PEG-6000 and NaCl increased slightly. TaCKX3 transcription was strongly up-regulated by 6-BA. This experiment indicated that TaCKX3 gene is salt and drought inducible and can be significantly up-regulated by 6-BA.
Discussion
As a class of plant hormones, endogenous CKs levels in plants are maintained by the balance between biosynthesis and metabolism. The CKX was thought by far to be the only enzyme in plants to catalyze the catabolism of the specific CKs (Hare and Van Staden 1994; Jones and Schreiber 1997; Galuszka et al. 2001). Since the activity of CKX was first discovered (Pačes et al. 1971), the molecular and biochemical characteristics of CKX were well analyzed in the previous studies (Jones and Schreiber 1997). However, the detailed functions of CKX in plant growth and development were still unclear. It has been made great progress to study the functions of CKX in the past few years since the first CKX gene was cloned from maize (Houba-Hérin et al. 1999; Morris et al. 1999). Overexpressing DSCKX1 orchid resulted in CKX activity increasing with a reduction of CK content. On the contrary, antisense transgenic plants showed higher endogenous CK content than wild-type plants (Yang et al. 2003a). In maize, though all of ZmCKX1∼5 were expressed at early stages of kernel development, each ZmCKX gene had its own spatial and temporal expression profile (Massonneau et al. 2004). In other words, the functional study of CKX genes has shown the direct relationship between CKX activity and CKs metabolism.
The results of previously cloned CKX genes showed that CKX genes belong to multigene family (Galuszka et al. 2004). There are 7 homologous CKX genes in Arabidopsis and at least 11 in rice and 5 in maize (Galuszka et al. 2004; Massonneau et al. 2004). The large volume of wheat genome implies that there may be more CKX genes in wheat. However, the full-length CKX genes were rarely cloned from wheat by now (Feng et al. 2008). In this work, we cloned a novel TaCKX3 gene from Hexaploid wheat. The cDNA nucleotide sequence of TaCKX3 was aligned with that of TaCKX1 using DNAMAN, and the result showed that they share 60% identity (data not shown). Nevertheless, the deduced proteins of the two TaCKX genes have lower identity.
As other known CKXs, the deduced amino acid polypeptide of TaCKX3 has two conserved domains for FAD binding and CK binding, respectively. However, the predicted sequence lacks signal peptide, which means that it may be a nonsecreted protein and function in the cytosol. There is generally only one CKX per plant genome that lacks a translocation signal (Šmehilová et al. 2009). By alignment analysis, the putative protein has low sequence homology with other CKX except ZmCKX10, which also has no signal peptide (Šmehilová et al. 2009). TaCKX3 and ZmCKX10 have significant identity scores and were distributed in one group in the phylogenetic tree, which suggests that they may have similar biochemical functions. Interestingly, AtCKX7 is also a cytoplasm protein without signal peptide (Schmülling et al. 2003), but it has low homology with TaCKX3 and ZmCKX10.
Environmental stress can change plant growth by altering gene expression and cellular metabolism. Under drought stress, the amount of CKX increased threefold in the roots of sunflower plants (Manju et al. 2001) and the enzyme activity had been enhanced in the stressed leaf tissue. Although many data of the biochemical properties of the enzyme were reported at present, the CKX gene expression and its regulation were still not clear. One study demonstrated that drought could induce CKX1 expression at the pedicel region of maize kernels on molecular level (Brugiere et al. 2003). Vaseva-Gemisheva et al. (2005) showed that PsCKX1 and PsCKX2 mRNA expression was both increased in leaves of drought stressed pea plants. Up to now, the study of CKX in responding to salt stress in plants is few reported.
In this work, the real-time quantitative reverse transcription PCR showed that TaCKX3 gene can be significantly induced by 6-BA in germinating embryos, and the gene expression increased slightly in germinating embryos treated by NaCl or PEG-6000 comparing with that treated by H2O. The results suggest that TaCKX3 is a salt and drought inducible gene. Many researches have demonstrated that CKX activity may be induced by CKs (Jones et al. 1992; Dietrich et al. 1995). In our study, the result showed that the TaCKX3 gene was enhanced by exogenous 6-BA. It is consistent with the previous studies of applying exogenous CKs resulted in the increasing of CKX expression (Wang et al. 2009). These results clearly show the positive response of the CKX transcription under CK induction on the molecular level. Because each CKX gene’s spatial and temporal expression profile are almost different in one plant (Massonneau et al. 2004), further work is required in order to illuminate the detailed functions of CKX in every stage of plant growth and development.
Abbreviations
- BLAST:
-
Basic Local Alignment Search Tool
- cDNA:
-
complementary DNA
- CKs:
-
cytokinins
- CKX:
-
cytokinin oxidase/dehydrogenase
- DNA:
-
deoxyribonucleic acid
- FAD:
-
flavin adenosine dinucleotide
- MW:
-
molecular weight
- NT:
-
nulli-tetrasomic
- PCR:
-
polymerase chain reaction
- pI :
-
isoelectric point
- RNA:
-
ribonucleic acid
- RT-PCR:
-
reverse transcription polymerase chain reaction
References
Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M (2005) Cytokinin oxidase regulates rice grain production. Science 309:741–745
Bilyeu KD, Cole JL, Laskey JG, Riekhof WR, Esparza TJ, Kramer MD, Morris RO (2001) Molecular and biochemical characterization of a cytokinin oxidase from maize. Plant Physiol 125:378–386
Brugiere N, Jiao SP, Hantke S, Zinselmeier C, Roessler JA, Niu XM, Jones RJ, Habben JE (2003) Cytokinin oxidase gene expression in maize is localized to the vasculature, and is induced by cytokinins, abscissic acid, and abiotic stress. Plant Physiol 132:1228–1240
Chatfield JM, Armstrong DJ (1986) Regulation of cytokinin oxidase activity in callus tissues of Phaseolus vulgaris L. cv. Great Northern. Plant Physiol 80:493–499
Dietrich JT, Kaminek M, Blevins DG, Reinbott TM, Morris RO (1995) Changes in cytokinins and cytokinin oxidase activity in developing maize kernels and the effects of exogenous cytokinin on kernel development. Plant Physiol Biochem 33:327–336
Feng DS, Wang HG, Zhang XS, Kong LR, Tian JC, Li XF (2008) Using an inverse PCR method to clone the wheat cytokinin oxidase/dehydrogenase gene TaCKX1. Plant Mol Biol Rep 26:143–155
Galuszka P, Frébort I, ebela M, Sauer P, Jacobsen S, Pe P (2001) Cytokinin oxidase or dehydrogenase? Mechanism of cytokinin degradation in cereals. Eur J Biochem 268:450–461
Galuszka P, Frébortová J, Werner T, Yamada M, Strnad M, Schmülling T, Frébort I (2004) Cytokinin oxidase/dehydrogenase genes in barley and wheat: cloning and heterologous expression. Eur J Biochem 271:3990–4002
Hare PD, Van Staden J (1994) Cytokinin oxidase: biochemical features and physiological significance. Physiol Plant 91:128–136
Hitoshi S (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57:431–449
Houba-Hérin N, Pethe C, d’Alayer J, Laloue M (1999) Cytokinin oxidase from Zea mays: purification, cDNA cloning and expression in moss protoplasts. Plant J 17:615–626
Huang S, Cerny RE, Qi Y, Bhat D, Aydt CM, Hanson DD, Malloy KP, Ness LA (2003) Transgenic studies on the involvement of cytokinin and gibberellin in male development. Plant Physiol 131:1270–1282
Jones RJ, Schreiber BMN (1997) Role and function of cytokinin oxidase in plants. Plant Growth Regul 23:123–134
Jones RJ, Schreiber BM, McNeil K, Brenner ML, Foxon G (1992) Cytokinin levels and oxidase activity during kernel development. In M Kamínek, DWS Mok, E Zažímalová (Eds.), Physiology and Biochemistry of Cytokinins in Plants. SPB Academic Publishing, The Hague, The Netherlands, pp 235–239
Kaminek M, Armstrong DJ (1990) Genotypic variation in cytokinin oxidase from Phaseolus callus cultures. Plant Physiol 93:1530–1538
Manju RV, Kulkarni MJ, Prasad TG, Sudashana L, Sashidar VR (2001) Cytokinin oxidase activity and cytokinin content in roots of sunflower under water stress. Ind J Exp Biol 39:786–792
Massonneau A, Houba-Hérin N, Pethe C, Madzak C, Falque M, Mercy M, Kopecny D, Majira A, Rogowsky P, Laloue M (2004) Maize cytokinin oxidase genes: differential expression and cloning of two new cDNAs. J Exp Bot 55:2549–2557
Mochida K, Yoshida T, Sakurai T, Ogihara Y, Shinozaki K (2009) TriFLDB: a database of clustered full-length coding sequences from Triticeae with applications to comparative grass genomics. Plant Physiol 150:1135–1146
Mok DWS, Mok MC (2001) Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol 152:89–118
Morris RO, Bilyeu KD, Laskey JG, Cheikh N (1999) Isolation of a gene encoding a glycosylated cytokinin oxidase from maize. Biochem Biophys Res Commun 255:328–333
Pačes V, Werstiuk E, Hall RH (1971) Conversion of N 6-(Δ2-isopentenyl) adenosine to adenosine by enzyme activity in tobacco tissue. Plant Physiol 48:775–778
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA space length polymorphisms in barley: Mendelian inheritance, chromosomal locations and population dynamics. Proc Natl Acad Sci USA 81:8014–8018
Schmülling T, Werner T, Riefler M, Krupková E, Bartrinay Manns I (2003) Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J Plant Res 116:241–252
Šmehilová M, Galuszka P, Bilyeu KD, Jaworek P, Kowalska M, Šebela M, Sedlářová M, English JT, Frébort I (2009) Subcellular localization and biochemical comparison of cytosolic and secreted cytokinin dehydrogenase enzymes from maize. J Exp Biol 60:2701–2712
Terrine C, Laloue M (1980) Kinetics of N 6-(Δ2-isopentenyl) adenosine degradation in tobacco cells: evidence of regulatory mechanism under the control of cytokinins. Plant Physiol 65:1090–1095
Vaseva-Gemisheva I, Lee D, Karanov E (2005) Response of Pisum sativum cytokinin oxidase/dehydrogenase expression and specific activity to drought stress and herbicide treatments. Plant Growth Regul 46:199–208
Wang Y, Luo JP, Wei ZJ, Zhang JC (2009) Molecular cloning and expression analysis of a cytokinin oxidase(DhCKX) gene in Dendrobium huoshanense. Mol Biol Rep 36:1331–1338
Werner T, Motyka V, Strnad M, Schmülling T (2001) Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA 98:10487–10492
Whitty CD, Hall RH (1974) A cytokinin oxidase in Zea mays. Can J Biochem 52:787–799
Yang SH, Yu H, Goh CJ (2003a) Functional characterisation of a cytokinin oxidase gene DSCKX1 in Dendrobium orchid. Plant Mol Biol 51:237–248
Yang SH, Yu H, Xu Y, Goh CJ (2003b) Investigation of cytokinin-deficient phenotypes in Arabidopsis by ectopic expression of orchid DSCKX1. FEBS Lett 555:291–296
Acknowledgments
The authors thank Dr. Bin Liu (Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles) for the critical review of the manuscript. They also thank Prof. Bin Wang (Institute of Genetics and Developmental Biology, Chinese Academy of Science) for sending us seeds of nulli-tetrasomic lines of “Chinese Spring.” This project was supported by the Scientific Research Award Foundation for Outstanding Young and Middle-Aged Scientists of Shandong Province of China (BS2009NY037) and the International Cooperation Program for excellent Lecture of 2009 by Shandong Provincial Education Department, P.R. China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Sequence data of TaCKX3 from this article have been deposited at GenBank under accession number GQ925404.
Rights and permissions
About this article
Cite this article
Ma, X., Feng, DS., Wang, HG. et al. Cloning and Expression Analysis of Wheat Cytokinin Oxidase/Dehydrogenase Gene TaCKX3 . Plant Mol Biol Rep 29, 98–105 (2011). https://doi.org/10.1007/s11105-010-0209-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-010-0209-x