Abstract
Homology in the promoter region leads to transcriptional gene silencing (TGS). This could be circumvented by designing synthetic promoters which are functionally similar but divergent in their sequence. In the present study, we asked whether such synthetic promoters can escape trans-inactivation when challenged with a silenced copy of the wild-type prototype. We tested the activity of two synthetic cauliflower mosaic virus 35S (35S) domain A promoters, 2mA and 3mA, with only 35% and 41% sequence identity with the wild type 35S (wtA) in the presence of silencing locus 271 of Nicotiana tabacum that inactivates any 35S promoter driven transgene through TGS. Based on comparison of β-glucuronidase (gus) activity under the transcriptional control of wtA, 2mA, and 3mA in the presence and absence of the silencing locus, we demonstrate that the inactivation of the 2mA and 3mA promoters is significantly delayed in comparison to that of wtA although microhomology at the transcription factor binding sites eventually leads to the silencing of these synthetic promoters. We further postulate that it is possible to design functional synthetic promoters that can escape gene silencing in the background of silencing locus if the cis-elements are smaller and do not contain methylation prone CG and CNG.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Homology dependent gene silencing (HDGS) in transgenic plants occurs when multiple copies of a transgene or a transgene with homology to an endogenous plant gene are present in the plant nucleus. Different types of silencing phenomena occur in plants depending on the region of homology, their arrangements, orientations, relative positions of the interacting homologous sequences, and the degree of somatic and germinal heritability. On the one hand, homology in the promoter region has been known to cause repression of transcription, which has been termed transcriptional gene silencing (TGS), and on the other hand, homology in the coding region causes mRNA degradation and is called as posttranscriptional gene silencing (PTGS). It has recently been demonstrated that HDGS is mediated by double-stranded RNA (dsRNA) and that both TGS and PTGS result in the methylation of homologous sequences at the target loci. The two pathways although distinct are mechanistically related (Sijen et al. 2001). TGS has been further classified into cis and trans-inactivation. Cis-inactivation occurs due to homology in intra-allelic repeats, closely linked on the same DNA molecule. This is caused by the insertion of multiple and rearranged copies of a transgene at a single locus or in the presence of closely linked copies of transgene that results in methylation through RNA-directed DNA methylation or heterochromatinization that suppresses transcription from promoters. These cis-inactivated loci, in many cases, can suppress another copy of the homologous sequence if brought into its background (in trans). Such loci have also been called “silencing loci”. Trans-inactivation requires interaction of the silencing locus with the target sequence, caused by ectopic DNA–DNA pairing between the loci. This results in a transfer of the silenced state from one locus to another, either by transfer of repressive chromatin states to targets or by de novo methylation of the target sequence. At the same time, reciprocal ectopic interaction of genes causes cosuppression of both the genes. In cosuppression or PTGS, silencing occurs at the RNA level. Once initiated against the RNA of a given transgene, PTGS leads to the suppression or degradation of homologous RNA from either the endogenous gene or the transgene, through the production of dsRNA. These phenomena have been extensively reviewed (Matzke and Matzke 1995; Meyer and Saedler 1996; Depicker and van Montagu 1997; Stam et al. 1997; Vaucheret et al. 1998; Wassenegger and Pélissier 1998; Fagard and Vaucheret 2000; Iyer et al. 2000; Vaucheret et al. 2001; Matzke et al. 2002; Vaucheret 2006 and Voinnet 2008).
Irrespective of the nature of interaction, homology among sequences is the key factor that initiates silencing. Silencing can be avoided by developing gene constructs that have minimum homology to each other or to endogenous genes. Homology can be easily reduced in coding regions by using codon degeneracy, in which coding regions can be modified for their DNA sequence without altering the amino acid sequence (Kumpatla et al. 1998). Synthetic binary vectors can also be developed where the region between the T-DNA borders has been modified to reduce/remove any sequence homology with the other available binary vectors (Arumugam et al. 2007).
To circumvent TGS due to homology in the promoter region, one can either use diverse promoters isolated from different plant and viral genomes or design synthetic promoters. The cauliflower mosaic virus (CaMV) 35S (35S) promoter is the most extensively used promoter to express transgenes in plants. However, only few of these promoters have expression level comparable to the 35S promoter. Although several other viral promoters have been identified to date (Medberry et al. 1992; Bhattacharyya-Pakrasi et al. 1993; Verdaguer et al. 1996; Maiti et al. 1997; Tzafir et al. 1998; Dey and Maiti 1999; Holmberg et al. 2002; Stavolone et al. 2003; Xie et al. 2003), 35S still remains the promoter of choice. These promoters could be used to drive expression of different genes in transgenic cassettes.
The challenge, however, lies in designing synthetic promoters, which would be functionally homologous but divergent in their sequence. Most of the synthetic promoters designed till date are either hybrids of two existing promoters (Comai et al. 1990; Ni et al. 1995) or are functional add-ons to a basal promoter (Rushton et al. 2002) rather than being de novo synthesized promoters. The only de novo construction of a complete synthetic promoter has been reported by Sawant et al. (2001). In an earlier study (Bhullar et al. 2003), we showed that synthetic promoters can be designed either by placing the known cis-elements in a divergent DNA sequence or by “domain swapping” wherein domains of a promoter can be exchanged with functionally equivalent domains from a heterologous promoter. We evaluated the first strategy using two domain A synthetic 35S promoters, Mod2A1T and Mod3A1T. Both the promoters were found to express at par with the 35S (Bhullar et al. 2003).
In the present study, we have tested whether synthetic promoters with limited sequence homology can escape silencing when placed in the background of a strong silencing loci (SL) for 35S, the “271 locus” (Vaucheret 1993). This locus consists of multiple copies of the tobacco nitrite reductase (NIR) sequence in antisense orientation (RIN) driven by the 35S promoter and the bacterial neomycin phosphotransferase (npt) sequence driven by CaMV 19S promoter. The 271 locus has been shown to elicit TGS and PTGS by producing aberrant small double-stranded RNAs corresponding to 19S, 35S promoter, and NIR sequences (Mourrain et al. 2007). It has been called the universal silencer of 19S or 35S. Vaucheret (1993) also reported that 90-bp homology is enough to trigger inactivation. Since in the Mod2A1T and Mod3A1T promoters developed in the earlier study (Bhullar et al. 2003), domain B (−91 to −343) is the same as that of the wild-type 35S promoter, we have used three truncated promoters corresponding to the domain A of wild-type 35S, Mod2A1T and Mod3A1T (hitherto called as wtA, 2mA, and 3mA, respectively) to test their activity in the presence (SL; 271.5.8) and absence of the silencing loci (nonsilencing locus (NSL) 271.5.9). We demonstrate that in the background of the silencing locus, inactivation of the 2mA and 3mA is significantly delayed when compared to that of wtA, although microhomology at the transcription factor binding sites eventually leads to the silencing of these synthetic promoters.
Materials and Methods
Assembly of Synthetic Promoters
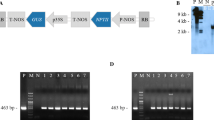
The synthetic domain A promoters were synthesized as described previously (Bhullar et al. 2003). Briefly, the promoters were assembled using recursive PCR, followed by subsequent cloning in pPCR-Script Amp SK(+) and then sequenced to confirm the fidelity of the sequence. To create truncated promoters 35S, Mod2A1T and Mod3A1T (Bhullar et al. 2003), domain A (−90 to +1) region was removed as EcoRV–NcoI fragment and cloned in a modified pMCS5 (MoBiTec, Gottingen, Germany), named ∆pMCS5 as EcoRV–NcoI fragment. ∆pMCS5 was created by deleting the region between MluI–EcoRI restriction sites. The domain A promoters were then removed as a EcoRV–NcoI fragment and cloned upstream to the β-glucuronidase (gus) reporter gene with a 35S poly(A) signal in pPCR-Script Amp SK(+) (Stratagene, La Jolla, CA, USA). All clonings followed standard protocols as described in Sambrook et al. (1989). The promoter-gus expression cassette thus developed was cloned as SacI–SalI fragment in binary vector pPZP200 (Hajdukiewicz et al. 1994) containing a nopaline synthase promoter (Pnos)-driven neomycin phosphotransferase (nptII) gene, as a selection marker for plant transformation.
Development of Transgenics and Crosses
Binary vectors were mobilized into the disarmed Agrobacterium strain GV2260 by electroporation. Agrobacterium sp.-mediated transformation of leaf disk in tobacco (Nicotiana tabacum cv. Xanthi) explants was carried out following the protocol of Svab et al. (1995). The tobacco transformants were selected on kanamycin (100 mg/l). With each construct, several independent transgenic lines were developed. Ten to 12 independent T 0 transgenic lines were grown in a green house (16:8 h day/night cycle, 28 ± 2°C, relative humidity 70%) along with the tobacco transgenic homozygous lines 271.5.8 (SL) and 271.5.9, which is an isogenic line without the transgene integration (NSL). Since in the lines containing SL loci the nitrate reductase activity is blocked, these lines were grown in medium B supplemented with 10 mM KCl and 10 mM ammonium succinate (Vaucheret 1993) and were watered with the nutrient of Coϊc and Lesaint (Vaucheret et al. 1992). Further, the SL lines become kanamycin sensitive, as the 19S gets inactivated (Mourrain et al. 2007).
The T 0 transgenic lines carrying the promoter constructs (wtA, 2mA, and 3mA) were selfed, as well as crossed to both SL and NSL lines. Seeds were collected from growth-chamber grown plants. The seeds were surface-sterilized with 70% alcohol followed by five to six washings with water and were germinated on medium B, supplemented with 10 mM KCl and 10 mM ammonium succinate (Vaucheret 1993) in the presence of kanamycin (100 mg/l).
Enzyme Assays for Estimation of Promoter Strength
The total protein present in the seedlings/tissues was extracted in the GUS extraction buffer (Jefferson 1987). Protein concentration was estimated following Bradford (1976). Fluorometric GUS assays using 4-methyl umbelliferyl β-d-glucuronide substrate were performed according to Jefferson (1987) on 15-day-old seedlings, 30-day-old seedlings, and on the root tissues of 3-month-old plantlets. The product released (methylumbelliferone, MU) was estimated with DyNA Quant 200 Fluorometer (Hoefer Pharmacia Biotech, San Francisco, CA, USA). GUS activity was expressed as picomoles MU per minute per milligram protein. The fold inactivation was calculated as the ratio of the promoter activity in the background of NSL to that of SL.
GUS Staining
For histochemical GUS staining, the samples were stained overnight in a GUS staining solution containing 0.5 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc) in 50 mM sodium phosphate (pH 7.0) at 37°C.
Genomic DNA Isolation and Southern Analysis
Plant Genomic DNA was isolated using cetyltrimethylammonium bromide protocol described by Rogers and Bendich (1994). About 10 μg of each DNA sample was digested with KpnI (New England Biolabs), electrophoresed on a 0.8% agarose gel, and blotted on nylon membrane (Hybond N+, Amersham Pharmacia Biotech). Blots were probed with the coding sequence of nptII gene to ascertain the presence of silencing locus. Probes were labeled with α-[32P]-dCTP using the Megaprime DNA Labelling System (Amersham Pharmacia Biotech). Hybridization and washing conditions were based on standard procedures as prescribed by Sambrook et al. (1989).
Statistical Analysis of Data
The data on the GUS activity obtained in transgenic lines in the background of SL and NSL with different constructs was analyzed using students’ unpaired T test. p values < 0.05 were accepted as significantly different.
Results
Experimental Design, Development of Transgenics, and Crosses with SL and NSL
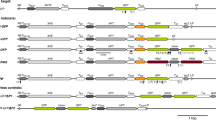
A truncated 35S promoter containing only domain A of the wild-type 35S promoter (wtA) or synthetic promoters 2mA and 3mA has been used to study the activity of these promoters in the presence of the silencing locus 271.5.8 (SL). The domain A (−90 to +1) of 2mA and 3mA promoters shows 65% and 59% divergence in its sequence in regions flanking the identified cis-elements from that of the 35S (Bhullar et al. 2003; Fig. 1). Although domain A is known to drive much lower expression levels than a full-length promoter, the activity can be quantified, especially in the root tissue, as this domain is largely responsible for expression in roots (Benfey et al. 1990). The present study deals with the expression of these synthetic promoters in the presence of the silencing locus 271.5.8 (SL).
Sequence comparison of the domain A (−90 to +1) of the 35S promoter (wtA) with that of the 2mA and 3mA promoters. The pentameric repeat of the as-1 element and TATA box are indicated in bold italics. The 35S promoter in the regions flanking the identified cis-elements shows a similarity of 35% and 41%, with that of 2mA and 3mA promoters, respectively. The region which is identical among the three promoters has been underlined
To test the activity of these promoters, they were cloned upstream to the gus gene. The binary vector developed also contained a Pnos driven nptII gene that served as a selection marker for plant transformation as well as a reference gene to monitor silencing.
Ten to 12 independent tobacco transgenic lines for each construct were generated and transferred to the soil in the green house, along with homozygous SL and NSL lines. The T 0 transgenic lines were selfed as well as crossed to both SL and NSL lines. The seeds from these crosses were harvested, and the promoter activity analyzed at various stages of seedling development viz., GUS analysis by histochemical staining in 7-day-old seedlings, quantification of GUS activity in 15- and 30-day-old seedlings and roots of 3-month-old plantlets. The effect of SL on domain A was analyzed by comparing the activity of the promoters in the background of SL to that of the background of NSL.
Segregation Analysis of T 1 Seeds
Selfed seeds of approximately ten independent lines for each of the constructs were used for segregation analysis. Seeds resistant and sensitive to kanamycin were scored after 30 days of germination. The results are summarized in Table 1. Based on chi-square analysis, it was observed that in most of the transgenic lines, except for those developed with 3mA, the integration of transgene at single locus occurred at a higher frequency. However, in case of 3mA, only one line had insertion at a single locus.
Expression of the gus Gene in the Presence (SL) and Absence (NSL) of Silencing Loci
In order to study the effect of silencing loci on the domain A promoter in 7-day-old seedlings, ∼50 F 1 seedlings obtained from crosses with SL and NSL lines were tested for GUS expression by histochemical staining (Fig. 2). The percentage of lines exhibiting GUS staining was calculated. These have been represented in Fig. 3.
Of the six lines tested in case of wtA, progeny from four of the lines (1.9, 2.5, 1.15, and 1.11) did not show GUS expression in any of the seedlings from crosses with SL. This shows that SL trans-inactivates the wild-type domain A of 35S promoter in wtA constructs as reported by Vaucheret (1993). In the case of 2mA, three out of the five lines tested escaped trans-inactivation. In the two lines showing trans-inactivation, the percentage of seedlings showing GUS expression was found to be more than that observed with wtA construct. This indicated a lesser extent of trans-inactivation in case of lines with 2mA construct. In 3mA, five of the seven lines escaped trans-inactivation, while in two lines, 1.20 and 1.17, there was ∼40% reduction in seedlings expressing the gus gene.
The activity of wild-type and synthetic domain A promoters was analyzed in 15-day-old kanamycin (KanR) seedlings. Approximately ten kanamycin resistant F 1 seedlings from crosses with NSL and SL lines were used for quantifying the activity of the GUS protein in three independent experiments with internal replicates. The results obtained are summarized in Table 2. The results are presented as the GUS activity calculated as the mean of three independent experiments, along with standard error in transgenic lines crossed with NSL as well as SL. Fold inactivation that reflects the drop in promoter activity in the background of SL as compared to that of NSL was also calculated.
In the case of wtA, the GUS activity in transgenic lines crossed with NSL and SL showed that in six out of the seven plants tested, the fold inactivation varied from 2.02 to 5.92. Lines showing at least 2-fold inactivation have been taken as suppressed, which is also reflected in their p values which range from 0 to 0.024 in these lines, indicating that these means are significantly different. Only one line, wtA 2.9, had p value of 0.051 that did not get silenced. Thus, in case of wtA, the domain A gets trans-inactivated in the presence of SL, but not in its absence, i.e., in NSL.
Out of the six lines tested in case of transgenics with 2mA promoter, two of the lines (1.21 and 1.29) demonstrated a significant drop in promoter activity in the background of SL, although the fold inactivation in the case of line 1.29 was 1.5-fold. Similarly, four of the six transgenic lines with 3mA promoter were observed to be active even in the presence of the silencing locus.
In the 30-day-old seedlings, line 2.9 containing the wtA promoter was also observed to be silenced. The fold inactivation for other transgenic lines with wtA was also relatively higher as compared to inactivation observed in the 15-day-old seedlings (Table 3). Further, all these lines showed greater than 10-fold fall in activity in the roots of 3-month-old plantlets, suggesting that trans-inactivation had set in. Interestingly, of the progenies obtained from crosses with SL, three lines each with 2mA and 3mA construct still escaped trans-inactivation in 30-day-old seedlings. Of these six lines, four of the lines (viz., 2mA 1.4, 2mA 1.9, 2mA 1.48, and 3mA 1.20) retained activity even in the roots of 3-month-old plantlets. The GUS activity of the lines which escaped trans-inactivation is summarized in Table 4.
The presence of the silencing locus in the F 1 progenies that escaped trans-inactivation was confirmed by Southern blot analysis (Fig. 4). The genomic DNA was digested with restriction enzyme KpnI and hybridized with a NcoI fragment of the nptII gene. The hybridization pattern as observed in the SL parental line was also observed in the F 1 progenies obtained from crosses between SL and wtA 2.9, 2mA 1.9 and 3mA 1.20.
Southern blot analysis to demonstrate the presence of silencing locus in representative wtA, 2mA, and 3mA lines crossed with silencing line (SL). a Restriction map of plasmid pRiN in the genome of Silencing locus. (Vaucheret 1993). b Ten micrograms of genomic DNA of SL and isogenic line NSL without the silencing locus, as well as representative wtA, 2mA, and 3mA lines crossed with SL and NSL line digested with BamHI and KpnI and probed with the NcoI fragment of the nptII gene. Except for lane 2 that contains DNA digested with BamHI, in all other lanes, digestion has been carried out with KpnI. Lane 1 NSL, lane 2 SL digested with BamHI, lane 3 SL digested with KpnI, lane 4 wtA 2.9 × SL, lane 5 wtA 2.9 × NSL, lane 6 2mA 1.9 × SL, lane 7 2mA 1.9 × NSL, lane 8 3mA 1.20 × SL, lane 9 3mA 1.20 × NSL
Effect of SL Line on the nptII Gene
F1 progenies obtained from crosses between the different synthetic promoter containing lines and SL or NSL were germinated in the presence of kanamycin. The percentage of KanR progenies obtained from crosses with the SL line was observed to be similar to those obtained from crosses with the NSL line in each case (Fig. 5). This indicated that the nptII gene driven by the Pnos present in the cassette was not silenced.
Discussion
Homology between promoter sequences can lead to homology-based gene silencing. Silencing loci like 271 loci in tobacco are known to trans-inactivate 35S promoter when brought in its genetic background (Mourrain et al. 2007). In this study, we investigated the possibility of circumventing silencing by reducing the similarity between the sequence of the target promoter and the inactivated promoter at the silencing locus. Using the 271 line developed by Vaucheret (1993) which inactivates 35S promoter-driven genes through TGS, we have demonstrated that synthetic domain A promoters 2mA and 3mA can escape trans-inactivation up to the seedling stage, and there is a significant delay in the induction of silencing.
In a previous study, we had developed synthetic 35S promoters by placing the identified cis-elements of the domain A in a variant DNA stretch. These synthetic promoters were shown to be functionally equivalent to the wild-type 35S promoter (Bhullar et al. 2003). The activity of the domain A of the wild-type 35S promoter (wtA) and the two synthetic promoters (2mA and 3mA) was analyzed against the background of a general silencer for the 35S promoter, i.e., line 271 (SL line 271.5.8). The activity of these promoters in the background of NSL (line 271.5.9) was used as a control.
Based on the expression of gus gene in F 1 progenies obtained from crosses between lines containing the target promoter constructs viz., wtA, 2mA, and 3mA, and the NSL line, it was observed that these promoters were functionally equivalent as reported in our earlier study (Bhullar et al. 2003). However, in the presence of the silencing locus, the wild-type 35S domain A promoter was found to be trans-inactivated even at the 15-day-old seedling stage in all lines, except one, i.e., wtA2.9. In all these cases, there was greater than 2-fold inactivation. The fold inactivation progressively increased in all the events including wtA2.9 in the 30-day-old seedlings as well as in the roots of 3-month-old plantlets. On the other hand, ∼67% of the transgenic lines with the synthetic 35S promoter constructs escaped trans-inactivation at the 15-day-old seedling stage. All these lines showed less than 2-fold inactivation. Although the transgene inactivation progressed in the older seedlings as well as in the roots, four of the lines escaped trans-inactivation, showing similar expression of the promoter driven gus gene in the background of SL line as that in the NSL line.
In all the lines studied, the nptII gene did not get trans-inactivated by the silencing loci which is in concurrence with the observation made by Vaucheret (1993) and Mourrain et al. (2007) that the silencing occurs because of the homology between promoters in the case of 271 line. Although the nptII gene is silenced in the 271 loci, it is not able to trans-inactivate the same in domain A constructs since in the silencing locus, the gene is under the control of 19S and in the latter it is under control of the Pnos.
The expression of the lines which escaped silencing in the roots of 3-month-old plantlets could not be studied in the older mature plants as in the silencing line 271, the endogenous nitrite reductase gene is inactivated and has to be nutritionally supplemented for its growth (Vaucheret et al. 1992). In our study, we observed that even after the necessary nutritional supplementation, the 271.5.8 line (SL) showed retarded growth in comparison to that of its near-isogenic line without the silencing loci (NSL, 271.5.9 line). This was also reflected in the F 1 generation from crosses with the SL locus. Although the seedlings grew well up to 1 month, at a later stage, their growth was retarded. This was also reflected in the difference observed in the chlorophyll content between F 1 lines generated from crosses with SL and NSL (data not shown). These reflected variations in the physiology of the two lines and as promoter activity is significantly influenced by the physiology of the cell, activity of the promoters could not be compared in older plants.
Our observations in the early stages of development clearly demonstrate that there is a significant delay in the progression of trans-inactivation of the synthetic promoters. The onset of transgene silencing is possibly due to the microhomology between the wild-type 35S promoter and the synthetic promoters at the transcription factor binding sites (Fig. 1). Although it has been previously shown that 90 bp of homology in the promoter sequence is enough for trans-inactivation (Vaucheret 1993), this study shows that even a smaller region (∼30 bp) can result in trans-inactivation although the progress is substantially delayed. In a previous study, Thierry and Vaucheret (1996) had demonstrated that Figwort mosaic virus 34S promoter does not get silenced (up to after 5 months of growth) in the presence of the 271 locus. The 34S promoter shares 63% overall identity with the domain A of the CaMV 35S but with never more than six contiguous identical nucleotides. In the synthetic promoters designed in the present study, we were unable to reduce the stretch of identity as changing the seven intervening bases between the TGACG repeats in the as-1 element led to a significant drop in promoter activity (Bhullar et al. 2003). Interestingly, the as-1 element in the 34S promoter has a sequence which is not identical to that in the 35S promoter. It would be interesting to check the functionality of the as-1 element present in the 34S promoter in the context of the synthetic promoter. If found to be functionally equivalent, such a promoter could possibly escape silencing in the background of 271 locus.
It has been demonstrated (Mourrain et al. 2007) that the mode of silencing brought about by the “271 transgene silencing locus” is via small double-stranded RNA corresponding to the 35S promoter. The production of aberrant RNAs that share homology with promoter sequences triggers TGS in trans. Mourrain et al. (2007) hypothesized that an endogenous promoter positioned at one end of the transgene sequence at 271 locus led to the accumulation of aberrant RNA that were converted into dsRNA, which when processed into siRNAs trigger silencing and methylation of homologous sequences in cis and trans. On comparison with the wild-type promoter, the synthetic promoters have a smaller target region for the siRNAs to act. There is thus a delay in the onset of silencing. However, as the targets are the cis-elements, the methylation at these sites would eventually lead to its silencing. While designing these synthetic promoters, methylation-prone CG and CNG were removed, but the cis-element in subdomain A, as-1 element, is a pentameric direct repeat TGACG, which contains two methylation prone CG dinucleotides. It has been also demonstrated earlier in a report by Diéguez et al. (1998) that cytosine methylation is not a prerequisite for the initiation of transcriptional gene silencing but is required for its maintenance. Keeping this in mind, it is possible that if the cis-elements are smaller and do not contain methylation prone CG and CNG, synthetic promoters could escape gene silencing.
References
Arumugam N, Gupta V, Jagannath A, Mukhopadhyay A, Pradhan AK, Burma PK, Pental D (2007) A passage through in vitro culture leads to efficient production of marker-free transgenic plants in Brassica juncea using the Cre-loxP system. Transgenic Res 16:703–712. doi:10.1007/s11248-006-9058-7
Benfey PN, Ren L, Chua N-H (1990) Tissue-specific expression from CaMV 35S enhancer subdomains in early stages of plant development. EMBO J 9:1677–1684
Bhattacharyya-Pakrasi M, Pen J, Elmer JS, Laco G, Shen P, Kaniewska MB, Kononowicz H, Wen F, Hodges TK, Beachy RN (1993) Specificity of a promoter from the rice tungro baciliform virus for expression in phloem tissues. Plant J 4:71–79. doi:10.1046/j.1365-313X.1993.04010071.x
Bhullar S, Chakravarthy S, Advani S, Datta S, Pental D, Burma PK (2003) Strategies for development of functionally equivalent promoters with minimum sequence homology for transgene expression in plants: cis-elements in a novel DNA context versus domain swapping. Plant Physiol 132:988–998. doi:10.1104/pp.103.020602
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi:10.1016/0003-2697(76)90527-3
Comai L, Moran P, Maslyar D (1990) Novel and useful properties of a chimeric plant promoter combining CaMV 35S and MAS elements. Plant Mol Biol 15:373–381. doi:10.1007/BF00019155
Depicker A, van Montagu M (1997) Post-transcriptional gene silencing in plants. Curr Opin Cell Biol 9:373–382. doi:10.1016/S0955-0674(97)80010-5
Dey N, Maiti IB (1999) Structure and promoter/leader deletion analysis of Mirabilis mosaic virus (MMV) full-length transcript promoter in transgenic plants. Plant Mol Biol 40:771–782. doi:10.1023/A:1006285426523
Diéguez MJ, Vaucheret H, Paszkowski J, Scheid OM (1998) Cytosine methylation at CG and CNG sites is not a prerequisite for the initiation of transcriptional gene silencing in plants, but it is required for its maintenance. Mol Gen Genet 259:207–215. doi:10.1007/s004380050806
Fagard M, Vaucheret H (2000) (Trans) gene silencing in plants: how many mechanisms? Annu Rev Plant Physiol Plant Mol Biol 51:167–194. doi:10.1146/annurev.arplant.51.1.167
Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25:989–994. doi:10.1007/BF00014672
Holmberg N, Harker M, Gibbard CL, Wallace AD, Clayton JC, Rawlins S, Hellyer A, Safford R (2002) Sterol C-24 methyltransferase type 1 controls the flux of carbon into sterol biosynthesis in tobacco seed. Plant Physiol 130:303–311. doi:10.1104/pp.004226
Iyer ML, Kumpatla SP, Chandrasekharan BM, Hall TC (2000) Transgene silencing in monocots. Plant Mol Biol 43:323–346. doi:10.1023/A:1006412318311
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405. doi:10.1007/BF02667740
Kumpatla SP, Chandrasekharan MB, Lyer LM, Li G, Hall TC (1998) Genome intruder scanning and modulation systems and transgene silencing. Trends Plant Sci 3:97–104. doi:10.1016/S1360-1385(97)01194-1
Maiti IB, Gowda S, Kierman J, Ghosh SK, Shepherd RJ (1997) Promoter/leader analysis and plant expression vectors with the Figwort mosaic virus (FMV) full-length transcript (FLt) promoter containing single and double enhancer domains. Transgenic Res 6:143–156. doi:10.1023/A:1018477705019
Matzke AM, Aufsatz W, Kanno T, Mette FM, Matzke MJA (2002) Homology-dependent gene silencing and host defence in plants. Adv Genet 46:235–273. doi:10.1016/S0065-2660(02)46009-9
Matzke MA, Matzke AJM (1995) How and why do plants inactivate homologous (trans)genes? Plant Physiol 107:679–685. doi:10.1104/pp.107.3.679
Medberry SL, Lockhart BEL, Olszewski NE (1992) The Commelina yellow mottle virus promoter is a strong promoter in vascular and reproductive tissues. Plant Cell 4:185–192. doi:10.1105/tpc.4.2.185
Meyer P, Saedler H (1996) Homology-dependent gene silencing in plants. Annu Rev Plant Phys 47:23–48. doi:10.1146/annurev.arplant.47.1.23
Mourrain P, van Blokland R, Kooter JM, Vaucheret H (2007) A single transgene locus triggers both transcriptional and post-transcriptional silencing through double-stranded RNA production. Planta 225:365–379. doi:10.1007/s00425-006-0366-1
Ni M, Cui D, Einstein J, Narasimhulu S, Vergara CE, Gelvin SB (1995) Strength and tissue specificity of chimeric promoters derived from the octopine and mannopine synthase genes. Plant J 7:661–676. doi:10.1046/j.1365-313X.1995.7040661.x
Rogers SO, Bendich AJ (1994) Extraction of total cellular DNA from plants, algae and fungi. In: Gelvin SB, Schilperoort RA (eds) Plant molecular biology manual, D1, 2nd edn. Kluwer Academic, Dordrecht, pp 1–8
Rushton PJ, Reinstadler A, Lipka V, Lippok B, Somssich IE (2002) Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell 14:749–762. doi:10.1105/tpc.010412
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Sawant S, Singh PK, Madanala R, Tuli R (2001) Designing of an artificial expression cassette for the high-level expression of transgenes in plants. Theor Appl Genet 102:635–644. doi:10.1007/s001220051691
Sijen T, Vijn I, Rebocho A, van Blokland R, Roelofs D, Mol JN, Kooter JM (2001) Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr Biol 11:436–440. doi:10.1016/S0960-9822(01)00116-6
Stam M, Mol NM, Kooter JM (1997) The silence of genes in transgenic plants. Ann Bot 79:3–12. doi:10.1006/anbo.1996.0295
Stavolone L, Kononova M, Pauli S, Ragozzino A, de Haan P, Milligan S, Lawton K, Hohn T (2003) Cestrum yellow leaf curling virus (CmYLCV) promoter: a new strong constitutive promoter for heterologous gene expression in a wide variety of crops. Plant Mol Biol 53:703–713. doi:10.1023/B:PLAN.0000019110.95420.bb
Svab Z, Hajdukiewicz P, Maliga P (1995) Generation of transgenic tobacco plants by cocultivation of leaf disks with Agrobacterium pPZP binary vectors. In: Maliga P, Klessig DF, Cashmore AR, Gruissem W, Varner JE (eds) Methods in plant molecular biology, a laboratory course manual. Cold Spring Harbour Laboratory, New York, pp 55–77
Thierry D, Vaucheret H (1996) Sequence homology requirements for transcriptional silencing of 35S transgenes and post-transcriptional silencing of nitrite reductase (trans)genes by the tobacco 271 locus. Plant Mol Biol 32:1075–1083. doi:10.1007/BF00041391
Tzafir I, Torbert KA, Locckhart BEL, Somers DA, Olszewski NE (1998) The sugarcane bacilliform badnavirus promoter is active in both monocots and dicots. Plant Mol Biol 38:347–356. doi:10.1023/A:1006075415686
Vaucheret H (1993) Identification of a general silencer for 19S and 35S promoters in a transgenic tobacco plant: 90 bp of homology in the promoter sequences are sufficient for trans-inactivation. C R Acad Sci Paris Life Sciences 316:1471–1483
Vaucheret H (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20:759–771. doi:10.1101/gad.1410506
Vaucheret H, Kronenberger J, Lepingle A, Vilaine F, Boutin J-P, Caboche M (1992) Inhibition of tobacco nitrite reductase activity by expression of antisense RNA. Plant J 2:559–569. doi:10.1046/j.1365-313X.1992.t01-25-00999.x
Vaucheret H, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel J-B, Mourrain P, Palauqui J-C, Vernhettes S (1998) Transgene-induced gene silencing in plants. Plant J 16:651–659. doi:10.1046/j.1365-313x.1998.00337.x
Vaucheret H, Christophe B, Fagard M (2001) Post-transcriptional gene silencing in plants. J Cell Sci 114:3083–3091
Verdaguer B, de Kochko A, Beachy RN, Fauquet C (1996) Isolation and expression in transgenic tobacco and rice plants, of the cassava vein mosaic virus (CVMV) promoter. Plant Mol Biol 31:1129–1139. doi:10.1007/BF00040830
Voinnet O (2008) Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci 13:317–328. doi:10.1016/j.tplants.2008.05.004
Wassenegger M, Pélissier T (1998) A model for RNA-mediated gene silencing in higher plants. Plant Mol Biol 37:349–362. doi:10.1023/A:1005946720438
Xie Y, Meng M, Chen L, Zhu Z (2003) Isolation and identification of a super strong plant promoter from cotton leaf curl Multan virus. Plant Mol Biol 53:1–14. doi:10.1023/B:PLAN.0000009257.37471.02
Acknowledgment
We thank Dr. Herve Vaucheret, INRA Centre de Versailles, France, for providing us the seed material for silencing line 271.5.8 and nonsilencing line 271.5.9. We thank Ms. Taru Gautam for her technical assistance and Dr. Anjana N. Dev for critically going through the manuscript. This work was supported by grants from Department of Biotechnology, Government of India and University of Delhi, India. SB and SD were supported by Research Fellowships from Council of Scientific and Industrial Research, Government of India.
Author information
Authors and Affiliations
Corresponding author
Additional information
Simran Bhullar and Sudipta Datta contributed equally to this study.
Rights and permissions
About this article
Cite this article
Bhullar, S., Datta, S. & Burma, P.K. Delayed Trans-inactivation of Synthetic Domain A 35S Promoters by “Tobacco 271 Locus” due to Reduced Sequence Homology. Plant Mol Biol Rep 29, 1–11 (2011). https://doi.org/10.1007/s11105-010-0202-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-010-0202-4