Abstract
The tropical spice crop ginger (Zingiber officinale Roscoe) is highly susceptible to soft rot disease caused by the necrotrophic oomycete Pythium aphanidermatum (Edson) Fitzp. However, Zingiber zerumbet (L.) Smith, a wild relative of ginger, is resistant to P. aphanidermatum and has been proposed as a potential donor for soft rot resistance to Z. officinale. We identified a member of the pathogenesis-related protein group 5 (PR-5) gene family in Z. zerumbet that is expressed constitutively but upregulated in response to infection by P. aphanidermatum. Expression of this gene was upregulated as early as 1.5 h post inoculation (hpi) with the pathogen, peaked at 6 hpi, declined by 9 hpi, and again peaked at 15 hpi before declining at 48 hpi. A cDNA of this PR-5 gene, designated as ZzPR5, encodes a 226-amino-acid predicted protein with a calculated pI of 5.05. The N terminus of this protein contains a 22-amino-acid signal peptide, suggesting that the protein may show apoplastic accumulation like other acidic PR-5 proteins. Phylogenetic analysis revealed high similarity between ZzPR5 and PR-5 proteins reported from other plant species, especially from other Zingiberales. Molecular modeling of ZzPR5 protein revealed an acidic surface cleft, a feature characteristic of glycoside hydrolases and antifungal PR-5 proteins. In molecular docking studies, a linear polymeric molecule of (1,3)-β-d-glucan, a major constituent of the oomycete cell wall, fitted favorably into the surface cleft of ZzPR5 and interacted with acidic amino acids known to be involved in glucan hydrolysis, suggesting a potential antioomycete activity for ZzPR5 protein. Elucidation of the molecular mechanism of ZzPR5 may provide important insight toward engineering soft rot resistance into the obligatory asexual ginger.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ginger (Zingiber officinale Roscoe; Zingiberaceae) is an important cash crop in tropical and subtropical countries, the rhizome being widely used as a spice and in the pharmaceutical industry (Lawrence 1984). However, soft rot disease caused by the soil-borne necrotrophic oomycete Pythium aphanidermatum (Edson) Fitzp continues to devastate much of the ginger crop. Zingiber zerumbet (L.) Smith, a wild congener of ginger is resistant to P. aphanidermatum (Kavitha and Thomas 2008a). Defense transcriptome profiling of Z. zerumbet following its inoculation with P. aphanidermatum provided strong evidence for transcriptional reprogramming of several genes known to be involved in host defense and signaling in other plants (Kavitha and Thomas 2008b). Further studies suggested that the resistance of Z. zerumbet against P. aphanidermatum is likely to be governed by a mechanism involving production of antimicrobial compounds that is independent of the hypersensitive response (HR) (Kavitha and Thomas 2008c).
Even though the gene-for-gene model, which underlies host defense against biotrophs, is known in considerable detail, relatively little is understood about the components of the basal defense mechanism that governs host defense against necrotrophs (Gomez-Gomez 2004; Okubara and Paulitz 2005; van Kan 2006). Mounting molecular evidence strongly suggests that plants interact with biotrophic and necrotrophic pathogens by using similar pathways but with opposite outcomes (Glazebrook 2005; van Kan 2006; Kliebenstein and Rowe 2008). For example, although HR can prevent the spread of an invading biotroph, it is actually beneficial to an invading necrotroph because it facilitates pathogen ingress through dead tissue, leading to host susceptibility (van Kan 2006; Kliebenstein and Rowe 2008).
Several lines of evidence suggest that the pathogenesis-related (PR)-5 subfamily of proteins (also known as thaumatin-like proteins [TLPs]) has a crucial role in host defense against oomycetes and necrotrophic pathogens (Vleeshouwers et al. 2000; van Loon et al. 2006; Asselbergh et al. 2007). First, overexpression of PR-5 genes has been shown to enhance pathogen resistance in plants (van Loon et al. 2006). Second, certain isoforms of PR-5 have strong affinity and hydrolytic activity toward (1,3)-β-d-glucans (Trudel et al. 1998; Grenier et al. 1999; Osmond et al. 2001), the major cell-wall constituents of most oomycetes (Latijnhouwers et al. 2003). Finally, high-resolution crystal structures of PR-5 proteins revealed the presence of a long, deep surface cleft, which is crucial to their antifungal activities (Koiwa et al. 1999; Osmond et al. 2001; Leone et al. 2006; Ghosh and Chakrabarti 2008).
Because ginger is obligatorily asexual and all extant ginger cultivars are uniformly susceptible to soft rot (Kavitha and Thomas 2008a, b), a transgenic approach using alien genetic resources seems the only plausible method to improve Pythium resistance in ginger. Z. zerumbet is a potential donor of soft rot resistance for the genetic improvement of ginger (Kavitha and Thomas 2008a) and our laboratory is engaged in characterizing the defense transcriptome of the Z. zerumbet–P. aphanidermatum pathosystem. In the present report, we have analyzed the expression pattern of a P. aphanidermatum-responsive PR-5 gene in Z. zerumbet, cloned and characterized its cDNA, and discussed the likely function of the protein encoded by this gene in governing defense mechanisms in Z. zerumbet.
Materials and Methods
Pathogen Inoculation, RNA Isolation, and Quantitative Real-Time PCR
A Z. zerumbet accession (accession number 2010-9), which is immune to P. aphanidermatum (Kavitha and Thomas 2008a), was used for pathogen inoculation. Rhizomes harvested from mature plants were sprouted in earthen pots in red earth/sand/leaf compost and the pseudostems were allowed to grow for 3 months before using them for inoculation. The collar region of the pseudostems was inoculated as described earlier (Kavitha and Thomas 2008a) with a field isolate of P. aphanidermatum (RGCB P117) obtained from the Indian Institute of Spices Research, Kozhikode, Kerala, India.
RNA was isolated from Z. zerumbet according to Salzman et al. (1999) at 1.5, 3, 6, 9, 12, 15, 24, and 48 h post inoculation (hpi) with P. aphanidermatum and just before inoculation (0 hpi). Total RNA was treated with RNase-free DNase I (Promega) to remove traces of genomic DNA, if any.
cDNA was synthesized from 1 µg of DNase I-treated total RNA using a SMART™ cDNA synthesis and library construction kit (BD Biosciences) and the reverse-transcribed products were diluted tenfold. Primers that are designed to the conserved regions of PR-5 genes reported from other plant species and capable of amplifying a 50-bp product were used for quantitative real-time polymerase chain reaction (qRT-PCR). The qRT-PCR was set up in a final volume of 20 µL containing 10 µL SYBR Green PCR Core Reagent (Applied Biosystems), 2 µL diluted cDNA, and 300 nM each of the designed primers (forward, 5′-agcaccgtcgtcttcacctgccccg-3′; reverse, 5′-ggcagaaggtgacactgtagttgg-3′). The PCR conditions were as follows: 50°C for 2 min initially followed by 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min in a real-time PCR machine (ABI 7500). The housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (forward, 5′-ggaattgtggaaggtttgatgac-3′; reverse, 5′-tcgtccacctctccagtcctt-3′) was used as an endogenous control. Two biological replicates were analyzed at each time point with three technical replicates for each biological sample. The comparative C T method was used for expression analysis, which mathematically transforms the threshold cycle (C T) into a relative expression level of the gene. The relative expression level of the PR-5 gene was normalized with that of the housekeeping genes. We set a threshold of twofold induction of the PR-5 gene in P. aphanidermatum-inoculated versus control (0 hpi) samples as being significant, after other studies of transcript profiling (Chen et al. 2007).
Isolation of Full-Length cDNA
To isolate full-length PR-5 cDNA, we pooled the DNA-free total RNA obtained from Z. zerumbet at four time points between 6 and 15 hpi, and 1 µg pooled sample was used to synthesize cDNAs for 5′ and 3′ rapid amplification of cDNA ends (RACE) using a SMART™ RACE cDNA amplification kit (Clontech) following the manufacturer's protocol, and the cDNA products were diluted 100-fold. Oligonucleotide primers (PR51, 5′-gccctgctctgtttcttccttttcc-3′; PR52, 5′-caagggcagaaggtgacactgtagt-3′) designed to the conserved regions of PR-5 genes cloned from other plant species were used for amplifying the cDNA ends. For 5′ RACE PCR, Universal Primer Mix A (UPM) provided with the kit and PR51 were used; for 3′ RACE PCR, UPM and PR52 were used. PCR amplification was programmed on a Mastercycler eps (Eppendorf) with 25 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 2 min. The RACE products were fractionated on agarose gels, eluted (GFX PCR DNA and Gel Band Purification Kit, Amersham Pharmacia), cloned using the pGEM-T Easy Vector System (Promega), and sequenced (ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit, Applied Biosystems) in an ABI Prism 3730 genetic analyzer (Applied Biosystems).
Primers (forward, 5′-atcatggctacatcaaccaccgccg-3′; reverse, 5′-aagggcagaaggtgacactgtagttggtt-3′) were designed based on sequence information of inserts from multiple clones derived from 5′ and 3′ RACE products and were used to amplify the full-length cDNA of PR-5 gene from Z. zerumbet using an Advantage 2 PCR Enzyme System (Clontech) according to the manufacturer's instructions. The full-length amplicon was eluted, cloned, and sequenced as described above. We designated the cDNA of PR-5 gene cloned from Z. zerumbet as ZzPR5. Sequence data from this article have been deposited at the National Center for Biotechnology Information (NCBI) under accession number FJ550342.
Sequence Analysis
Homology searches were performed with BLAST and BLASTX algorithms to confirm sequence identity. Sequence characteristics of the ZzPR5 product were assessed following multiple alignment of deduced amino acid sequences of ZzPR5 and the PR-5 genes from Hordeum vulgare (AJ001268), Musa acuminata (1Z3Q), Thaumatococcus daniellii (1RQW), and soybean P21 protein (P25096) using CLUSTALW. Phylogenetic analysis was performed on ZzPR5 and the PR-5 gene products of other plant species identified following BLASTX analysis using the neighbor-joining method implemented in the software MEGA version 4 (Tamura et al. 2007). Robustness of clustering was checked by bootstrapping 1,000 replicates.
Molecular Modeling
The three-dimensional (3D) structure of ZzPR5 was generated using the software MODELLER. The aim of our comparative modeling was to generate the most probable structure of the ZzPR5 protein through alignment with template sequences that simultaneously satisfied spatial restraints and local molecular geometry. A BLASTX search of the Protein Databank demonstrated high sequence identity (65%) between ZzPR5 and M. acuminata PR-5 protein (1Z3Q), and models of ZzPR5 protein were constructed based on the crystal structure coordinates of 1Z3Q obtained at a resolution of 1.7 Å resolution (Leone et al. 2006). The structure generated for ZzPR5 was improved further by refinement of the loop conformations after assessing the compatibility of its amino acid sequences with the 1Z3Q structure using the Protein Health module in Discovery Studio (DS Modeling 1.1; Accelrys, San Diego, CA, USA). All simulation experiments were carried out using a DELL PowerEdge T300 Workstation with Quad Dual Intel Xeon 3.2 GHz processors.
Backbone conformation of the refined structure was evaluated by inspection of the Psi/Phi angles in a Ramachandran plot generated using the software PROCHECK version 3.5 (Laskowski et al. 1993) and comparing the Z score of ZzPR5 and the template (1Z3Q), estimated using the software Prosa2003 (Sippl 1993). Deviation of the modeled structure from the template was assessed by computing root mean square deviation (RMSD) values for positional differences between equivalent atoms following the superposition of Cα traces and backbone atoms of the model onto the template. The protein structure image of the model was illustrated using the software PyMOL.
Docking Simulations Between (1,3)-β-d-Glucan and ZzPR5
To examine the affinity between ZzPR5 and (1,3)-β-d-glucan, we performed molecular docking experiments. The ZzPR5 homology model was used as the template and docking was carried out by introducing protein flexibility into the docking protocol through the generation of an ensemble of protein conformations using molecular dynamic (MD) simulations performed in the Discovery Studio Simulation module (Discovery Studio, Accelrys). Calculations were performed using the CHARMm Force-Fields PARM22 and the polysaccharide-specific CHEAT95 (Kouwizjer and Grootenhuis 1995), as implemented in Discovery Studio. The simulation was performed for 500 ps with a time step of 0.001 and the temperature set to 300 K. The production type was set to NPT ensemble (constant temperature and pressure dynamics) with a radius of nonbonded interaction of 14.0 Å. For the estimation of protein–ligand complexes, LigandFit score was employed as a scoring function. Docking characteristics of ZzPR5–(1,3)-β-d-glucan complex was also examined using the software AutoDock 4.1 (Morris et al. 1998).
Results
Expression Profile of the PR-5 Gene of Zingiber zerumbet
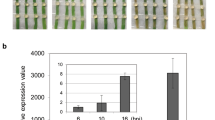
While investigating changes in expression of genes in Z. zerumbet in response to P. aphanidermatum inoculation, we identified a member of the PR-5 protein gene family that was strongly induced following pathogen treatment. Figure 1 shows the expression pattern for this PR-5 gene, determined by qRT-PCR analysis, at different time points following P. aphanidermatum inoculation. The gene was found to be expressed constitutively in Z. zerumbet. Upregulation of this gene was detected as early as 1.5 hpi with P. aphanidermatum and the mRNA level was significantly elevated (greater than twofold the basal level) at 3 hpi. The qRT-PCR analysis recorded two expression maxima for this gene, one at 6 hpi and the other between 15 and 24 hpi, before declining at 48 hpi (Fig. 1).
Cloning and Sequence Analysis of ZzPR5
A full-length cDNA of the PR-5 gene of Z. zerumbet was cloned using RACE PCR; the cDNA was designated as ZzPR5. ZzPR5 is 894 bp in length and has an open reading frame of 680 nucleotides. A GenBank BLASTp search revealed the conceptual gene product to have the highest identity (65%) with the M. acuminata PR-5 protein (accession number 1Z3Q). ZzPR5 encodes a precursor protein of 226 amino acid residues with a predicted signal peptide of 22 amino acid residues at the N terminus (Fig. 2). The conceptual protein has an apparent molecular weight of 57.5 kDa and a theoretical pI of 5.05. Multiple sequence alignment revealed 16 conserved cysteine residues (Fig. 2), which participate in eight disulfide bonds characteristic of most PR-5 proteins (Campos et al. 2002).
Multiple sequence alignment of the deduced ZzPR5 protein with PR-5 proteins from other taxa viz., M. acuminata (1Z3Q_A), T. danielli (1RQW), P21 protein from Glycine max (P25096), and H. vulgare (AJ001268). Asterisks indicate 16 conserved cysteine residues and the arrow indicates the cleavage site between the signal peptide and mature protein
BLASTp searches using ZzPR5 sequences retrieved altogether 30 PR-5 proteins (or TLPs) reported from different plant species, and we examined the phylogenetic relationship between ZzPR5 and these protein sequences. The resulting tree is presented in Fig. 3. ZzPR5 was found to be closest to PR-5 proteins isolated from two other Zingiberales, M. acuminata (1Z3Q) and T. danielli (1THW; Fig. 3). High bootstrap values supported the robustness of the clustering.
Molecular Modeling
The crystal structure of M. acuminata PR-5 (1Z3Q) was used as template for homology modeling of ZzPR5. The 3D models obtained for ZzPR5 are shown in Fig. 4a, b. The main chain conformations for 97.8% of the amino acid residues were within the favored or allowed regions of the Ramachandran plot. The G-factor values, indicating the quality of covalent bond lengths and bond angles, were −0.86 for dihedrals and −0.25 for covalent bonds with an overall value of −0.57. Superposition of the Cα atoms of the template (1Z3Q) and target protein (ZzPR5) gave a calculated RMSD of 0.083 Å. Prosa2003 analysis yielded a Z score of −5.11 for ZzPR5 and −5.23 for the template (1Z3Q). Analysis of the secondary structure revealed the presence of five α-helices, 14 β-strands, and 28 turns in ZzPR5.
3D model of the ZzPR5 protein. a Ribbon representation showing the three domains: green domain I, orange domain II, and magenta domain III. Structure obtained by energy minimizing the average conformation over the last 1,000 fs of a MD simulation. b Molecular surface showing electrostatic potential on a scale that varies from red to blue, representing negative and positive potential, respectively. The arrow indicates the interdomain acidic cleft
Three distinct domains are evident in the 3D model of ZzPR5 (Fig. 4a). Domain I, which corresponds to the N terminus of the protein, consists of two bundles of antiparallel β-sheet connected by loops to form a flattened β-sandwich. Domain II comprises a main α-helix associated with shorter helical segments, and domain III consists of β-strands linked by a loop. A prominent deep cleft that transverses the protein surface is found at the interface of domains I and II (Fig. 4b). The side chains of residues Arg49, Tyr76, Asn81, Glu85, Ser100, Val102, Cys145, Thr146, Cys154, and Tyr177 intrude into the cleft. Calculation of the electrostatic potential of the ZzPR5 model revealed the cleft to be highly acidic (Fig. 4b).
Molecular Docking Studies
We undertook docking simulations between ZzPR5 and polymeric (1,3)-β-d-glucan, the cell-wall constituent of most oomycetes (Latijnhouwers et al. 2003), to investigate the glucan binding properties and glucanase potential of the former. Due to conformational constraints in accommodating insoluble glucans in docking studies (Osmond et al. 2001), we used a single chain consisting of five (1,3)-β-d-glucan units. The model produced by Discovery Studio Simulation predicts that eight amino acid residues, Arg49, Tyr76, Glu85, Asp98, Ser100, Asp143, Thr146, and Cys154, are involved in hydrogen bonding with the docked carbohydrate (Fig. 5). Docking studies using AutoDock 4.1 predicted the involvement of four of these residues (Tyr76, Glu85, Asp98, and Ser100) in hydrogen bonding with the docked molecule (data not shown). In the docked complex, (1,3)-β-d-glucan passed through the acidic interdomain cleft of ZzPR5 with Glu85 and Asp98 on either side. The calculated distance between the carboxylic oxygen atoms of the Glu85–Asp98 pair is 3.34 Å, which is well within the distance of 5 Å required between the two catalytic residues for glucanase activity (Zechel and Withers 2001).
Discussion
The PR-5 gene identified in Z. zerumbet was found to be expressed constitutively. Constitutively expressed defense-related genes have a vital role in the defense machinery of plants because they help the host to block the pathogen at the attempted point of penetration (Vleeshouwers et al. 2000; Zhang et al. 2007; Fung et al. 2008). For example, Solanum tuberosum cultivars with very high constitutive expression of PR genes, including PR-5, were resistant to the oomycete pathogen Phytophthora infestans, whereas cultivars with no or very low levels were susceptible (Vleeshouwers et al. 2000). Similarly, quick and robust induction of pathogenesis-related genes is another factor deciding the host's effectiveness in suppressing an invading pathogen (Pritsch et al. 2000; Ge et al. 2007). Thus, the very early (3 hpi with P. aphanidermatum) and robust (nearly 2.5-fold) accumulation of PR-5 transcripts further indicates a crucial role for this gene in the P. aphanidermatum resistance machinery of Z. zerumbet.
Induction of the PR-5 gene in Z. zerumbet following P. aphanidermatum inoculation is biphasic, and similar expression profiles have been reported for other defense-related genes (Alignan et al. 2006; Adhikari et al. 2007). The early expression peak (3 to 6 hpi) could reflect a quick reinforcement of defense machinery at the attempted site of pathogen ingress, perhaps by sensing the physical force exerted by the pathogen as hypothesized recently by Hardham et al. (2008). The second expression peak (15 to 24 hpi) is likely to be a result of a heightening of basal defense governed by a signaling cascade.
Several lines of evidence suggest that HR and its associated defense signaling, as well as PR-5 protein accumulation—all common defense mechanisms against biotrophs (Glazebrook 2005)—actually increase the host's vulnerability against necrotrophs (Glazebrook 2005; van Kan 2006; Kliebenstein and Rowe 2008). Because the resistance mechanism in Z. zerumbet against P. aphanidermatum is HR-independent (Kavitha and Thomas 2008c), a signaling process which circumvents HR may have elicited the secondary induction of this PR-5 gene.
The ZzPR5 protein is acidic and possesses an N-terminal signal peptide, suggesting that it accumulates in the apoplast, as do other acidic PR-5 proteins (Thompson et al. 2006). ZzPR5 also contains 16 conserved cysteine residues (Campos et al. 2002) that form eight disulfide bridges believed to confer on PR-5 proteins resistance to proteases and pH-induced or heat-induced denaturation (Breiteneder 2004).
Molecular modeling revealed that ZzPR5 possesses the 3D folds and core structures that are conserved in other PR-5 proteins (Koiwa et al. 1999; Osmond et al. 2001; Thompson et al. 2006; Ghosh and Chakrabarti 2008). ZzPR5 possesses a prominent interdomain surface cleft, a feature characteristic of most PR-5 proteins with antifungal activity (Koiwa et al. 1999; Ghosh and Chakrabarti 2008) and glycoside hydrolase enzymes (Osmond et al. 2001). Indeed, many PR-5 proteins hydrolyze or at least bind (1,3)-β-d-glucans (Trudel et al. 1998; Grenier et al. 1999; Osmond et al. 2001), and this cleft is the candidate region for (1,3)-β-d-glucan binding (Osmond et al. 2001; Ghosh and Chakrabarti 2008). The level of affinity calculated in silico between the cleft of a PR-5 protein and a (1,3)-β-d-glucan molecule is a good predictor of the protein's β-1,3-glucanase activity and antifungal efficacy in vivo (Osmond et al. 2001; Ghosh and Chakrabarti 2008). The high affinity calculated in the docking studies and the favorable orientation of the Glu–Asp pair in the docked complex, as found in glucanases (Zechel and Withers 2001; Ghosh and Chakrabarti 2008), signifies a hydrolytic potential for ZzPR5 on (1,3)-β-d-glucans. PR-5 proteins are believed to disrupt proper assembly of the fungal cell wall during hyphal extension by binding to nascent (1,3)-β-d-glucan molecules (Osmond et al. 2001). Accumulation of ZzPR5 in the apoplast, inferred from its N-terminal signal peptide, would allow the protein to engage the pathogen at its initial site of penetration (Ellis et al. 2006) and where the invading oomycetes secrete effector substances (Song et al. 2009).
PR-5 proteins have diverse functions, and thus the regulation of different PR-5 genes also varies (Breiteneder 2004; van Loon et al. 2006). Similarly, the affinity of PR-5 proteins for carbohydrates varies between proteins (Trudel et al. 1998; Grenier et al. 1999; Osmond et al. 2001), so that different PR-5 isoforms have different antifungal specificities, able to inhibit the growth of certain fungal species but not others (Osmond et al. 2001). With the premise that the nature and quantity of carbohydrate constituents of cell walls vary from pathogen to pathogen, Osmond et al. (2001) opined that plants might have coevolved different PR-5 isoforms capable of inhibiting evolving pathogen populations that have different cell-wall carbohydrate compositions. The ZzPR5 protein of Z. zerumbet identified in this study likely possesses hydrolytic activity against the (1,3)-β-d-glucans of P. aphanidermatum.
In conclusion, our study identified a potential component of the basal defense of Z. zerumbet against P. aphanidermatum. Elucidation of its molecular mechanism in governing resistance of Z. zerumbet against P. aphanidermatum may provide valuable insights for engineering soft rot resistance into the obligatory asexual ginger.
Abbreviations
- hpi:
-
hours post inoculation
- HR:
-
hypersensitive response
- PR:
-
pathogenesis-related
- qRT-PCR:
-
quantitative real-time polymerase chain reaction
- RACE:
-
rapid amplification of cDNA ends
- RMSD:
-
root mean square deviation
- ROS:
-
reactive oxygen species
- TLP:
-
thaumatin-like protein
- ZzPR5:
-
Zingiber zerumbet pathogenesis-related protein 5
References
Adhikari TB, Balaji B, Breeden J, Goodwin SB (2007) Resistance of wheat to Mycosphaerella graminicola involves early and late peaks of gene expression. Physiol Mol Plant Pathol 71:55–68
Alignan M, Hewezi T, Petitprez M, Dechamp-Guillaume G, Gentzbittel L (2006) A cDNA microarray approach to decipher sunflower (Helianthus annus) responses to the necrotrophic fungus Phoma macdonaldi. New Phytol 170:523–536
Asselbergh B, Curvers K, Franca SC, Audenaert K, Vuylsteke M, Breusegem FV, Hofte M (2007) Resistance to Botrytis cinerea in sitiens, an abscisic acid-deficient tomato mutant, involves timely production of hydrogen peroxide and cell wall modifications in the epidermis. Plant Physiol 144:1863–1877
Breiteneder H (2004) Thaumatin-like proteins-a new family of pollen and fruit allergens. Allergy 59:479-481
Campos MA, Ribeiro SG, Rigden DJ, Monte DC, Grossi de Sa MF (2002) Putative pathogenesis-related genes within Solanum nigrum L. var. americanum genome: isolation of two genes coding for PR5-like proteins, phylogenetic and sequence analysis. Physiol Mol Plant Pathol 61:205–216
Chen LR, Chen YJ, Lee CY, Lin TY (2007) MeJA-induced transcriptional changes in adventitious roots of Bupleurum kaoi. Plant Sci 173:12–24
Ellis J, Catanzariti AM, Dodds P (2006) The problem of how fungal and oomycetes avirulence proteins enter plant cells. Trends Plant Sci 11:61–63
Fung RWM, Gonzalo M, Fekete C, Kovacs LG, He Y, Marsh E, McIntyre LM, Schachtman DP, Qiu W (2008) Powdery mildew induces defense-oriented reprogramming of the transcriptome in a susceptible but not in a resistant grapevine. Plant Physiol 146:236–249
Ge X, Li GJ, Wang SB, Zhu H, Zhu T, Wang X, Xia Y (2007) AtNUDT7, a negative regulator of basal immunity in Arabidopsis, modulates two distinct defense response pathways and is involved in maintaining redox homeostasis. Plant Physiol 145:204–215
Ghosh R, Chakrabarti C (2008) Crystal structure analysis of NP24-I: a thaumatin-like protein. Planta 228:883–890
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227
Gomez-Gomez L (2004) Plant perception systems for pathogen recognition and defence. Mol Immunol 41:1055–1062
Grenier J, Potvin C, Trudel J, Asselin A (1999) Some thaumatin-like proteins hydrolyse polymeric β-1,3-glucans. Plant J 19:473–480
Hardham AR, Takemoto D, White RG (2008) Rapid and dynamic subcellular reorganization following mechanical stimulation of Arabidopsis epidermal cells mimics responses to fungal and oomycete attack. BMC Plant Biol 8:63
Kavitha PG, Thomas G (2008a) Population genetic structure of the clonal plant Zingiber zerumbet (L.) Smith (Zingiberaceae), a wild relative of cultivated ginger, and its response to Pythium aphanidermatum. Euphytica 160:89–100
Kavitha PG, Thomas G (2008b) Defence transcriptome profiling of Zingiber zerumbet (L.) Smith by mRNA differential display. J Biosci 33:81–90
Kavitha PG, Thomas G (2008c) Expression analysis of defense-related genes in Zingiber (Zingiberaceae) species with different levels of compatibility to the soft rot pathogen Pythium aphanidermatum. Plant Cell Rep 27:1767–1776
Kliebenstein DJ, Rowe HC (2008) Ecological costs of biotrophic versus necrotrophic pathogen resistance, the hypersensitive response and signal transduction. Plant Sci 174:551–556
Koiwa H, Kato H, Nakatsu T, Oda J, Yamada Y, Sato F (1999) Crystal structure of tobacco PR-5d protein at 1.8 Å resolution reveals a conserved acidic cleft structure in antifungal thaumatin-like proteins. J Mol Biol 286:1137–1145
Kouwizjer MLCE, Grootenhuis PDJ (1995) Parametrization and application of CHEAT95, an extended atom force field for hydrated oligosaccharides. J Phys Chem 99:13426–13436
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemistry of protein structures. J Appl Crystallgr 26:283–291
Latijnhouwers M, de Wit PJGM, Govers F (2003) Oomycetes and fungi: similar weaponry to attack plants. Trends Microbiol 11:462–469
Lawrence BM (1984) Major tropical spices—ginger (Zingiber officinale Rosc.). Perfum Flavor 9:1–40
Leone P, Menu-Bouaouiche L, Peumans WJ, Payan F, Barre A, Roussel A, Van Damme EJ, Rougé P (2006) Resolution of the structure of the allergenic and antifungal banana fruit thaumatin-like protein at 1.7 Å. Biochimie 88:45–52
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J Comput Chem 19:1639–1662
Okubara PA, Paulitz TC (2005) Root defense responses to fungal pathogens: a molecular perspective. Plant Soil 274:215–226
Osmond RI, Hrmova M, Fontaine F, Imberty A, Fincher GB (2001) Binding interactions between barley thaumatin-like proteins and (1,3)-β-D-glucans. Kinetics, specificity, structural analysis and biological implications. Eur J Biochem 268:4190–4199
Pritsch C, Muehlbauer GJ, Bushnell WR, Somers DA, Vance CP (2000) Fungal development and induction of defense response genes during early infection of wheat spikes by Fusarium graminearum. Mol Plant Microbe Interact 13:159–169
Salzman RA, Fujita T, Zhu-Salzman K, Hasegawa PM, Bressan RA (1999) An improved RNA isolation method for plant tissues containing high levels of phenolic compounds or carbohydrates. Plant Mol Biol Rep 17:11–17
Sippl MJ (1993) Recognition of errors in three-dimensional structures of proteins. Proteins 17:355–362
Song J, Win J, Tian M, Schornack S, Kaschani F, Ilyas M, van der Hoorn R, Kamoun S (2009) Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc Natl Acad Sci U S A 106:1654–1659
Tamura K, Dudley J, Nei M, Kumar S (2007) Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson CE, Fernandes CL, de Souza ON, Salzano FM, Bonatto SL, Freitas LB (2006) Molecular modeling of pathogenesis-related proteins family 5. Cell Biochem Biophys 44:385–394
Trudel J, Grenier J, Potvin C, Asselin A (1998) Several thaumatin-like proteins bind to β-1,3-glucans. Plant Physiol 118:1431–1438
van Kan JAL (2006) Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci 11:247–253
van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162
Vleeshouwers VGAA, Dooijeweert WV, Govers F, Kamoun S, Colon LT (2000) Does basal PR gene expression in Solanum species contribute to non-specific resistance to Phytophthora infestans? Physiol Mol Plant Pathol 57:35–42
Zechel DL, Withers SG (2001) Dissection of nucleophilic and acid-base catalysis in glycosidases. Curr Opin Chem Biol 5:643–649
Zhang X, Dai Y, Xiong Y, DeFraia C, Li J, Dong X, Mou Z (2007) Overexpression of Arabidopsis MAP kinase kinase 7 leads to activation of plant basal and systemic acquired resistance. Plant J 52:1066–1079
Acknowledgments
ANR and KAG gratefully acknowledge the Council for Scientific and Industrial Research (CSIR), Government of India for the research fellowships (F. No. 09/716/(0090)/2007/EMR-I to ANR and F. No. 09/716/(0103)/2008/EMR-I to KAG) and GT gratefully acknowledges the Department of Biotechnology (DBT), Government of India for the research grant (Grant No. BT/PR2211/Agr/08/162/2001).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Aswati Nair, R., Kiran, A.G., Sivakumar, K.C. et al. Molecular Characterization of an Oomycete-Responsive PR-5 Protein Gene from Zingiber zerumbet . Plant Mol Biol Rep 28, 128–135 (2010). https://doi.org/10.1007/s11105-009-0132-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-009-0132-1