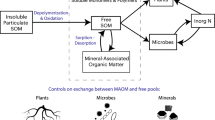
Abstract
Background and aims
The amount and type of root exudates can influence P availability in the rhizosphere directly by desorption or dissolution of soil minerals, or indirectly by decomposition of soil organic matter (SOM). This study aimed to determine the mechanisms by which specific root exudates influence the distribution and availability of P in soils with low P availability.
Methods
Water, glucose, alanine, and oxalate were delivered through a simulated root into soils for 15 days. Zymography and planar optodes were used to image potential phosphatase activity, and O2 and pH distribution, respectively. Soils were analyzed for resin extractable inorganic P (Pi), dissolved organic C (DOC), water soluble Fe, and Al, and microbial community structure. Characterization of SOM and P were conducted using ultra-high resolution mass spectrometry and 31P solution nuclear magnetic resonance (NMR), respectively.
Results
The addition of oxalate resulted in the greatest resin extractable Pi, DOC, and water-soluble Fe, and Al compared to the other exudates suggesting destabilization of mineral associated organic matter (MAOM) and release of organic P (Po). Both 31P solution NMR and ultra-high resolution mass spectrometry analysis provided evidence of mineralization of Po released from the destabilization of MAOM.
Conclusion
The study demonstrates the important role microbial and plant-derived metal chelating ligands play in destabilizing MAOM, releasing SOM and importantly Po, that when mineralized may contribute to increasing Pi availability in soils with low P availability.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Phosphorus is an essential macronutrient for proper plant and microorganism growth and function. It strongly binds to soil minerals (such as Al- and Fe-oxide-hydroxides) making it the least mobile and least available nutrient for plants especially in highly weathered tropical soils. Generally, under P limiting conditions plants can increase P availability and acquisition using a variety of physical and chemical mechanisms such as alteration of root system architecture, secretion of organic anions, acidification of the rhizosphere, and production of extracellular enzymes (Jones 1998). Plants may also indirectly enhance P acquisition via symbiotic interactions with rhizosphere microorganisms (e.g., arbuscular mycorrhizal fungi) or through the release of root exudate compounds that promote the growth of P mobilizing bacteria (Rodríguez et al. 2006; Bulgarelli et al. 2013).
Root exudates are organic compounds released from intact root cells and generally divided into low molecular weight (LMW; < 600 Da) compounds (Lehmann and Kleber 2015), root secretions, and root excretions (McNear 2013). The large concentration gradient of LMW organic compounds between the cytosols of epidermal root cells and the rhizosphere promotes outward diffusion of organic compounds. Plants can release about 5─10% of net fixed C as exudates and sugars making up the largest component of the exudate pool with a significant proportion as glucose (~ 40 − 50%) (Hütsch et al. 2002; Jones et al. 2004). Low molecular weight organic anions are usually present in the form of di- and tri- carboxylate anions such as malate, citrate, and oxalate. They may contain varying amounts of pH-dependent negative charge facilitating complexation with metal cations that can destabilize mineral-organic complexes and release bound anions (such as Pi) into soil solution. Low molecular weight organic anions destabilize mineral-organic complexes via direct abiotic interactions with mineral surfaces and MAOM. Destabilization of mineral-organic complexes mobilizes formally protected MAOM, thus making organic matter available for microbial degradation and assimilation (Clarholm et al. 2015). For example, several researchers have found that destabilization of mineral-organic complexes by simulated root exudates (e.g., oxalate) increased the bioavailability of OM leading to greater C mineralization (Keiluweit et al. 2015) and N utilization (Yuan et al. 2018). It is also possible that destabilization of mineral-organic complexes may increase Po accessibility for microbial degradation as it does with organic nitrogen (Daly et al. 2021; Jilling et al. 2021). Several studies have found that destabilization of organic-mineral complexes by organic anions releases Po bound to soil minerals (Fox et al. 1990; Fox and Comerford 1992; Wei et al. 2010) however, the fate of the released Po was unclear. The Po released from the destabilization of MAOM likely resulted in greater Pi availability via enzymatic hydrolysis and may represent a potential pathway of increasing P availability for plants.
Plants release a mixture of complex organic compounds and exudation of organic compounds are influenced by several factors (e.g., plant species and cultivar, plant growth stage, plant nutrition, microorganisms, environmental conditions, etc.) making it difficult to generalize about the composition of organic compounds present in root exudates (Grayston et al. 1997; Rovira 1969). Different chemical classes of exudates have been identified (Jones et al. 2009; Guo et al. 2015) and their roles in nutrient acquisition, signaling, pathogen resistance, symbiotic associations, etc. determined (Geurts and Franssen 1996; Paterson 2003; Marschner 2008). Depending on the type of exudate compound and their carbon use efficiency (CUE), microbes can utilize these compounds rapidly and efficiently as a source of C or N or both. Among exudate compounds, glucose is an easily available source of C for microorganisms with high CUE followed by amino acids which provide N in addition to C. Although organic anions have low CUE, it has been shown that microorganisms can readily take up and utilize organic anions for both cell growth and respiration (McAllister and Lepo 1983; Jones et al. 1996). Glucose released as exudate can provide readily available C for microorganisms as an energy supply which is then used to enhance degradation of SOM, in a process described as rhizosphere priming, improving access to other nutrients such as N (Dijkstra et al. 2009) and possibly P (Joshi et al. 2021). In N limited soil, the microbial decomposer community utilizes root exudates as a C source and acquires N via decomposition of SOM (i.e., priming) (Kuzyakov et al. 2000). It is generally considered that C and P cycling in soils are decoupled (McGill and Cole 1981) and thus, P limitation in soil does not trigger priming. However, recent studies suggest coupled microbial cycling of organic C and P, and plants may release exudate compounds to induce greater microbial mineralization of Po as a strategy to increase Pi availability (Spohn and Kuzyakov 2013; Spohn et al. 2013; Joshi et al. 2021).
The objective of this study is to investigate the mechanisms by which specific root exudates enhance the availability of mineral-bound Po from a soil with low P availability. We used three common components of root exudates differing in their mode of action (microbial C-source (i.e., glucose), microbial C and N source (i.e., alanine), and metal chelate (i.e., oxalate)) and tested their influence on the biotic and abiotic processes driving P availability in soil with naturally lesser available P. We hypothesized that glucose would serve primarily as a microbial C source inducing the greatest SOM mineralization in order to access P (and N), while alanine (containing both C and N) would result in lesser mineralization of SOM. Oxalate is not an efficient microbial C source but is an effective metal chelate; therefore, we hypothesized that P availability will increase by both the release of mineral bound Pi and mineralization of released mineral associated Po by enzymatic hydrolysis.
Materials and methods
Experimental setup
Rhizoboxes (14 cm × 10 cm × 2 cm, Fig. SI 1 a) were prepared by first filling with 125 g of Sadler silt loam soil (Table SI 1) to a final soil bulk density of 1.2 g cm−3. A simulated root consisting of a 7.5 cm long, 3 mm diameter, 0.22 µm porous wick material (Rhizotron™, Rhizosphere Research Products, Wageningen, Netherlands) was then inserted into the center of the rhizobox and the open face of the rhizobox covered with a transparent acrylic plate and then a black acrylic plate to omit light during the experiment. Boxes were then equilibrated for 1 week at 75% gravimetric water holding capacity. After equilibration, glucose, alanine, and oxalate were added to individual boxes on a total C basis using syringe pumps at a rate of 15 µmol C cm−2 d−1 per rhizobox at 1 mL d−1 assuming that approximately 10 g of soil surrounding the root are affected (Keiluweit et al. 2015). To each rhizobox, 120 µmol C d−1 was added for a total of 1.8 mmol of C over the 15 days experiment. Glucose, oxalate (Sigma Aldrich catalog number G8270 and 241172, respectively), and alanine (Acros Organics catalog number 102830250) were chosen as representative root exudates for their distinctly different acidity (pKa), microbial CUE, and sorptive affinity (Table 1). Glucose and Alanine were prepared as is (pH ~ 7) while oxalate was prepared by adjusting the pH of oxalic acid from two to four with dropwise addition of NaOH producing equal proportions of HC2O4− and C2O42− (oxalate) anions. Control boxes received 1 mL d−1 of water (control, hereafter) to test for physical effects from simulated root placement and the effects of wetting on SOM degradation and mineralization that occurred in the absence of rhizosphere priming. Triplicate rhizoboxes were used for each treatment and destructively harvested for analysis on day 15. Because the methods used to evaluate rhizosphere processes were destructive, the experiment was repeated three times. Each repeated experiment consisted of 12 rhizoboxes (3 replicates × 4 root exudate treatments (water, glucose, alanine, oxalate)) arranged in a randomized complete block format. The first experimental repeat was used to visualize the spatial distribution of potential acid phosphatase activities using zymography. The second was used to determine the in-situ distribution of pH and O2 using planar optodes. The third experimental repeat was used to collect soil samples 0 − 4 mm (rhizosphere soil), 5 − 12 mm (intermediate soil), and 13 − 50 mm (bulk soil) from each side of the simulated roots for biogeochemical analysis.
In-situ measurement of potential phosphatase activity
Zymography was used to determine the spatial distribution of potential phosphatase enzyme activities around the simulated roots following the protocol described in (Razavi et al. 2016). Briefly, a polyamide membrane filter paper with diameter of 20 cm and pore size of 0.45 µm was cut into 10 × 10 cm2 and saturated with a 12 mmol L−1 4-methylumbelliferyl-phosphate substrate prepared in MES buffer and pH adjusted to soil pH i.e., ~ 5.54. At the time of sampling, rhizoboxes were opened and the saturated membrane applied directly to the soil surface covering rhizosphere, intermediate and bulk soils (Fig. SI 1 b). 4-methylumbelliferyl-phosphate on the membrane is hydrolyzed to 4-methylumbelliferyl by any phosphatase enzyme present in the soil sample providing an estimate of the distribution of the enzyme. After 1 h incubation in the dark, the membrane was carefully removed from the soil surface and any adhering soil particles gently removed using tweezer. The membrane was then immediately placed in a dark cabinet under UV light (λem = 355 nm) and the fluorescent distribution of 4-methylumbelliferyl imaged (λem = 460 nm) using a digital camera (Canon Powershot SX10IS, Melville, NY) set at a fixed distance between the UV light source and the sample. To correlate fluorescence intensity in the zymographs with concentration, a standard calibration was obtained by imaging 4 cm2 membranes soaked in 25, 50, 100, 250, 500, or 1000 µM solution of 4-methylumbelliferyl substrate (Razavi et al. 2016). On an area basis, the amount of 4-methylumbelliferyl was calculated from the concentration of the solution, size, and the volume of solution taken up by the membrane. Images were processed and analyzed using the open-source software Fiji (Schindelin et al. 2012; Spohn and Kuzyakov 2014).
In-situ measurement of soil pH and O2 concentration
Planar optodes (PreSens Precision Sensing GmbH, Regensburg, Germany) were used to visualize the spatial distribution of soil pH and O2 concentration after addition of exudates and water. Three self-adhesive pH sensitive and three O2 sensitive foils (10 cm × 1 cm each) were alternately attached to the inside of the transparent acrylic sheet covering the open face of the rhizobox (Fig. SI 1 c). The plate was attached to the box using double sided tape with the sensor foils in direct contact with the rhizosphere, intermediate, and bulk soils. The transparent plate bearing the sensor foils was then covered with a black, opaque plate to omit light during the experiment. Measurements were taken for spatially distributed pH and O2 concentration at 15 days using the VisiSens TD Basic System with the Big Area Imaging Kit (PreSens Precision Sensing GmbH, Regensburg, Germany). The resulting images were relativized against calibration curves using the PreSens Scientific software (PreSens Precision Sensing GmbH, Regensburg, Germany). For O2, a two-point calibration curve was created using O2 free water and the highest observed O2 content from the control boxes. Oxygen free conditions were created by adding sodium sulfite to water until all measurable O2 was depleted as determined by a dissolved O2 meter. A six- point calibration curve ranging from pH 4.99–7.52 in 0.5 pH unit increments was created for the analysis of pH using the PreSens CaliPlate system (PreSens Precision Sensing GmbH, Regensburg, Germany), where all pH solutions were maintained at a standard ionic strength as recommended by the manufacturer’s protocol. The open-source platform Fiji was then used to generate plots showing the end-to-end distribution of pH or O2 across the foil (Schindelin et al. 2012).
Determination of resin extractable Pi, water-soluble Fe, Al, and DOC
Rhizosphere soils were extracted using water to determine resin extractable Pi (resin-Pi, hereafter), DOC, Fe and Al. Briefly, a HCO3−1 saturated 4 × 4 cm2 resin strip (anion exchange membrane, VWR International) was added to a 50 mL falcon tube containing 0.5 g of fresh rhizosphere soil and 30 mL of water and shaken for 16 h. Resin strips were then removed, washed in water, and placed in 10 mL of 0.2 N HNO3 for 12 h to desorb Pi. The molybdate reactive P concentration in the solution was measured by using the phosphomolybdate blue method (Murphy and Riley 1962).The tube with remaining soil suspension was centrifuged and the supernatant collected to quantify water soluble Fe and Al using ICP-OES and the DOC content using a Flash Elemental Analyzer 1112 (Thermo Fisher Scientific Inc., Waltham, MA).
Extraction and characterization of phosphorus by 31P solution NMR
Phosphorus forms in soil samples were extracted and characterized using 31P solution NMR. First, P was extracted by shaking 1.5 g of rhizosphere soils with 30 mL of a solution containing 0.25 mol L−1 NaOH and 0.05 mol L−1 EDTA for 4 h at 20 °C (Cade-Menun and Preston 1996). After extraction, the samples were centrifuged at 10,000 g for 30 min to separate and remove the solution from the soil. Then an aliquot was collected to determine total P extracted and the Fe concentration. The remaining solution was frozen immediately, lyophilized, and ground to a fine powder. Before the analysis, 1 g of lyophilized sample was dissolved in 1.5 mL of 1 mol L−1 NaOH and then centrifuged at 10,000 g for 10 min to remove any fine particles. Then 630 µL of sample supernatant was mixed with 70 µL D2O (i.e., 10%) and the mixture transferred into a 5 mm NMR tube for measurement. To aid in identifying P species in soil extracts, spiking experiments were conducted using a set of stock solutions of selected P compounds, based on recommendations outlined in (Cade-Menun 2015) (see supporting information for more detail).
NMR measurements were performed on a Bruker Avance III spectrometer operating at a field strength of 17.6 T (303.70 MHz, 31P) and equipped with a Bruker BBO broadband tunable, direct detect 5 mm probe. All experiments were conducted at a regulated temperature of 20 °C (Cade-Menun and Liu 2013). Parameters used to collect sample spectra included a spectral width of 100 ppm, 90° pulse excitation, an acquisition time of 0.8 s (49,278 total points), and a relaxation delay of 7.5 s (recycle delay of 8.3 s based on the measured T1 of 1.66 s of the orthophosphate resonance). No proton de-coupling was employed. The number of transients acquired for each spectrum was 3,072 and post-acquisition processing included zero-filling to 131,072 points and multiplication by a decaying exponential function (line-broadening, typically between 2—10 Hz) prior to Fourier transform.
Spectra were processed in Bruker’s TopSpin 3.6.2 and included aligning spectra by setting the orthophosphate peak at 6.0 ppm (unit of the chemical shift) for comparison during analysis with values in the 31P compound spectral library in (Cade-Menun 2015). The actual orthophosphate peak position referenced to externally measure 85% H3PO4 was about 6.5 ppm. Spectra were baseline corrected and integrated based on regions where visible signal intensity was observed in at least one spectrum. The regions integrated included orthophosphate between 6.70 and 5.51 ppm, orthophosphate monoesters between 5.50 and 2.87 ppm, two separate diester regions, based on observed signals, between 1.38 and -0.12 ppm (diester 1) and -0.54 to -1.73 ppm (diester 2), and pyrophosphate between -3.29 and -4.08 ppm. Spectral deconvolution using the line fitting utility in MestReNova 14.0.1 was also performed to estimate contributions of individual monoester species identified within the region that included α-glycerolphosphate, phytate (myo-inositolhexakisphosphate (IHP)) scyllo-IHP, β-glycerolphosphate, and ribonucleic acid (RNA) mononucleotides (Fig. SI 4).
Extraction and characterization of soil organic matter by Fourier-transform ion cyclotron resonance mass spectrometry (FTICR MS)
Extraction of SOM was performed following the sequential fractionation method described in (Tfaily et al. 2017) with slight modification. Briefly, 1 g of rhizosphere soil sample was first suspended in 10 mL water for 4 h with shaking to extract polar compounds. The samples were then centrifuged, and supernatants were collected and saved for analysis of water extractable organic matter (WEOM) fraction. An aliquot of the WEOM was collected, and the DOC concentration was measured on a Flash Elemental Analyzer 1112 (Thermo Fisher Scientific Inc., Waltham, MA). Before samples were injected into the mass spectrometer, the WEOM fraction was cleaned and desalted using solid phase extraction and eluted in methanol (Dittmar et al. 2008). Ultra-high resolution mass spectra of the WEOM fraction was collected using a 12 Tesla Bruker SolariX FTICR spectrometer located at the Pacific Northwest National Labs, Environmental Molecular Science Laboratory following the method described in (Tfaily et al. 2017).
Determination of soil microbial community structure
Microbial community composition in rhizosphere and bulk soils after supplying water and exudates for 15 days were determined using a high-throughput 96 well plate based phospholipid fatty acid (PLFA) protocol (Buyer and Sasser 2012; Joshi et al. 2021). PLFA provides estimates of the total viable bacterial and fungal biomass including their categorization into representative biomarker groups such as gram positive (G +) bacteria, gram negative (G-) bacteria, arbuscular mycorrhizal (AM) fungi, general fungi, protists, and actinobacteria using the peak naming table accompanying the MIDI system (MIDI Inc., Newark, DE, USA).
Statistical analysis
Prior to analysis of variance, data were checked for normal distribution. Analysis of variance was conducted on soil physicochemical properties using a general linear model in JMP version 13.0 (SAS Institute, Cary, NC) testing for the effect of root exudate compounds on the various parameters measured. Where significant, differences between the means were determined using a least square means Student’s t-test at a probability level of p < 0.05.
For analysis of the PLFA data, concentrations and proportional abundance of the microbial biomarker groups (G+ bacteria, G- bacteria, actinobacteria, AM fungi, fungi and protists) were treated as continuous response variables and analyzed using a generalized linear mixed model (PROC GLIMMIX, SAS v.9.3) following the procedure described in (Joshi et al. 2021). Where the influence of root exudates was found significant, differences between the means were determined using a least square means Student’s t-test at a probability level of p < 0.05. To test how root exudates influenced microbial community structure, microbial biomarker group concentrations were first Hellinger transformed (Ramette 2007) and then using Sorensen (Bray-Curtis) distances and slow and thorough settings a non-metric multidimensional scaling analysis was performed using PC-ORD (version 6.0, MjM Software, Gleneden Beach, OR). A multi-response permutation procedure (MRPP) in PC-ORD was then used to determine if microbial community composition differed significantly between exudate treatments with the hypothesis being that these compounds will not be different. P values were adjusted for multiple comparisons using a Bonferroni's correction. In MRPP, a small p-value indicates that the predefined grouping variables are more different than expected by chance. The effect size is reflected in the A-value, the chance corrected within-group agreement, which indicates the similarity of samples within a group. A = 1 if the samples in a group are identical, and A is closer to zero if their heterogeneity is higher than expected by chance. An A value > 0.3 for ecological data is considered high (McCune et al. 2002). Non-metric multidimensional scaling was also used to determine any relationships between soil physicochemical parameters, the microbial biomarker group concentrations, and community structure by correlating PLFA biomarker group concentrations with axis scores in the non-metric multidimensional scaling ordination. Those parameters that had an associated r2 value of 0.300 or greater were overlaid on the non-metric multidimensional scaling ordination as a biplot. The direction and length of the bi-plot vectors indicate the direction (positive or negative) and strength of the correlation while the angle between vectors indicates the correlation between biomarker group concentrations (small angles = higher correlation). Principal component analysis (PCA) was used to determine the soil biogeochemical parameters differentiating the exudate treatments using the relative abundance of water extractable compound classes determined via FTICR-MS, phosphorus fractions determined via NMR, and the concentrations of resin-Pi, water-soluble Fe and Al, DOC, and microbial biomarker groups.
Results
Spatial distribution of phosphatase activities around simulated roots
Compared to control, glucose, alanine, and oxalate increased phosphatase enzyme activities in soils around the simulated roots and their spatial distribution around the simulated root differed depending on the type of exudates (Fig. 1). Alanine induced the greatest enzyme activities where the distribution appeared as discrete hot spots with high activities surrounding (~ 1─2 cm) the root. Oxalate, on the other hand, induced slightly higher enzyme activities compared to glucose with an area of low activity at the center surface and a pattern of activity that was more diffuse and distributed further (~ 2─3 cm) from the root.
a) 2-dimensional distribution of potential phosphatase activities around a simulated root after delivering water, glucose, alanine, and oxalate for 15 days. The size of each enzyme foil is 10 × 10 cm2 and the simulated root is in the center of the foil (5 cm). b Plot profiles of potential phosphatase activities around a simulated root generated by integrating all vertical pixel intensities from left to right across the foil. Dashed line shows the approximate location of the simulated root
Spatial distribution of soil pH and O2 concentration around simulated roots
Background pH in bulk soils of all treatments ranged from ~ 6.2 to 6.4 (Fig. 2). In the control treatment receiving water only (water pH = 6.8) there was a slight increase in rhizosphere pH to ~ 6.5 compared to the background soils (Fig. 2a). Supply of glucose, alanine, and oxalate for 15 days had a distinct influence on the distribution of soil pH (Fig. 2) and O2 concentration (Fig. SI2) in the soil around the simulated roots. Compared to the control, the supply of glucose decreased rhizosphere pH by ~ 0.5 pH units up to ~ 1 cm from the simulated root (Fig. 2b), while those receiving alanine and oxalate increased the rhizosphere pH by ~ 1 pH unit up to 2 cm away from the simulated root. Oxygen concentrations around the simulated root delivering alanine and oxalate were reduced by ~ 3% while the glucose treatment resulted in ~ 10% decrease relative to the control (Fig. SI 2 and Fig. SI 3).
a 2-dimensional distribution of soil pH around a simulated root after delivering water, glucose, alanine, and oxalate for 15 days. Each pH strip was 10 (length) × 1 (breadth) cm. b Plot profiles of pH around a simulated root generated by integrating all vertical pixel intensities from left to right across the foil. Dashed line shows the approximate location of the simulated root
Concentration of resin-Pi, water-soluble Fe and Al, and DOC
Supply of glucose, alanine, and oxalate had distinctly different effects on the mobilization of resin-Pi, water soluble Fe, and Al, and DOC. Compared to the control, oxalate released the greatest, and glucose the least, amount of water-soluble Fe and Al, resin-Pi, and DOC (Table 2). Rhizosphere soils in the oxalate treatment had more than twice the amount of Fe and Al compared to those receiving glucose and alanine and an overall increase in resin-Pi of more than 2 mg kg−1. While all three exudates significantly increased DOC concentrations relative to the control, the oxalate treatment notably resulted in 2─3 times greater DOC (e.g., 43 mg L−1 for oxalate compared to 14 mg L−1 for water) relative to all other treatments.
Soil phosphorus forms—31P solution NMR
The largest proportion of the total integrated area across all samples was for the orthophosphate monoesters signal region (~ 60%) followed by the peak corresponding to orthophosphate (~ 33%) with the remainder split between orthophosphate diesters (two signal regions, diester 1 and 2) and the pyrophosphate peak (~ 4% for both) (Fig. 3, Fig. SI4, and Table 3). No signals were detected from other P forms such as polyphosphates and phosphonates after 3,072 transients. The means of the integrated areas for each of the aforementioned signal regions or peaks (Fig. 3) of each exudate treatment were compared to the means of the control and tested for significant differences using Student’s t-test assuming equal variances (n = 3). Significant differences (p < 0.05) were observed in the means for oxalate’s orthophosphate (p = 0.006), diester 2 (p = 0.02), and pyrophosphate (p = 0.006) regions with relative percent changes of 3.4 (± 0.9), -34 (± 13), and 21 (± 6) %, respectively, as well as the orthophosphate monoester region for both the alanine and glucose treatments (p = 0.04), with relative percent changes for both of 1.7 (± 0.8) %.
Stack plot of representative 31P NMR spectra of NaOH/EDTA extracts from rhizosphere soils around a simulated root after delivering, water, glucose, alanine, and oxalate for 15 days. The spectra are plotted with 10 Hz line broadening to accentuate the diester region signals around 0 ppm. The expanded region of the oxalate (blue, top) spectrum is shown to highlight identified compounds in the orthophosphate monoester region from spiking experiments or literature reported values and is displayed with 2 Hz line broadening. The letter coded assignments are as follows: a) myo-IHP (phytate), b) α-glycerolphosphate, c) β-glycerolphosphate, d) RNA mononucleotides, and e) scyllo-IHP
In comparing the ratios of all Po to Pi forms, the oxalate treatment had the lowest Po/Pi ratio overall and its mean was significantly different from the control with a ratio of 1.61 versus 1.74, respectively (p = 0.001) (Table 3). The same trend was observed when looking at the ratio of Po to only orthophosphate with ratio means of 1.81 and 1.93 for oxalate and water, respectively (p = 0.003). The means of the ratios for alanine and glucose treatments were greater than the control but not significantly different.
The spiking experiments (Fig. SI 4) permitted identification of species within the heavily overlapped orthophosphate monoester region and included α-glycerolphosphate (~ 4.65 ppm), myo-IHP (phytate) (peaks at ~ 5.13, 4.35, 3.96, and 3.78 ppm), β-glycerolphosphate (4.32 ppm observed in spiked spectra), RNA mononucleotides, degraded from intact RNA (broad overlapping signals with peaks at approximately 4.26 and 4.19 ppm observed in the spiked spectra), and scyllo-IHP (3.50 ppm) (the latter assigned from its position up-field relative to spiked-in phosphocholine chloride (3.62 ppm) (Cade-Menun 2015; Turner and Richardson 2004). Using spectral deconvolution, the proportions of α-glycerolphosphate, myo-IHP, RNA mononucleotides, and scyllo-IHP in the monoester region could be reasonably estimated but reliable results for β-glcyerolphosphate were not obtainable. The remaining integrated area of the monoester region, after accounting for the identified species, was collectively referred to as “unassigned monoester area” in Table 3. In all cases, the means of the replicates for each treatment were compared to the means for the water as above. Within the monoester region, a significant difference was only identified for an apparent increase in the proportion of scyllo-IHP in the alanine treatment relative to the water with a relative percent difference of + 11 (± 5) % (p = 0.04).
Although it would be impossible to determine the exact proportion of diester degradation products whose signal area should be redistributed to the diester region given the amount of overlap, uncertainty in the fits, and potential contributions from other yet unidentified monoester compounds, we applied a conservative estimate based on one of three correction scenarios (Schneider et al. 2016) in which all of the area corresponding to α-glycerolphosphate (β-glycerolphosphate would also be included but was not fit here) and one-half of the area of the RNA mononucleotides was shifted back to the diester region. For comparison, the results of the total monoesters and diesters before and after the correction was applied are provided in Table 3. With the correction applied, no significant differences in the means were noted as they were for the monoesters for the alanine and glucose treatments and for diester deoxyribonucleic acid (DNA) specifically in the oxalate treatment before correction.
Changes in soil organic matter composition and P containing compounds –FTICR MS
Compared to the control, the WEOM fraction from rhizosphere soils receiving glucose, alanine, and oxalate had decreased percent relative abundance of amino-sugar-, lignin-, protein-like compounds, and CHOP containing compounds whereas there was an increased in condensed hydrocarbon-, tannin-like, and CHONSP containing compounds (Fig. 4). Among the exudate compounds, supply of oxalate resulted in the greatest decrease in percent relative abundance of amino-sugar-, protein-like, and CHOP containing compounds by ~ 3, 8, and 3%, respectively, compared to the control, with a significant increase in the percent relative abundance of condensed hydrocarbon-, tannin-like, and CHONSP containing compounds by ~ 15, 5, and 2%, respectively. Furthermore, compared to the control, the oxalate treatment resulted in the appearance of a greater number of peaks in the CHONP region (bottom left corner of Van Krevelen diagram) and a greater disappearance of peaks in the CHOP region (top center of Van Krevelen diagram) (Fig. 5).
The mass distribution, nominal oxidation state of carbon, and Gibbs free energy of reaction of the WEOM fraction from rhizosphere soils after supply of oxalate were distinctly different from the control, glucose, and alanine treatments (Fig. 6). The mass distribution of organic compounds in the oxalate treatment had two visually different regions (R1 and R2) compared to other treatments. The regions R1 and R2 had lower and higher intensity of ridgelines of masses between ~ 300 to 450 Da and ~ 500 to 700 Da, respectively (Fig. 6a). Compared to the control, the oxalate treatment had the lowest intensity of nominal oxidation state of carbon ridgelines between -1 to 0 (Fig. 6 b). Similarly, the lowest intensity of Gibbs free energy of reaction ridgelines, ~ 60 to 100 kJ (mol C)−1, was measured in the oxalate treatment compared to control, glucose, and alanine (Fig. 6c).
Organic compounds mass (a), nominal oxidation state of carbon (b), and Gibbs energy of reaction distribution (c) of WEOM in rhizosphere soils around a simulated root after delivering (1) water, (2) glucose, (3) alanine, and (4) oxalate for 15 days. Dashed lines in the figure show the selected region where R1 (250–400 Da) and R2 (500–700 Da) are two distinct regions within the mass of organic compounds
Changes in microbial community structure – PLFA
Root exudates had a significant effect on microbial community composition (Table 2). The non-metric multidimensional scaling ordination for bulk and rhizosphere soils resulted in a 2-dimensional solution with a final stress of 2.95 after 40 iterations (Fig. 7). Microbial community structure in the rhizosphere soils receiving glucose, alanine, and oxalate separated along axis 1 and were distinctly different from the control and bulk soils, which were similar. Microbial community structure in rhizosphere soils receiving oxalate were distinctly different from those receiving either glucose (MRPP A = 0.45091, p = 0.00178) or alanine (MRPP A = 0.34909, p = 0.00245) both of which were similar (MRPP A = 0.00909, p = 0.37482). Alanine and glucose microbial communities are correlated with greater concentrations of all biomarker group concentrations and total PLFA biomass. Analysis of the microbial biomarker group concentrations showed that exudate treatments had a significant effect on the concentration of AM Fungi, general Fungi, protists, G + bacteria, and G- bacteria biomarker groups and total PLFA (Table 2). The concentrations of all biomarker groups were significantly greater in the rhizosphere soils from the alanine and glucose treatments compared to the control, while in the oxalate treatment all but G- were less than alanine and glucose and only AMF, G-, and total PLFA were significantly greater than the control.
Nonmetric Multidimensional Scaling plot showing the difference in microbial community structure between bulk (B; filled shapes) and rhizosphere (R; open shapes) soils around a simulated root after delivering, water, glucose, alanine, and oxalic anions for 15 days. Correlations between PLFA microbial biomarker group concentrations and the axis score with an r2 > 0.300 are displayed as vectors indicating the strength (vector length) and direction of the relationship. Angles between vectors indicate their relative correlation
Relationships among exudates and soil biogeochemical parameters – PCA
The first two components of the PCA generated using soil biogeochemical parameters accounted for 44.8% (PC1) and 24.1% (PC 2), respectively, representing 68.9% of the total variance (Fig. 8). The root exudate treatments were distinctly different from the water-only control (Fig. 8a). Among root exudate treatments glucose and alanine were similar and were positively correlated with the microbial biomarker group (i.e., AM Fungi, general Fungi, protists, G + bacteria, and G- bacteria) concentrations, lignin, carbohydrate, and lipids (Fig. 8). The Oxalate treatment was different from glucose and alanine, separating along PC1, and was positively correlated with condensed hydrocarbon, DOC, resin-Pi, and water-soluble Fe and Al concentrations.
Score (a) and loadings (b) plots from the principal component analysis using FTICR MS biochemical classes of water extractable organic matter, microbial biomarker group concentrations, resin-Pi, water soluble Fe and Al, and DOC of rhizosphere soils around a simulated root after supplying water (control, blue circles), glucose (green inverted triangles), alanine (red triangles), and oxalate (orange diamonds) for 15 days
Discussion
In this study we hypothesized that root exudate compounds with specific modes of action will result in differences in Po mobilization and P cycling in a soil with lesser P availability. To test our hypothesis, we introduced glucose (as microbial C source), alanine (a microbial C and N source), and oxalate (a metal chelator) individually via a simulated root for 15 days at a rate of 15 µmol C cm−2 d−1 into a soil with low total P availability and performed a variety of analyses. Changes in the microbial community and physicochemical environment of the rhizosphere depended on N availability and CUE of the root exudates supplied. An increase in total microbial biomass together with a decrease in rhizosphere O2 concentration, more so for glucose and alanine than oxalate (Fig. SI 2 and Fig. SI 3), is indicative of the greater predicted CUE for glucose and alanine compared to oxalate (Table 1). Rhizosphere pH was also lower in the glucose treatment (Fig. 2), notably, like enzyme activity and O2 concentrations, around the top center of the artificial root forming a hotspot of microbial activity (Kuzyakov and Blagodatskaya 2015). Glucose decreased both soil pH and O2 concentrations while alanine and oxalate increased the pH and decreased O2 concentration in the rhizosphere. Although, both biotic and abiotic processes affect soil pH, the changes in pH in the glucose treatment are most likely due to microbial metabolism (i.e., growth and respiration). When microbes use glucose as a C source, carboxylic groups produced during glycolysis may dissociate protons that then lower the pH of the surrounding soil (Yan et al. 1996; Reed et al. 2003). Concomitantly, greater microbial respiration increases the CO2 concentration (i.e. decreased O2 concentration) that can then react with soil solution to release protons, lowering pH. This result is consistent with the study by Keiluweit et al. (2015) who found the supply of glucose lowered soil pH by ~ 0.5 pH unit. In the case of alanine and oxalate, an increase in soil pH was attributed to proton consuming microbial decarboxylation processes (Barekzai and Mengel 1993; Yan et al. 1996) or ligand exchange between hydroxyl groups of Al or Fe hydroxides and oxalate, respectively (Hue and Amien 1989). Other studies have reported a similar increase in soil pH after addition of oxalate (Srinivasan and Mahadevan 2010; Keiluweit et al. 2015; Menezes-Blackburn et al. 2016).
The chemical form of root exudate had a specific effect on the intensity and spatial distribution of potential phosphatase activities. In general, release of the three exudates increased potential enzyme activities in the rhizosphere and, depending on the type of exudate, also stimulated microbial growth (Renella et al. 2007). The low available P concentration in the soils used in this study means that the microorganisms are lacking sufficient P (and C) for optimal growth as is evidenced by the treatment receiving water (control) having the least total microbial biomass and potential acid phosphatase activity. Triggered by the introduction of exudate C, the microbes synthesized phosphatase enzymes needed to hydrolyze Po in order to access the needed Pi (Nannipieri 1994; Wasaki et al. 2003). Alanine served as a source of C and, importantly, N (Hamer and Marschner 2005) for microorganisms that when added to the soils with lesser P availability resulted in the greatest total microbial biomass (Table 2) and potential phosphatase activity (Fig. 1). As microbial synthesis of phosphatase requires substantial investment of N, the presence of N in alanine is the most likely reason for higher potential enzyme activities compared to glucose and oxalate. This finding is consistent with a previous study where addition of alanine as a root exudate increased phosphatase activities (Spohn et al. 2013). Similarly, application of N fertilizers or atmospheric N deposition have both been shown to correlate with increases in phosphatase activity (Allison et al. 2007). However, an increase in phosphatase activity does not necessarily mean greater mineralization of Po because of limited substrate availability for hydrolysis of Po, particularly non-phytate phosphomonoesters in soil (Jarosch et al. 2019). Unlike alanine, addition of oxalate resulted in a distinctly different, diffuse pattern of enzyme activity (Fig. 1). Due to rapid adsorption of enzymes to clay minerals and soil oxides, the enzymes can be easily immobilized on soil particles and become part of MAOM (Burns 1982). These immobilized enzymes are generally protected from degradation and thus retain their catalytic capacity (Burns et al. 2013). Oxalate as a metal chelate may destabilize mineral organic complexes and release mineral associated enzymes along with MAOM resulting in the diffuse pattern of enzyme activity.
Greater DOC, resin-Pi, and notably water-soluble Fe and Al compared to the glucose and alanine treatments provide evidence for MAOM destabilization by oxalate (Table 2). Both alanine and oxalate raised rhizosphere pH (Fig. 2), however, only in the oxalate treatment was the increase in pH accompanied by 2–3 times greater water soluble Fe, Al and DOC. Oxalate can destabilize both poorly crystalline (ferrihydrite and am-Al(OH)3) and crystalline Fe- and Al-oxide-hydroxide (goethite and gibbsite) as well as aluminosilicate (e.g. montmorillonite) minerals (Ramos et.al. 2014; Olsen and Rimstidt 2008; Golubev et al. 2006). Surface complexation of oxalate on ferrihydrite, for example, is effective at polarizing and destabilizing metal–oxygen bonds leading to mineral dissolution (Schwertmann 1991; Stumm and Furrer 1987). Oxalate enhances the dissolution rate of montmorillonite by both formation of Al-oxalate complexes in solution (changing the equilibrium conditions) and with the aluminol edges of the mineral (Ramos et al. 2014). The dissolution rate was shown to increase moving from pH 4 to 7.5 where both of these processes (proton- and ligand-promoted dissolution) were involved. Therefore, it is likely that formation of both soluble and surface complexes with oxalate promoted mineral dissolution and the subsequent release of associated organic matter (Li et. al. 2021). Oxalate may also directly destabilize the free and previously mineral associated OM by inducing breakage of metal (e.g. Ca, Al and Fe) bridges shown to hold the supramolecular aggregate structure of SOM together (Clarholm et al. 2015).
Increased DOC concentrations after supplying oxalate suggests mobilization of MAOM into soil solution thus making OM containing both N and P accessible for microbial degradation. The DOC released by oxalate contained a complex mixture of organic molecules of different sizes dominated by larger molecules (region R2 ~ 500 – 700 Da) such as tannins, condensed hydrocarbon-like, and CHONSP containing compounds (Figs. 4 and 6a). A greater decrease in the percent relative abundance of amino sugar-like, protein-like, and CHOP containing compounds with masses of ~ 300 − 400 Da (Figs. 4 and 6a) provides direct evidence for mineralization of the released OM. Increased accessibility of released OM via direct destabilization of MAOM (Jilling et al. 2018) has been shown to result in SOM mineralization and increased N availability (Keiluweit et al. 2015; Li et al. 2021).
Supplying oxalate increased the resin-Pi fraction, which is generally considered a plant available P pool (Sibbesen 1983). It is well known that LMW organic anions such as oxalate destabilize organic-mineral complexes and release Pi. Several studies have also found that LMW organic anions release Po bound to soil minerals (Fox et al. 1990; Fox and Comerford 1992; Wei et al. 2010 Menezes-Blackburn et al. 2021). Therefore, it is also possible that enzymatic hydrolysis of previously mineral stabilized Po, potentially enhanced by the concomitant release of previously mineral stabilized enzymes, could have contributed to the increase of Pi in soil solution. Although Po released due to destabilization of mineral-organic complexes was not directly measured in this study, the decrease in % relative abundance of complex CHOP compounds (Figs. 4 and 6a), an increase in potential phosphatase activities (Fig. 1), together with greater resin-Pi, and concomitant decrease in Po/Pi ratio determined via NMR (Table 2 and 3) indirectly suggest enzymatic hydrolysis of Po released from destabilization of MAOM. It appears then that both processes, direct destabilization and release of Pi and mineralization of Po, may be responsible for greater Pi in soil solution. Our findings are consistent with a proposed three-step mechanism for nutrient (i.e., N and P) acquisition from SOM as follows: 1) LMW organic anions produced by roots, bacteria, and fungi destabilized SOM from mineral-organic complexes, 2) enzymatic hydrolysis of destabilized and accessible Po and release of Pi in a bioavailable form, and 3) uptake of Pi by roots and microorganisms (Clarholm et al. 2015).
The greater CUE of glucose and alanine (Table 1) increased PLFA biomarker group concentrations (i.e., total microbial biomass) resulting in distinctly different microbial community structure (Table 2 and Fig. 7). Principal component analysis emphasized the effect of CUE and mode of action of individual root exudate compounds on rhizosphere biogeochemistry (Fig. 8). The association of carbohydrates, lipids and amino sugars in the soils receiving glucose and alanine corresponds with the greater microbial biomass in these treatments suggesting that soil microorganism used the easily available C to synthesize C storage molecules (e.g. glycogen, lipids, or starch) (Wilson et al. 2010). The supply of easily available C resulted in greater bacterial biomass, likely attributable to the rapid growth of generalist microorganisms (i.e. r-strategists) (Fontaine et al. 2003) that correlated with an increase in protists (Table 2). Correlation between increasing protist and bacterial biomass may be due to shared preference for the high C, low pH environment or an increase in bacterivorous protists (Oliverio et al. 2020). Like glucose, alanine had greater protist concentrations relative to the oxalate and water treatment, but significantly less than the glucose treatment. The alanine treatment had significantly more AMF biomarkers than all other treatments suggesting that the presence of both C and N in alanine provided more resources to support AMF during the pre-symbiotic phase that may have implications for both P and N assimilation by infected plants (Graham et al. 1981; Monther and Kamaruzaman 2012). Since the CUE of oxalate is significantly lower than glucose and alanine (Table 1), it is not an easily available C source for microorganisms; although it can increase some specialized slow growing microbial species (e.g., K-strategist) (Malý et al. 2009). However, it mobilized OM (containing both N and P) into soil solution by destabilization of mineral-organic complexes making it accessible for microbial degradation as evidenced by the 3% reduction in O2 concentrations relative to the water-only treatment. Microorganisms are generally limited by C, as such the mineralization of Po is most likely caused by microbial C demand (Spohn and Kuzyakov 2013; Spohn et al. 2013) with the concomitant release of additional Pi.
Conclusions
Depending on the specific mode of action and CUE, release of glucose, alanine, and oxalate, resulted in distinctly different biochemical processes altering P availability. A key outcome of this study is that Po released by oxalate-induced destabilization of mineral-organic complexes can be hydrolyzed via microbial produced or previously mineral-stabilized enzymes. This process may be a potential strategy used by plants to increase P availability particularly in forest soils with low Pi but greater Po concentrations (Achat et al. 2010). These results add to the growing call to improve soil carbon models by including biotic activity (root exudates, enzymes) and the influence of nutrient availability as a direct cause of mineral destabilization and C release.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- SOM:
-
Soil organic matter
- Pi :
-
Inorganic P
- DOC:
-
Dissolved organic C
- NMR:
-
Nuclear magnetic resonance
- Po :
-
Organic P
- MAOM:
-
Mineral associated organic matter
- LMW:
-
Low molecular weight
- CUE:
-
Carbon use efficiency
- IHP:
-
Myo-inositolhexakisphosphate
- RNA:
-
Ribonucleic acid
- DNA:
-
Deoxyribonucleic acid
- WEOM:
-
Water extractable organic matter
- PLFA:
-
Phospholipid fatty acid
- AM:
-
Arbuscular mycorrhizal
- G + :
-
Gram positive
- G-:
-
Gram negative
- MRPP:
-
Multi-response permutation procedure
- OM:
-
Organic matter
- FTICR MS:
-
Fourier-transform ion cyclotron resonance mass spectrometry
- PCA:
-
Principle component analysis
References
Achat DL, Bakker MR, Zeller B et al (2010) Long-term organic phosphorus mineralization in Spodosols under forests and its relation to carbon and nitrogen mineralization. Soil Biol Biochem 42:1479–1490. https://doi.org/10.1016/j.soilbio.2010.05.020
Allison VJ, Condron LM, Peltzer DA et al (2007) Changes in enzyme activities and soil microbial community composition along carbon and nutrient gradients at the Franz Josef chronosequence, New Zealand. Soil Biol Biochem 39:1770–1781. https://doi.org/10.1016/j.soilbio.2007.02.006
Apostel C, Dippold MA, Bore E, Kuzyakov Y (2017) Sorption of alanine changes microbial metabolism in addition to availability. Geoderma 292:128–134. https://doi.org/10.1016/j.geoderma.2017.01.016
Barekzai A, Mengel K (1993) Effect of microbial decomposition of mature leaves on soil pH. Zeitschrift Für Pflanzenernährung Und Bodenkunde 156:93–94. https://doi.org/10.1002/jpln.19931560115
Brant JB, Sulzman EW, Myrold DD (2006) Microbial community utilization of added carbon substrates in response to long-term carbon input manipulation. Soil Biol Biochem 38:2219–2232. https://doi.org/10.1016/j.soilbio.2006.01.022
Bulgarelli D, Schlaeppi K, Spaepen S et al (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838
Burns RG (1982) Enzyme activity in soil: Location and a possible role in microbial ecology. Soil Biol Biochem 14:423–427. https://doi.org/10.1016/0038-0717(82)90099-2
Burns RG, DeForest JL, Marxsen J et al (2013) Soil enzymes in a changing environment: current knowledge and future directions. Soil Biol Biochem 58:216–234. https://doi.org/10.1016/j.soilbio.2012.11.009
Buyer JS, Sasser M (2012) High throughput phospholipid fatty acid analysis of soils. Appl Soil Ecol 61:127–130. https://doi.org/10.1016/j.apsoil.2012.06.005
Cade-Menun BJ (2015) Improved peak identification in 31P-NMR spectra of environmental samples with a standardized method and peak library. Geoderma 257–258:102–114. https://doi.org/10.1016/j.geoderma.2014.12.016
Cade-Menun BJ, Liu CW (2013) Solution phosphorus-31 nuclear magnetic resonance spectroscopy of soils from 2005 to 2013: a review of sample preparation and experimental parameters. Soil Sci Soc Am J 78:19–37. https://doi.org/10.2136/sssaj2013.05.0187dgs
Cade-Menun BJ, Preston CM (1996) A comparison of soil extraction procedures for 31P NMR spectroscopy. Soil Sci 161(11):770–785
Clarholm M, Skyllberg U, Rosling A (2015) Organic acid induced release of nutrients from metal-stabilized soil organic matter – the unbutton model. Soil Biol Biochem 84:168–176. https://doi.org/10.1016/j.soilbio.2015.02.019
Daly AB, Jilling A, Bowles TM, Buchkowski RW, Frey SD, Kallenbach DM, Keiluweit M, Mooshammer M, Schimel JP, Grandy AS (2021) A holistic framework integrating plant-microbe-mineral regulation of soil bioavailable nitrogen. Biogeochemistry 154(2):211–229. https://doi.org/10.1007/s10533-021-00793-9
Dijkstra FA, Bader NE, Johnson DW, Cheng W (2009) Does accelerated soil organic matter decomposition in the presence of plants increase plant N availability? Soil Biol Biochem 41:1080–1087. https://doi.org/10.1016/j.soilbio.2009.02.013
Dittmar T, Koch B, Hertkorn N, Kattner G (2008) A simple and efficient method for the solid-phase extraction of dissolved organic matter (SPE-DOM) from seawater. Limnol Oceanogr Methods 6:230–235. https://doi.org/10.4319/lom.2008.6.230
Fontaine S, Mariotti A, Abbadie L (2003) The priming effect of organic matter: a question of microbial competition? Soil Biol Biochem 35:837–843. https://doi.org/10.1016/S0038-0717(03)00123-8
Fox TR, Comerford NB (1992) Influence of oxalate loading on phosphorus and aluminum solubility in spodosols. Soil Sci Soc Am J 56:290–294. https://doi.org/10.2136/sssaj1992.03615995005600010046x
Fox TR, Comerford NB, McFee WW (1990) Kinetics of phosphorus release from spodosols: effects of oxalate and formate. Soil Sci Soc Am J 54:1441–1447. https://doi.org/10.2136/sssaj1990.03615995005400050038x
Frey SD, Lee J, Melillo JM, Six J (2013) The temperature response of soil microbial efficiency and its feedback to climate. Nat Clim Chang 3:395–398. https://doi.org/10.1038/nclimate1796
Geurts R, Franssen H (1996) Signal transduction in rhizobium-induced nodule formation. Plant Physiol 112:447 LP – 453. https://doi.org/10.1104/pp.112.2.447
Golubev SV, Bauer A, Pokrovsky OS (2006) Effect of pH and organic ligands on the kinetics of smetite dissolution at 25oC. Geochim Cosmochim Acta 70(17):4436–4451. https://doi.org/10.1016/j.gca.2006.06.1557
Graham JH, Leonard RT, Menge JA (1981) Membrane-mediated decrease in root exudation responsible for phorphorus inhibition of vesicular-arbuscular mycorrhiza formation. Plant Physiol 68:548–552
Grayston SJ, Vaughan D, Jones D (1997) Rhizosphere carbon flow in trees, in comparison with annual plants: the importance of root exudation and its impact on microbial activity and nutrient availability. Appl Soil Ecol 5:29–56. https://doi.org/10.1016/S0929-1393(96)00126-6
Guo J, McCulley RL, McNear DH Jr (2015) Tall fescue cultivar and fungal endophyte combinations influence plant growth and root exudate composition. Front Plant Sci 6:183. https://doi.org/10.3389/fpls.2015.00183
Hamer U, Marschner B (2005) Priming effects in different soil types induced by fructose, alanine, oxalic acid and catechol additions. Soil Biol Biochem 37:445–454. https://doi.org/10.1016/j.soilbio.2004.07.037
Hue NV, Amien I (1989) Aluminum detoxification with green manures. Commun Soil Sci Plant Anal 20:1499–1511. https://doi.org/10.1080/00103628909368164
Hütsch BW, Augustin J, Merbach W (2002) Plant rhizodeposition — an important source for carbon turnover in soils. J Plant Nutr Soil Sci 165:397–407. https://doi.org/10.1002/1522-2624(200208)165:4%3c397::AID-JPLN397%3e3.0.CO;2-C
Jagadamma S, Mayes MA, Phillips JR (2012) Selective sorption of dissolved organic carbon compounds by Temperate Soils. PLoS ONE 7:e50434
Jarosch KA, Kandeler E, Frossard E, Bünemann EK (2019) Is the enzymatic hydrolysis of soil organic phosphorus compounds limited by enzyme or substrate availability? Soil Biol Biochem 139:107628. https://doi.org/10.1016/j.soilbio.2019.107628
Jilling A, Keiluweit M, Contosta AR et al (2018) Minerals in the rhizosphere: overlooked mediators of soil nitrogen availability to plants and microbes. Biogeochemistry 139:103–122. https://doi.org/10.1007/s10533-018-0459-5
Jilling A, Keiluweit M, Gutknecht JLM, Grandy AS (2021) Priming mechanisms providing plants and microbes access to mineral-associated organic matter. Soil Biol Biochem 158:108265. https://doi.org/10.1016/j.soilbio.2021.108265
Jones DL (1998) Organic acids in the rhizosphere – a critical review. Plant Soil 205:25–44. https://doi.org/10.1023/A:1004356007312
Jones DL, Prabowo AM, Kochian LV (1996) Kinetics of malate transport and decomposition in acid soils and isolated bacterial populations: the effect of microorganisms on root exudation of malate under Al stress. Plant Soil 182:239–247. https://doi.org/10.1007/BF00029055
Jones DL, Hodge A, Kuzyakov Y (2004) Plant and mycorrhizal regulation of rhizodeposition. New Phytol 163:459–480. https://doi.org/10.1111/j.1469-8137.2004.01130.x
Jones DL, Nguyen C, Finlay RD (2009) Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant Soil 321:5–33. https://doi.org/10.1007/s11104-009-9925-0
Joshi SR, Morris JW, Tfaily MM et al (2021) Low soil phosphorus availability triggers maize growth stage specific rhizosphere processes leading to mineralization of organic P. Plant Soil 459:423–440. https://doi.org/10.1007/s11104-020-04774-z
Keiluweit M, Bougoure JJ, Nico PS et al (2015) Mineral protection of soil carbon counteracted by root exudates. Nat Clim Chang 5:588–595. https://doi.org/10.1038/nclimate2580
Kuzyakov Y, Blagodatskaya E (2015) Microbial hotspots and hot moments in soil: concept & review. Soil Biol Biochem 83:184–199. https://doi.org/10.1016/j.soilbio.2015.01.025
Kuzyakov Y, Friedel JK, Stahr K (2000) Review of mechanisms and quantification of priming effects. Soil Biol Biochem 32:1485–1498. https://doi.org/10.1016/S0038-0717(00)00084-5
Lehmann J, Kleber M (2015) The contentious nature of soil organic matter. Nature 528:60–68. https://doi.org/10.1038/nature16069
Li H, Bolscher T, Winnick M, Tfaily MM, Cardon ZG et al (2021) Simple plant and microbial exudates destabilize mineral-associated organic matter via multiple pathways. Environ Sci Technol 55(5):33–3398. https://doi.org/10.1021/acs.est.0c04592
Malý S, Královec J, Hampel D (2009) Effects of long-term mineral fertilization on microbial biomass, microbial activity, and the presence of r- and K-strategists in soil. Biol Fertil Soils 45:753–760. https://doi.org/10.1007/s00374-009-0388-5
Marschner P (2008) The role of rhizosphere microorganisms in relation to P uptake by plants BT - the ecophysiology of plant-phosphorus interactions. In: White PJ, Hammond JP (eds). Springer Netherlands, Dordrecht, pp 165–176
McAllister CF, Lepo JE (1983) Succinate transport by free-living forms of Rhizobium japonicum. J Bacteriol 153:1155–1162
McCune B, Grace JB, Urban D (2002) Analysis of ecological communities. MjM Software, Gleneden Beach, Oregon
McGill WB, Cole CV (1981) Comparative aspects of cycling of organic C, N, S and P through soil organic matter. Geoderma 26:267–286. https://doi.org/10.1016/0016-7061(81)90024-0
McNear DH Jr (2013) The rhizosphere-roots, soil and everything in between. Nat Educ Knowl 4:1
Menezes-Blackburn D, Paredes C, Zhang H et al (2016) Organic acids regulation of chemical–microbial phosphorus transformations in soils. Environ Sci Technol 50:11521–11531. https://doi.org/10.1021/acs.est.6b03017
Menezes-Blackburn D, Bol R, Klumpp E et al (2021) Citric acid effect on the abundance, size and composition of water-dispersible soil colloids and its relationship to soil phosphorus desorption: a case study. J Soil Sci Plant Nutr 21:2436–2446. https://doi.org/10.1007/s42729-021-00534-9
Monther MT, Kamaruzaman S (2012) Arbuscular mycorrhizal fungi and plant root exudates bio-communications in the rhizosphere. Afr J Microbiol Res 6:7295–7301
Murphy J, Riley JP (1962) A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta 27:31–36. https://doi.org/10.1016/S0003-2670(00)88444-5
Nannipieri P (1994) The potential use of soil enzymes as indicators of productivity, sustainability and pollution, soil biota management in sustainable farming systems. In: Pankhurst CE, Doube BM, Gupta VVSR, Grace PR (eds). CSIRO, East Melbourne, pp 238–244
Oliverio AM, Geisen S, Delgado-Baquerizo M et al (2020) The global-scale distributions of soil protists and their contributions to belowground systems. Sci Adv 6:eaax8787. https://doi.org/10.1126/sciadv.aax8787
Olsen AA, Rimstidt JD (2008) Oxalate-promoted forsterite dissolution at low pH. Geochim Cosmochim Acta 72(7):1758–1766. https://doi.org/10.1016/j.gca.2007.12.026
Paterson E (2003) Importance of rhizodeposition in the coupling of plant and microbial productivity. Eur J Soil Sci 54:741–750. https://doi.org/10.1046/j.1351-0754.2003.0557.x
Ramette A (2007) Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 62:142–160. https://doi.org/10.1111/j.1574-6941.2007.00375.x
Ramos ME, Garcia-Palma S, Rozalen M, Johnston CT, Huertas FJ (2014) Kinetics of montmorilloite dissolution an experimental study of the effect of oxalate. Chem Geology 363:283–292. https://doi.org/10.1016/j.chemgeo.2013.11.014
Razavi BS, Zarebanadkouki M, Blagodatskaya E, Kuzyakov Y (2016) Rhizosphere shape of lentil and maize: spatial distribution of enzyme activities. Soil Biol Biochem 96:229–237. https://doi.org/10.1016/j.soilbio.2016.02.020
Reed JL, Vo TD, Schilling CH, Palsson BO (2003) An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol 4:R54. https://doi.org/10.1186/gb-2003-4-9-r54
Renella G, Landi L, Valori F, Nannipieri P (2007) Microbial and hydrolase activity after release of low molecular weight organic compounds by a model root surface in a clayey and a sandy soil. Appl Soil Ecol 36:124–129. https://doi.org/10.1016/j.apsoil.2007.01.001
Rodríguez H, Fraga R, Gonzalez T, Bashan Y (2006) Genetics of phosphate solubilization and its potential applications for improving plant growth-promoting bacteria. Plant Soil 287:15–21
Rovira AD (1969) Plant root exudates. Bot Rev 35:35–57. https://doi.org/10.1007/BF02859887
Schindelin J, Arganda-Carreras I, Frise E et al (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. https://doi.org/10.1038/nmeth.2019
Schneider KD, Cade-Menun BJ, Lynch DH, Voroney RP (2016) Soil phosphorus forms from organic and conventional forage fields. Soil Sci Soc Am J 80(2):328–340. https://doi.org/10.2136/sssaj2015.09.0340
Schwertmann U (1991) Solubility and dissolution of iron oxides. Plant and Soil 130:1–25. https://www.jstor.org/stable/42937281. Accessed 5 Nov 2023
Sibbesen E (1983) Phosphate soil tests and their suitability to assess the phosphate status of soil. J Sci Food Agric 34:1368–1374. https://doi.org/10.1002/jsfa.2740341209
Sokol NW, Sanderman J, Bradford MA (2019) Pathways of mineral-associated soil organic matter formation: Integrating the role of plant carbon source, chemistry, and point of entry. Glob Chang Biol 25:12–24. https://doi.org/10.1111/gcb.14482
Spohn M, Kuzyakov Y (2013) Phosphorus mineralization can be driven by microbial need for carbon. Soil Biol Biochem 61:69–75. https://doi.org/10.1016/j.soilbio.2013.02.013
Spohn M, Kuzyakov Y (2014) Spatial and temporal dynamics of hotspots of enzyme activity in soil as affected by living and dead roots—a soil zymography analysis. Plant Soil 379:67–77. https://doi.org/10.1007/s11104-014-2041-9
Spohn M, Ermak A, Kuzyakov Y (2013) Microbial gross organic phosphorus mineralization can be stimulated by root exudates – a 33P isotopic dilution study. Soil Biol Biochem 65:254–263. https://doi.org/10.1016/j.soilbio.2013.05.028
Srinivasan K, Mahadevan R (2010) Characterization of proton production and consumption associated with microbial metabolism. BMC Biotechnol 10:2. https://doi.org/10.1186/1472-6750-10-2
Stumm W, Furrer G (1987) The dissolution of oxides and aluminum silicates: Examples of surface-coordination- controlled kinetics. In: Stumm W (ed) Aquatic surface chemistry. Wiley, Hoboken, pp 197–219
Tfaily MM, Chu RK, Toyoda J et al (2017) Sequential extraction protocol for organic matter from soils and sediments using high resolution mass spectrometry. Anal Chim Acta 972:54–61. https://doi.org/10.1016/j.aca.2017.03.031
Turner BL, Richardson AE (2004) Identification of scyllo-inositol phosphates in soil by solution phosphorus-31 nuclear magnetic resonance spectroscopy. Soil Sci Soc Am J 68(3):802–808. https://doi.org/10.2136/sssaj2004.8020
Wasaki J, Yamamura T, Shinano T, Osaki M (2003) Secreted acid phosphatase is expressed in cluster roots of lupin in response to phosphorus deficiency. Plant Soil 248:129–136. https://doi.org/10.1023/A:1022332320384
Wei L, Chen C, Xu Z (2010) Citric acid enhances the mobilization of organic phosphorus in subtropical and tropical forest soils. Biol Fertil Soils 46:765–769. https://doi.org/10.1007/s00374-010-0464-x
Wilson WA, Roach PJ, Montero M et al (2010) Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol Rev 34:952–985. https://doi.org/10.1111/j.1574-6976.2010.00220.x
Yan F, Schubert S, Mengel K (1996) Soil pH increase due to biological decarboxylation of organic anions. Soil Biol Biochem 28:617–624. https://doi.org/10.1016/0038-0717(95)00180-8
Yuan Y, Zhao W, Zhang Z et al (2018) Impacts of oxalic acid and glucose additions on N transformation in microcosms via artificial roots. Soil Biol Biochem 121:16–23
Acknowledgements
We thank Joe Kupper, James W. Morris, and Martin Vandiviere for their help with soil sampling and analysis.
Funding
This work was supported in part by NIFA-AFRI award # 2016-67019-25281. A portion of this research was performed on a project award (https://doi.org/10.46936/genr.proj.2017.50047/60006261) from the Environmental Molecular Sciences Laboratory, a DOE Office of Science User Facility sponsored by the Biological and Environmental Research program under Contract No. DE-AC05-76RL01830.
Author information
Authors and Affiliations
Contributions
The study was conceptualized and designed by Sunendra R. Joshi and David H. McNear Jr. Material preparation, data collection and analysis were performed by Sunendra R. Joshi, David H. McNear Jr., Robert P. Young, and Malak M. Tfaily. The first draft of the manuscript was written by Sunendra R. Joshi with all authors providing review and comments. All authors read and approved of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Responsible Editor: N. Jim Barrow.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 1229 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Joshi, S.R., Tfaily, M.M., Young, R.P. et al. Root exudates induced coupled carbon and phosphorus cycling in a soil with low phosphorus availability. Plant Soil 498, 371–390 (2024). https://doi.org/10.1007/s11104-023-06442-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-023-06442-4