Abstract
Background and aims
Legumes can obtain nitrogen from mineral nitrogen and symbiotic nitrogen; however, the regulatory mechanisms by which legume plants respond to various nitrogen sources are still unclear.
Methods
Here, the flavonoid metabolism profiles of Robinia pseudoacacia (black locust) in response to different nitrogen sources during the early growth stages were performed using the hydroponics method.
Results
The flavonoids were accumulated specifically responding to different nitrogen sources (symbiotic nitrogen and NO3−) in the roots and root exudates of R. pseudoacacia and showed growth-stage-dependent changes in the plant growth process. More flavonoids were synthesized and secreted under nitrogen deficiency. The differential flavonoids among different treatments were significantly enriched in the flavone and flavonol biosynthesis pathway and isoflavonoid biosynthesis pathway. Liquiritigenin, naringenin, apigenin and kaempferol were not only the metabolic nodes of these two metabolic pathways but also the key nodes in the differential flavonoid network. Therefore, these four flavonoids may be the key flavonoids of R. pseudoacacia responding to different nitrogen sources.
Conclusion
Our findings provide an important insight into the adaptability of legumes to different nitrogen environments and are of great significance for understanding the molecular mechanisms underlying nitrogen utilization in legumes and will contribute to the development of sustainable agricultural practices and crop production.
Similar content being viewed by others

Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
It is well known that secondary metabolites are vital molecules in plant life, acting as protective agents against environmental factors. In particular, flavonoids, phenylpropanoid-derived compounds, play important roles in plant growth and development, as well as in resistance to biotic and abiotic stress. For example, gibberellin boosted root growth by directly decreasing flavonol biosynthesis in Arabidopsis (Tan et al. 2019). The synthesis of anthocyanin rhamnoside improved the cold resistance of A. thaliana (Li et al. 2017). The accumulation of flavonoids elevated the resistance to the brown planthopper in rice (Dai et al. 2019). More than 8000 distinct flavonoids have been discovered to date and the majority of these flavonoids can be divided into six major categories, namely flavones, flavonols, flavanones, isoflavones, flavanols and anthocyanins (Wang et al. 2022). Among them, isoflavones such as daidzein and genistein widely exist in legumes.
Legumes are one of the most important crop plants for humans because they provide raw materials for industry, feed for livestock and sources of food (Mathesius 2018). Legumes are important for sustainable agriculture because of their ability in fixing nitrogen from the atmosphere and ultimately help reduce the use of mineral fertilizers in the agro-ecosystems. Under low available nitrogen conditions, the neighboring plants can also stimulate flavonoid production in the roots and root exudates of the legumes. For example, the legumes co-cultivated with cereals like wheat have been shown to increase flavonoid biosynthesis and secretion in the roots (Leoni et al. 2021). The specific flavonoids secreted by the legume roots recruit rhizobia to colonize the rhizosphere. These flavonoids are perceived and bound by NodD, a transcriptional activator of the LysR family from the compatible rhizobia. Subsequently, NodD activates the expression of bacterial nod genes, resulting in the synthesis and secretion of the specific lipo-chitooligosaccharide signaling compounds, Nod factors (Peck et al. 2006). The perception of specific Nod factors triggers a signaling cascade in the host to initiate the nodule formation. Flavonoids play a role in different stages of nodule formation. Firstly, flavonoids are able to act as the selectors for compatible symbiotic organisms. Flavonoids can be used as the inducers of the nod gene expression of compatible rhizobia and as the phytoalexins inhibiting the infection of incompatible rhizobia (Liu and Murray 2016). Daidzein induced the expression of the nod genes in Bradyrhizobium japonicum (nodulating soybean), while it inhibited that of those from Rhizobium leguminosarum (nodulating clover or peas) (Begum et al. 2001). Secondly, flavonoids are essential for nodule initiation. It was confirmed that the flavonols could inhibit polar auxin transport in Medicago truncatula roots, thereby promoting the local accumulation of auxin in the initial site of root nodules and leading to the initiation of root nodules primordium (Wasson et al. 2006; Zhang et al. 2009). Finally, flavonoids may also be involved in the systemic regulation of the nodule number, for example, the exogenous supply of either daidzein or coumestrol increased the nodule number (Abd-Alla 2011). Although the molecular mechanisms that regulate the production of flavonoids during nodulation in plants are not well understood, the interconnection between the availability of nitrogen and the metabolism of flavonoids has been demonstrated. Nitrogen supply has a negative effect on flavonoid biosynthesis in plants. An obvious inverse relationship between nutrient availability and flavonoid accumulation was exhibited in Arabidopsis and tomato, with nitrogen limitation causing the maximum increase of flavonoids (Stewart et al. 2001).
Robinia pseudoacacia (black locust), one of the leguminous plants, can establish symbiosis with various rhizobia from different genera, endowing black locust with strong adaptability to the environment. Because of its prominent symbiotic nitrogen fixation ability, fast growth, the capacity of soil and water conservation and other advantages, R. pseudoacacia has become an important ecological afforestation pioneer tree species in China (Fan et al. 2018). Black locust also is used to develop new biological products for multi-directional uses, including lubricants, polymers, as well as bioenergy and fuels (Tyśkiewicz et al. 2019). In addition, it has also been reported that black locust has pharmacological benefits, owing to the activities of its flavonoids in living organisms (Tyśkiewicz et al. 2019). The main flavonoids in R. pseudoacacia are quercetin, rutin, kaempferol, robinin and acacetin (Guo et al. 2019; Tyśkiewicz et al. 2019; Veitch et al. 2010), which play vital roles in resistance to environmental stresses (Khalid et al. 2019; Zhao et al. 2016).
Flavonoids attracted Aeromonas sp. to colonize the roots of Arabidopsis and thus improved the ability of plants to resist dehydration (He et al. 2022). The higher concentration of quercetin in root exudates of the invasive plant Triadica sebifera enhanced the colonization of arbuscular mycorrhizal fungal and promoted its growth (Tian et al. 2021). Although the ecological functions of flavonoids have been investigated, the response of synthesis and secretion of flavonoids to different nitrogen nutrition from different sources during early growth stages in the leguminous tree species remains largely unexplored. In recent years, metabonomics methodology has become an effective tool for the comprehensive and efficient study of complex metabolic components, which has been widely used to investigate the detailed metabolite profile in plants (Yang et al. 2021; Zhao et al. 2020). However, due to the complexity of soil and plant interactions, current methods for collecting root exudates are limited. Hydroponics, the main method to study plant metabolism, allows accurate control over the chemical background of the plant growth, facilitates the collection of a sufficient amount of freshly produced exudates, avoids problems related to non-uniform mineral adsorption of exudate components and minimizes the microbial transformation of exuded metabolites (Zhalnina et al. 2018). Consequently, we initiated a study on the metabolism of flavonoids mediated by different nitrogen sources in the roots and root exudates of R. pseudoacacia during early growth stages by hydroponics. We hypothesized that R. pseudoacacia could regulate the synthesis and secretion of key compounds in the flavonoid metabolic pathway to adapt different nitrogen environments.
Materials and methods
Plant growth and bacterial cultivation
R. pseudoacacia seeds (Northwest A&F University, Yangling, China) were treated with 75% ethanol for 1 min, rinsed twice with sterile water, and sterilized with 50% sodium hypochlorite for 10 min. Then the seeds were washed with sterile water several times. The surface-sterilized seeds were placed in an Erlenmeyer flask containing 70℃ aseptic hot water and incubated overnight at 28℃ with shaking at 150 rpm. Finally, the sterile seeds were transferred to aseptic petri dishes covered with moistened sterile filter papers and incubated in the dark at 28℃ for 72 h.
The germinated seedlings were transferred to the cracks between the sterile filter paper rolls and the holders, which were placed in the sterile trays containing the nitrogen-free one-quarter strength Hoagland’s nutrient solution (in brief, the nutrient solution was modified by substituting 5 mM KCl and CaCl2 for 5 mM KNO3 and Ca(NO3)2, respectively) (Hoagland and Arnon 1950). Plants were cultivated in a glasshouse with a cycle of 16 h light at 24℃ and 8 h dark at 18℃. During this period, the nutrient solution was changed every 5 days.
The rhizobia strain used in this study, Mesorhizobium amorphae CCNWGS0123, was isolated from the nodules of R. pseudoacacia grown in heavy metal tailings in Gansu Province, China (Mohamad et al. 2012), and the strain specifically nodulated R. pseudoacacia (Chen et al. 2013; Wang et al. 2019). Mesorhizobium amorphae CCNWGS0123 was cultured in tryptone-yeast extract (TY) liquid medium at 28℃ with shaking at 200 rpm for 2–3 days. The bacteria were collected by centrifugation at 3300 g for 6 min. Rhizobial cell pellets were resuspended in sterile double-distilled water and collected by centrifugation and then repeated this step three times. In the end, rhizobial cell pellets were resuspended and adjusted to OD600 ≈ 0.3 with sterile double-distilled water.
Fifteen-day-old seedlings were treated with three nutrient solutions. The treatment regimes were: (1) the sterile nitrogen-free nutrient solution (nitrogen free, NF); (2) the nitrogen-free nutrient solution containing M. amorphae CCNWGS0123 suspension (symbiotic nitrogen, SN); (3) the sterile one-quarter strength Hoagland’s nutrient solution (mineral nitrogen, MN).
Measurements of growth phenotype and nitrogen content
The plants were sampled at 1 (infection initiation: refers to the rhizobial attachment to root hairs, I), 3 (nodule primordium initiation and formation, N), 15 (early stage of nitrogen-fixing nodules, E), and 28 (mature nitrogen-fixing nodules, M) days post-treatment (dpt). The sampling time was determined based on previous studies (Chen et al. 2013; Haynes et al. 2004; Vega-Hernández et al. 2001; Wang et al. 2019; Xiao et al. 2014). Chlorophyll content (Chl), the root length (RL), the shoot length (SL), the dry weights of both root and shoot (RDW and SDW) were determined. Ten plants were collected per treatment at each time point. The chlorophyll meter (SPAD-502 Plus, Konica Minolta, Japan) was used to measure the chlorophyll content of leaves. The length of each root and shoot were measured with a ruler. Plants were washed with water and dried to constant weight at 65℃. The nitrogen content of both shoot and root (ShN and RN) was measured. Three biological replicates were conducted for per treatment at each time point, and each biological replicate contained at least five individual plants. Dry samples of the plants were ground into powder and then digested with sulfuric acid and hydrogen peroxide to determine the nitrogen content (Sun et al. 2021).
Preparation and extraction of the roots and root exudates
On each sampling date, the roots were washed with sterile deionized water and freeze-dried using a vacuum freeze-dryer (Scientz-100 F). Three biological replicates were performed for per treatment, and each biological replicate contained mixed roots from at least five different plants. The freeze-dried samples were crushed by a mixer mill (MM 400, Retsch, Haan, Germany) with a zirconia bead for 1.5 min at 30 Hz. Samples (100 mg of lyophilized powder) were dissolved in 1.2 ml of 70% methanol solution, vortexed for 30 s every 30 min for 6 times in total, and then stored at 4℃ overnight. The following mixtures were centrifuged at 13,201 g for 10 min at 4℃. The extracts were filtrated by SCAA-104 with a 0.22 μm pore size (ANPEL, Shanghai, China) and stored in a sample flask for the ultra performance liquid chromatography tandem mass spectrometry (UPLC-MS/MS) analysis.
On each sampling date, root exudates were collected according to a modification of the method of Lv et al. and Sun et al. (Lv et al. 2020; Sun et al. 2016). Briefly, the whole plants were washed with sterile deionized water and then put the roots into a conical flask with 30 ml of sterile deionized water for 6 h. The conical flasks were covered with aluminium foil to create a dark environment for the roots. Then the root exudates were filtered by a 0.22 μm pore size (Millipore) to remove root debris and microorganisms. The filtrates were collected, frozen with liquid nitrogen, and freeze-dried using a vacuum freeze-dryer (Scientz-100 F). Three biological replicates were conducted for per treatment, and each biological replicate contained mixed root exudates from at least five different plants. The freeze-dried root exudates were extracted with 500 µl of 70% methanol solution and vortexed for 3 min. The following mixtures were centrifuged at 13,201 g for 10 min at 4℃. The extracts were filtrated by SCAA-104 with a 0.22 μm pore size (ANPEL, Shanghai, China) and stored in a sample flask for UPLC-MS/MS analysis.
UPLC-MS/MS conditions
The extracts were analyzed using a UPLC-ESI-MS/MS system (UPLC, SHIMADZU Nexera X2; MS, Applied Biosystems 4500 Q TRAP) equipped with an Agilent SB-C18 column (1.8 μm, 2.1 mm × 100 mm). The mobile phase was consisted of solvent A, pure water with 0.1% formic acid, and solvent B, acetonitrile with 0.1% formic acid. The gradient program was performed as follows: 95:5 V/V for 0 min, 5:95 V/V for 9.0 min, 5:95 V/V for 10.0 min, 95:5 V/V for 11.1 min, and 95:5 V/V for 14.0 min. The flow rate and injection volume were 0.35 ml min−1 and 4 µl, respectively. The column oven was set to 40℃. The effluent was alternatively connected to an ESI-triple quadrupole-linear ion trap (Q TRAP)-MS.
A triple quadrupole-linear ion trap mass spectrometer (Q TRAP), AB4500 Q TRAP UPLC/MS/MS system, equipped with linear ion trap (LIT) and triple quadrupole (QQQ) scans were used to detect metabolites. This system was equipped with an ESI turbo ion-spray interface operating in positive and negative ion mode and controlled using Analyst 1.6.3 software (AB Sciex). The ESI source operation parameter was an ion source (turbo spray, 550℃, 5500 V/-4500 V). Gas I, gas II, and curtain gas were set at 50, 60, and 25.0 psi, respectively. The collision-activated dissociation (CAD) was high. In the QQQ and LIT modes, instrument tuning and mass calibration were carried out with 10 and 100 µmol/l polypropylene glycol solutions, respectively. QQQ scans were acquired as MRM experiments with collision gas (nitrogen) set to medium. Declustering potential (DP) and collision energy (CE) were done with further optimization for individual MRM transitions. According to the metabolites eluted within this period, a specific set of MRM transitions were monitored for each period.
Metabolite analyses
Qualitative analysis of primary and secondary MS data was performed by comparison of the retention time (RT), the accurate precursor ions (Q1), product ions (Q3) values and the fragmentation patterns with those obtained by injecting standards using the same conditions if the standards were available (Sigma-Aldrich, St. Louis, MO, USA). The data were analyzed using a self-compiled database, MWDB (MetWare Biological Science and Technology Co., Ltd., Wuhan, China) and publicly available metabolite databases if the standards were unavailable. Metabolite quantification was performed using the MRM mode. The QQQ-MS eliminated the interference of non-target ions and screened the characteristic ions of each metabolite to obtain the signal strengths. After getting the mass chromatographic analysis data of metabolites in different samples, the mass chromatographic peaks of all substances were integrated by peak area, and the mass chromatographic peaks of the same metabolite in different samples were integrated and corrected. The corresponding relative metabolite contents were expressed as chromatographic peak area integrals. The identification of metabolites was performed using isotopic distribution, the accurate mass of metabolites, secondary fragments (MS2) and retention time (RT) of MS2 fragments. Through the intelligent secondary spectrum matching method independently developed by MetWare, the secondary spectrum and RT of the metabolites in the samples were matched intelligently with the secondary spectrum and RT of the MWDB database one by one. The MS tolerance and MS2 tolerance were set to 20 ppm (parts per million), respectively.
Principal component analysis (PCA), hierarchical clustering analysis (HCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) were executed on the metabolites of 72 samples using R software to study metabolite accession-specific accumulation. PCA and HCA were performed using the “vegan” package and “ComplexHeatmap” package, respectively (Gu et al. 2016), significance for PCA analyses was checked with “Adonis” (999 permutations). OPLS-DA was conducted based on the variable importance in projection (VIP) value in the test model by the “MetaboAnalystR” package (Chong and Xia 2018). Differential metabolites define as metabolites with P-value < 0.05, fold change (FC) values ≥ 2.0 or ≤ 0.5 and VIP scores > 1.0. The differential metabolites among the three treatment groups (NF, SN and MN) were analyzed by the Kyoto Encyclopedia of Genes and Genomes (KEGG) database with a P-value < 0.01. Network analysis was carried out to investigate the interactions of the differential metabolites using R software and the interactive platform Gephi. The relative content exceeding 0.01% was opted to calculate Pearson’s correlation between differential metabolites. An interaction was considered to be robust when Pearson’s correlation coefficient was > 0.6 and the P-value was < 0.01.
Statistical analysis
Before conducting statistical analysis, all data were investigated for normality using the Shapiro-Wilk test and were tested for the homogeneity of variances using Bartlett’s test. All statistical analyses were performed with R software (v 3.5.0). Unless stated, visualization of data relied on the ggplot2 package. The significance among the three treatments was tested by one-way analysis of variance (ANOVA) and Tukey’s HSD post hoc test in the stats and multcomp packages of R software (v 3.5.0) and significance was accepted at P-value < 0.05. The data of growth phenotype and nitrogen content were expressed as mean ± standard deviation (SD) and were plotted using Origin 8.0 software.
Results
The analyses of growth phenotype and nitrogen content under different nitrogen conditions
To explore the effect of three treatments (nitrogen free (NF), symbiotic nitrogen (SN) and mineral nitrogen (MN)) on the growth of the plant, the plants of all groups at 1 (I), 3 (N), 15 (E) and 28 (M) days post-treatment (dpt) were collected for analysis. Unsurprisingly, the mature nodules only appeared on SN roots at stage M (Fig. 1A), indicating that other treatments were not contaminated with rhizobia, hence the subsequent experimental results were credible. The growth of the plant among different treatments showed significant differences (P < 0.05) at the E and M stages (Fig. 1B). The leaf chlorophyll content (Chl), the shoot length (SL) and the shoot dry weight (SDW) in MN were dramatically higher than that in NF and SN at the E and M stages, and they were also markedly higher in SN than those in NF at stage M (Fig. 1B). However, the root length (RL) and the root dry weight (RDW) were significantly higher in NF than those in MN at the M stage (Fig. 1B). The nitrogen content of both shoot and root (ShN and RN) showed significant differences among different treatments at four stages (P < 0.05, Fig. 1C). ShN and RN in MN were significantly higher than those in the other two treatments (Fig. 1C). Also, ShN and RN in SN were significantly higher than those in NF at the I and M stages, as well as RN in SN at stage N (Fig. 1C). In addition, both ShN and RN in NF gradually decreased with plant growth, while ShN and RN in SN showed a decline in the first three stages and increased at stage M (Fig. 1C). In MN, ShN decreased gradually in the first three stages and increased at the M stage, and RN initially increased and then gradually decreased from the N stage (Fig. 1C). Overall, the nitrogen content in SN was lower than that in MN and was comparable to that in NF in the first three stages.
Plant phenotype and morphological indexes at four growth stages under different nitrogen conditions. A Phenotypic photos of the plants. B Statistical results of growth phenotype. C Statistical results of nitrogen content in the shoots and roots (n = 3). Data are presented as the mean ± standard deviation (SD). A statistical Tukey test was used, and means with different letters were statistically different at P < 0.05 in three treatments at the same stage. NF, nitrogen free. SN, symbiotic nitrogen. MN, mineral nitrogen. I, 1 day post-treatment (dpt). N, 3 dpt. E, 15 dpt. M, 28 dpt. (M), magnification of the roots of SN in M. Chl, the chlorophyll content. SL, the shoot length. SDW, the shoot dry weight. RL, the root length. RDW, the root dry weight. ShN, the shoot nitrogen content. RN, the root nitrogen content
Flavonoid metabolism profile responding to different nitrogen sources
Given that the special role of flavonoids in legumes responding to environmental nitrogen alterations, the flavonoid metabolism of the roots and root exudates of the three groups of plants described above were analyzed. A total of 295 flavonoids were detected in the roots, including 20 chalcones, 36 flavanones, 7 flavanonols, 5 anthocyanidins, 99 flavones, 59 flavonols, 21 flavonoid carbonosides, 3 flavanols, 3 tannins, 1 biflavones, and 41 isoflavones. In total, 242 flavonoids were identified in root exudates, containing 16 chalcones, 35 flavanones, 5 flavanonols, 2 anthocyanidins, 80 flavones, 40 flavonols, 20 flavonoid carbonosides, 2 flavanols, 2 tannins, 1 biflavones, and 39 isoflavones (Fig. 2A; Tables S1 and S2). It’s worth noting that some of these identifications, particularly those marked as Level 3 in the supplemental table, are less accurate and can be considered tentative identifications. PCA results showed that the metabolic changes in R. pseudoacacia roots and root exudates over time could be indicated by the time-dependent shifts at the level of the first principal component (PC1), which accounted for 28.69% and 46.59% of the total variation, respectively (Fig. 2B). Furthermore, the metabolite profiles of three treatments were separated by the second principal component (PC2), accounting for 15.22% and 18.36% of the total variance, respectively. Adonis analyses revealed that the flavonoid metabolism profiles in the roots (R2 = 47.84%, P = 0.001) and root exudates (R2 = 49.02%, P = 0.001) changed significantly with time. The flavonoid metabolism profiles in the roots (R2 = 19.73%, P = 0.001) and root exudates (R2 = 24.10%, P = 0.001) also showed significant differences among treatments. The four main clusters were obtained by hierarchical clustering analysis based on the relative differences in flavonoid accumulation patterns in the roots and root exudates (Fig. 2C). Flavonoids in cluster 1 were accumulated more in the inoculated roots at 28 dpt (SNM) and in the root exudates of all three treatments at 28 dpt (SNM, NFM and MNM). The flavonoids in cluster 2 were significantly enriched in the roots of nitrogen-free treatment and inoculation treatment at 15 and 28 dpt (SNE, NFE, SNM and NFM) and in the root exudates from nitrogen-free treatment and inoculation treatment at 15 dpt (SNE and NFE). In the roots and root exudates of all three treatments, the accumulation of flavonoids in cluster 3 was higher at 1 dpt (SNI, NFI and MNI). Flavonoids in cluster 4 were obviously enriched in the roots of nitrogen-free treatment at 3 and 15 dpt (NFC and NFE) and in the inoculated roots at 3 dpt (SNC), and in the root exudates from the three treatments at 1 dpt (SNI, NFI and MNI). Then, the differences in metabolites among the three treatments in the roots and root exudates were preliminarily screened with VIP > 1 (Fig. 3). In the three pairwise comparison groups (NF vs. MN, NF vs. SN, and MN vs. SN), up- and down-regulated flavonoids are for the latter. In NF vs. MN, 226 and 209 differential flavonoids (DFs) were derived from the roots and root exudates, respectively (Fig. 3A). Biflavones, flavanols, flavanonols and tannin were down-regulated in MN roots and root exudates, as well as anthocyanidins in MN roots and chalcones, flavonoid carbonoside, and isoflavones in MN root exudates (Fig. 3B). In NF vs. SN, 137 and 162 DFs were obtained from the root and root exudates, respectively (Fig. 3A). Biflavones were up-regulated, while flavanols and tannin were down-regulated in SN roots. Biflavones and flavanonols were down-regulated in SN root exudates (Fig. 3B). Besides, anthocyanidins, flavanols and tannin were not DFs in NF vs. SN (Fig. 3B). In MN and SN, 212 and 214 DFs were identified in the roots and root exudates, respectively (Fig. 3A). Biflavones, flavanols and flavanonols were up-regulated in SN roots (Fig. 3B). Flavanols and flavanonols and tannin were up-regulated, while biflavones were down-regulated in SN root exudates (Fig. 3B). Additionally, flavanones, flavones and flavonols exhibited both up-regulation and down-regulation within the same comparison group (Fig. 3B). In NF vs. MN and NF vs. SN, the number of down-regulated DFs in the roots and root exudates of MN and SN is greater than the number of up-regulated DFs, while in MN vs. SN, the number of up-regulated DFs in SN roots and root exudates is greater than the number of down-regulated DFs (Fig. 3B). These results showed that in the roots and root exudates, the accumulation patterns of flavonoids were different in response to distinct nitrogen sources and showed growth-stage-dependent changes.
Flavonoid metabolism profiles of the roots and root exudates of the plants subjected to three treatments during different growth stages (n = 3). A Classification of flavonoids. B Principal component analysis (PCA). C Heat map visualization. The profiles of all types of flavonoids were normalized to complete the linkage hierarchical clustering. NFI, NFN, NFE, and NFM, nitrogen-free treatment at 1, 3, 15 and 28 days post-treatment (dpt), respectively. SNI, SNN, SNE, and SNM, inoculation treatment at 1, 3, 15 and 28 dpt, respectively. MNI, MNN, MNE, and MNM, nitrogen addition treatment at 1, 3, 15 and 28 dpt, respectively
Differential flavonoid metabolism profiles of the roots and root exudates of the plants subjected to three pairwise comparison groups in all stages. A The number of differential flavonoids. B The number of up- and down-regulated differential flavonoids. NF, nitrogen free. SN, symbiotic nitrogen. MN, mineral nitrogen. In all pairwise comparison groups, up- and down-regulated compounds are for the latter
Metabolic changes of flavonoids at different growth stages
According to the above results, we speculated that in the growth process, plants would produce and secrete more flavonoids in the case of nitrogen deficiency, while produce and secrete less flavonoids in the case of sufficient nitrogen nutrition. To verify this conjecture, we next investigated the significant differences in metabolites among the three treatments in the plant growth process, and volcano plots were used to recognize the down-regulated (FC ≤ 0.5, VIP > 1, and P-value < 0.05) or up-regulated (FC ≥ 2, VIP > 1, and P-value < 0.05) flavonoids (Fig. 4).
Volcano plots of the roots (A) and root exudates (B) of the plants subjected to different comparison groups during different growth stages. NFI, NFN, NFE, and NFM, nitrogen-free treatment at 1, 3, 15 and 28 days post-treatment (dpt), respectively. SNI, SNN, SNE, and SNM, inoculation treatment at 1, 3, 15 and 28 dpt, respectively. MNI, MNN, MNE, and MNM, nitrogen addition treatment at 1, 3, 15 and 28 dpt, respectively. In all pairwise comparison groups, up- and down-regulated compounds are for the latter and are shown with red and green dots, respectively, and grey dots represent insignificant compounds
Firstly, the DFs in the roots and root exudates during the plant growth process were analyzed. In NF vs. MN, there were 16, 42, 71 and 102 DFs in the roots (Fig. 4A) and 100, 133, 141 and 88 DFs in the root exudates (Fig. 4B) at the four sampling time points, respectively, indicating that the number of DFs increased gradually with plant growth in the roots and increased gradually in the first three stages but decreased at stage M in the root exudates. In NF vs. SN, there were 16, 3, 10 and 28 DFs in the roots (Fig. 4A) and 55, 55, 57 and 65 DFs in the roots exudates (Fig. 4B) at the four stages, respectively, suggesting that in the roots and root exudates, the number of DFs increased gradually from stage N. In MN vs. SN, there were 29, 32, 81 and 66 DFs in the roots (Fig. 4A) and 74, 120, 118 and 90 DFs in the root exudates (Fig. 4B) at the four sampling time points, respectively, indicating that the number of DFs increased gradually in the first three stages but decreased at stage M in the roots and decreased gradually from the N stage in the root exudates.
Then, the cases of up-regulated and down-regulated DFs with plant growth were analyzed. In NF vs. MN, the number of up-regulated DFs in NF roots (2, 35, 68, and 101) gradually increased with the plant growth process, while the number of up-regulated DFs in MN roots (14, 7, 3, and 1) gradually decreased over time (Fig. 4A). Similarly, in NF vs. MN, the number of up-regulated DFs in NF root exudates (94, 125, 135, and 64) showed a progressive increase in the first three stages and decreased at stage M, whereas the number of up-regulated DFs in MN root exudates (6, 8, 6, and 24) fluctuated with plant growth (Fig. 4B). In NF vs. SN, the number of up-regulated DFs in both NF roots (8, 1, 4, and 20) and SN roots (8, 2, 6, and 8) showed the trend of first decline and then rise (Fig. 4A), and the number of up-regulated DFs in both NF root exudates (52, 42, 48, and 21) and SN root exudates (3, 13, 9, and 44) fluctuated with plant growth (Fig. 4B). In addition, in MN vs. SN, the number of up-regulated DFs in MN roots (25, 5, 3, and 2) gradually declined with plant growth, whereas the number of up-regulated DFs in SN roots (4, 27, 78, and 64) gradually increased in the first three stages and decreased at stage M (Fig. 4A). And the number of up-regulated DFs in SN root exudates (58, 113, 111, 76) was higher than that in MN root exudates (16, 7, 7, 14) at four stages (Fig. 4B).
As anticipated, the plants produced more flavonoids under nitrogen-free conditions than the plants inoculated with rhizobia or supplied with NO3− in the plant growth process. Generally, a nodule capable of fixing nitrogen is formed about 15 to 20 days after rhizobia infection; until then, the plants are deficient in nitrogen. Thus, compared with that in the plants supplied with NO3−, the inoculated plants produced and secreted more flavonoids during the first three stages, similar to the NF treatment, after which (15 days after inoculation) the synthesis and secretion of flavonoids were decreased. Hierarchical clustering analysis showed the same results (Fig. S1). The data provide strong support for the conjecture described above.
Enrichment analysis of DFs
To understand the case of enrichment of all the above DFs in the metabolic pathway, the KEGG pathway enrichment analysis was performed. The results showed that in the plant growth process, the metabolic pathways involved in the DFs significantly enriched responding to different nitrogen sources were mainly the biosynthesis of secondary metabolites, the flavonoid biosynthesis and the isoflavonoid biosynthesis in the roots, while were mainly the flavone and flavonol biosynthesis, and the isoflavonoid biosynthesis in the root exudates (Fig. S2).
Then, the metabolic profiles involved in the DFs significantly enriched (46 in the roots, 32 in the root exudates) with plant growth were summarized (Fig. 5), and whether each DFs was up-regulated or down-regulated in each pairwise comparisons (NF vs. MN, NF vs. SN, and MN vs. SN) was also shown in Fig. 5. In the roots and root exudates, some of the DFs were present in all three pairwise comparisons (shown in the large boxes with three colors in Fig. 5), suggesting that the synthesis and secretion of these flavonoids (7 in the roots, 14 in the root exudates) could be related to the changes in the environmental nitrogen status and the plant nitrogen nutrient status, including formononetin, calycosin, pseudobaptigenin, eriodictyol, dihydroquercetin, luteolin and dihydrokaempferol in the roots (Fig. 5A); and daidzein, calycosin, pseudobaptigenin, prunetin, biochanin A, formononetin-7-O-glucoside, biochanin A-7-O-glucoside, apigenin, formononetin-7-O-glucoside-6″-O-malonate, cosmosiin, rhoifolin, vitexin, nicotiflorin and apigenin-7-O-[β-D-apiosyl-(1->2)-(6-malonyl-β-D-Glc)] in the root exudates (Fig. 5B).
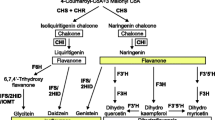
Metabolic pathways of the differential flavonoids in the roots (A) and in the root exudates (B). The large gray boxes indicate that the substance was not detected. The large boxes with different colors represent that the substance in the box was detected in two or more pairwise comparisons. Different comparison groups are shown with different colors in the boxes (pink represents NF vs. MN, yellow represents NF vs. SN, and blue represents MN vs. SN). The four small boxes of the same color arranged vertically represent different stages of plant growth, with stage I at the top and stage M at the bottom. The red and green borders of the small boxes indicate the up-regulation and down-regulation of compounds, respectively. In all pairwise comparison groups, up- and down-regulated compounds are for the latter. Note: flavanones liquiritigenin and naringenin are metabolic nodes at the intersection of the flavanone biosynthesis and the isoflavonoid biosynthesis; apigenin (flavone) is a metabolic node at the intersection of the flavanone biosynthesis and the flavone biosynthesis; kaempferol (flavonol) is a metabolic node at the intersection of the flavanonol biosynthesis and the flavonol biosynthesis. Purple star, metabolic nodes. NF, nitrogen free. SN, symbiotic nitrogen. MN, mineral nitrogen. I, 1 day post-treatment (dpt). N, 3 dpt. E, 15 dpt. M, 28 dpt. Solid arrows, single biosynthetic steps. Dotted arrows, multiple steps
The responses of DFs to different nitrogen sources were further analyzed. Some flavonoids showed significant changes in only one comparison group of roots or root exudates, and most of them changed significantly during the E and M stages, which was consistent with the variation in the plant growth phenotypes (Fig. 1). For example, in NF vs. MN, 2′-hydroxygenistein and biochanin A-7-O-glucoside at the E and M stages, and prunetin at stage E were significantly down-regulated in MN roots (Fig. 5A), demonstrating that the three flavonoids could be produced responding to nitrogen deficiency at the E and M stages; isovitexin was markedly up-regulated in MN root exudates at stage E (Fig. 5B), suggesting isovitexin could be involved in nitrogen uptake at stage E. In NF vs. SN, chrysin at stage E, pinocembrin and prunin at the E and M stages, and naringin at stage M were dramatically up-regulated in SN roots (Fig. 5A), indicating that they could be specifically interrelated to symbiotic nitrogen fixation; xanthohumol at the E and M stages and vitexin at stage M in SN roots were down-regulated (Fig. 5A), suggesting that these compounds could be produced responding to nitrogen deficiency at the E and M stages. Importantly, in MN vs. SN, six flavonoids, containing glycitein-7-O-glucoside at stage M, astragalin at the N and E stages, and nicotiflorin at stage N in the roots (Fig. 5A), and isoliquiritigenin, isosakuranetin and astragalin in the root exudates at stage N (Fig. 5B), were significantly up-regulated in SN, demonstrating that they could be mainly associated with the symbiotic infections. Besides, some flavonoids were present in multiple comparison groups and fluctuated with plant growth, suggesting that their response to different nitrogen nutrition of different sources were related to plant growth and development, such as genistein, isoliquiritigenin and eriodictyol in the roots (Fig. 5A), and prunetin and biochanin A in the root exudates (Fig. 5B).
Finally, two main metabolic pathways were identified (Fig. 5), including isoflavonoid biosynthesis, and flavone and flavonol biosynthesis. Concurrently, four compounds are metabolic nodes at the intersection of different flavonoid metabolic pathways, including liquiritigenin, naringenin, kaempferol and apigenin. In the plant growth process, node flavonoids were significantly enriched in response to different nitrogen sources. Compared with those in MN, liquiritigenin and naringenin were markedly down-regulated in both the 1-dpt NF and 1-dpt SN roots and were up-regulated in the 3-dpt NF roots, 28-dpt SN roots and in the root exudates of both NF and SN at 3 and 28 dpt (Fig. 5). Moreover, compared with those in MN, liquiritigenin was dramatically up-regulated in the 28-dpt NF roots and was down-regulated in the 15-dpt SN root exudates (Fig. 5), while naringenin was up-regulated in the 15-dpt NF roots and in the 15-dpt SN root exudates (Fig. 5). Apigenin was dramatically up-regulated in both NF roots and SN roots at 15 and 28 dpt, in the 1-dpt and 15-dpt NF root exudates and in the 15-dpt SN root exudates as compared to that in MN (Fig. 5). Compared with that in MN, kaempferol was significantly down-regulated at 1 dpt and subsequently up-regulated at 3 and 15 dpt in the SN roots, and also was up-regulated in the 1-dpt and 15-dpt NF root exudates (Fig. 5). Compared with that in NF, apigenin was markedly down-regulated in the 1-dpt, 3-dpt and 15-dpt SN root exudates, while kaempferol was dramatically down-regulated in the 15-dpt SN root exudates (Fig. 5). These results indicated that liquiritigenin, naringenin, apigenin and kaempferol were synthesized and secreted mainly responding to nitrogen deficiency and were associated with symbiotic nodulation, among which apigenin and kaempferol seemed to have a stronger response to nitrogen deficiency.
Network analysis of DFs in significantly enriched metabolic pathways
To further obtain the key DFs in response to different nitrogen sources, network analysis was performed on DFs in Fig. 6. In the network, nodes represent individual compounds. Edges represent pairwise correlations among nodes in the network, indicating biologically or biochemically meaningful interactions. There were 38 nodes and 97 edges in the roots network (Fig. 6A), and 32 nodes and 179 edges in the root exudates network (Fig. 6B), suggesting that the interactions among flavonoids in root exudates were stronger than that in the roots. There were 6 key nodes (degree > 10 and closeness centrality > 0.36) in the roots network, and the corresponding flavonoids were liquiritigenin, naringenin, daidzein-7-O-glucoside, formononetin-7-O-glucoside-6″-O-malonate, genistein and formononetin-7-O-glucoside (Fig. 6A). There were 10 key nodes (degree > 15 and closeness centrality > 0.58) in the root exudates network, and the corresponding flavonoids were 3,9-dihydroxypterocarpan, apigenin, cosmosiin, calycosin, daidzein-7-O-glucoside, genistein, kaempferol, luteoloside, rhoifolin, liquiritigenin (Fig. 6B). Two key nodes (liquiritigenin and naringenin) from the roots network and 3 key nodes (apigenin, kaempferol and liquiritigenin) from the root exudates network were consistent with metabolic nodes in Fig. 5, suggesting that plants may respond to different nitrogen sources by regulating the synthesis and secretion of these four key flavonoids.
Relative contents of key flavonoids
To obtain detailed information on the response of four key flavonoids described above (liquiritigenin, naringenin, apigenin, kaempferol) to different nitrogen sources, their relative contents were analyzed (Fig. 7). The content changes of liquiritigenin and naringenin in the roots and liquiritigenin in the root exudates under the different treatments varied with plant growth but showed similar trends under the same treatment (Fig. 7). For instance, the accumulation of liquiritigenin and naringenin in SN roots and liquiritigenin in SN root exudates fluctuated with plant growth and the trends of their content variation in SN were consistent with those in NF (Fig. 7). In MN, the accumulations of liquiritigenin and naringenin in the roots and liquiritigenin in the root exudates were at their highest level at stage I and much higher than those of other treatments, and then decreased rapidly with the plant growth (Fig. 7A). By contrast, the contents of liquiritigenin and naringenin in the roots and liquiritigenin in the root exudates of NF and SN were highest at stage N and were higher than those in MN at the last three stages (Fig. 7). These results indicated that the synthesis and secretion of liquiritigenin and naringenin could be mainly in response to nitrogen deficiency and symbiotic nitrogen fixation. In the root exudates, the content changes of apigenin and kaempferol under the different nitrogen conditions varied with plant growth but showed similar trends (Fig. 7B). In NF root exudates, the contents of apigenin and kaempferol were higher than or comparable to that of other treatments at the different stages of plant growth and their highest levels at the I stage (Fig. 7B), suggesting that apigenin and kaempferol were secreted to respond to nitrogen deficiency. Taken together, these results were similar to those of the metabolic nodes in Fig. 5.
Discussion
As a major component of protein, nucleic acids, and enzyme cofactors, nitrogen is an important macronutrient and plays a crucial role in plant growth and development (Chen et al. 2020). In general, nitrogen deficiency generally reduces plant leaf Chl concentration, yield, and biomass (Zhao et al. 2005). In our study, nitrogen deficiency inhibited the growth of aboveground parts of the plants, while the root nodule symbiosis and nitrogen addition can alleviate the inhibition of plant growth caused by nitrogen deficiency (Fig. 1). Furthermore, RL and RDW were significantly higher in NF than those in MN at stage M (Fig. 1B), which is an important adaptive strategy for plants in response to nitrogen deficiency (Sun et al. 2020). Nitrogen fixation cannot be performed in the early stages of the symbiotic relationship between legumes and rhizobia, so the ShN and RN of SN and NF should be comparable in the first three stages. However, the values of ShN at stage I and RN at the I and N stages of SN were markedly higher than those in NF, though the differences were not as significant as in the last stage (Fig. 1C), which may potentially be due to the release of nitrogen-containing compounds for plant growth after the breakdown of inoculated bacteria. Mature nodules provide nitrogen to plants, but cannot reach the comparable level as nitrate supply in a short period, so the ShN and RN of MN were significantly higher than those in SN at four stages (Fig. 1C). In NF, where external nitrogen sources were lacking, plants relied on internal nitrogen reservoirs to meet their growth demands, resulting in a gradual decline of both ShN and RN with plant growth (Fig. 1C). Unlike NF, the plants can acquire nitrogen through symbiotic nitrogen fixation by rhizobia in SN. However, in the initial stage of symbiotic organogenesis, young nodules do not have the ability to fix nitrogen until the nodules mature. Therefore, the substantial increase in ShN and RN at stage M may be associated with the nitrogen-fixation ability of the mature nodules (Fig. 1C). In MN, mineral nitrogen was acquired by the plants through root uptake and the long-distance transport between roots and shoots. Thus, the RN showed the trend of first rise and then decline (Fig. 1C). In summary, the changes of both ShN and RN in NF, SN and MN could be attributed to the different responses of plants to nitrogen availability and utilization during various growth stages.
The flavonoids secreted by plants into the rhizosphere have various ecological effects. For instance, medicarpin, pisatin and quercetin inhibited the growth of plant pathogens (Guenoune et al. 2001; Parvez et al. 2004; Wu and VanEtten 2004). Certain flavonoids acted as repellents for specific nematode species (Wuyts et al. 2006). Several flavonoids in root exudates of forage legume Desmodium uncinatum significantly inhibited the post-germination and attachment of the parasitic weed Striga (Hooper et al. 2010; Khan et al. 2010). Flavonoids are also classified as antimicrobials, insecticides and herbicides to promote plant growth based on their ecological effects in the rhizosphere. It has been identified that there were the flavonoids including acacetin, robinin, quercetin, rutin, kaempferol, apigenin and luteolin in R. pseudoacacia (Guo et al. 2019; Tyśkiewicz et al. 2019; Veitch et al. 2010), and apigenin, naringenin and isoliquiritigenin were found in its root exudates (Scheidemann and Wetzel 1997). In the current study, the flavonoid metabolism of the roots and root exudates in black locust were detected based on UPLC-MS/MS, and 295 and 242 flavonoids were characterized, respectively (Fig. 2A). These flavonoids were categorized into 11 major classes, including chalcones, flavanones, flavanonols, anthocyanidins, flavones, flavonols, flavonoid carbonoside, flavanols, tannin, biflavones, and isoflavones. This greatly expands our understanding of the specialized flavonoids in the roots and root secretions of R. pseudoacacia in response to different nitrogen environments.
The accumulation of specialized metabolites is one of the characteristics of various nutrient deficiencies in plants and the up-regulation of specialized metabolic pathways is considered to be the stress response mechanisms in plants, such as nutrient stress, drought stress and salt stress (Cao et al. 2019). Flavonoids, as one of these specialized metabolites, play a crucial role in resisting environmental stresses. It is worth mentioning that although different plant species often develop specific branches to adapt to diverse environmental conditions, they share a common core flavonoid pathway in higher plants (García-Calderón et al. 2020). The dynamic change analysis of flavonoids also showed that the synthesis and secretion of flavonoids varied with the different growth stages of plants in this study, which is consistent with the result that the secretion of isoflavones in soybeans showed growth-stage-dependent changes under normal conditions (Fig. 2B) (Sugiyama et al. 2016).
The flavonoid synthesis or flavonoid content was negatively correlated with increasing nitrogen supply. For instance, the levels of rutin and quercetin (flavonols), luteolin (flavones) and catechin (flavanols) were significantly increased due to nitrogen deficiency in inflorescences of Coreopsis tinctoria Nutt. (Li et al. 2021). Genistein and daidzein (isoflavones) secretions from soybean roots were significantly increased under nitrogen-deficient growth conditions, and glycitein (isoflavones) and isorhamnetin (flavonols) were accumulated to higher levels in soybean roots and shoots under low nitrogen conditions (Nezamivand-Chegini et al. 2022; Sugiyama et al. 2016). We found that more flavonoids were synthesized and secreted under nitrogen deficiency (Fig. 4). Due to the limitation of protein synthesis under nitrogen-deficient conditions, the availability of phenylalanine as a synthetic precursor of flavonoids increases that leading to the increased synthesis of flavonoids. Our results also provide evidence for previous research findings that nitrogen supply has a negative effect on flavonoid biosynthesis in plants.
As one of the responses to low nitrogen provision, plants produce and secrete flavonoids to increase nitrogen acquisition. For example, the flavonoids secreted by maize roots under nitrogen deprivation predominantly recruited Oxalobacteriaceae, which induced lateral root formation and thus facilitated nitrogen uptake from soil (Yu et al. 2021). Here, some flavonoids were identified to be markedly accumulated under nitrogen-free conditions compared to the two nitrogen treatments (Fig. 5), such as isoflavones (daidzein at N and M stages and formononetin at stage M), flavones (apigenin at stage E and cosmosiin at stage N) and flavanones (liquiritigenin at N and M stages and naringenin at stage N), suggesting these flavonoids were produced and secreted to respond to nitrogen deficiency and could be involved in nitrogen or other nutrition acquisition. In addition to getting nutrients, flavonoids are secreted by plants to defend against biotic and abiotic stresses by recruiting beneficial microorganisms and inhibiting harmful microorganisms. Acting as a repellent, daidzein (isoflavone) was secreted by soybean roots into the soils to induce the formation of a healthy microbial population (Okutani et al. 2020). Kaempferide 3-O-[2G-β-D-glucopyranosyl]-β-rutinoside (flavonols / flavanonols) from Dianthus caryophyllus had an obvious defense effect against Fusarium oxysporum (Curir et al. 2005). Therefore, in this study, the flavonoids synthesized and secreted by black locust under nitrogen-free conditions may also recruit beneficial microorganisms contributing to nitrogen acquisition, which needs to be further explored.
In addition to inhibiting harmful microorganisms, flavonoids can also promote the absorption of nutrient elements. The flavonoids, such as hesperetin (flavanones), luteolin and chrysin (flavones), enhanced mycorrhizal colonization in the roots, which led to the solubilization of mineral phosphate and may contribute to the mineralization of nutrients such as nitrogen (Bag et al. 2022). Flavonols (such as kaempferol, nicotiflorin, and rutin) also acted as metal-chelating agents, making it easier for plants to obtain certain trace elements (Gupta and Chakrabarty 2013). In the rhizosphere, flavonoids including genistein (isoflavones), quercetin and kaempferol (flavonols), improved Fe utilization by chelating and converting oxides of iron from Fe3+ to Fe2+ (Cesco et al. 2010). We also found some flavonoids such as isovitexin (flavonoid carbonoside) at stage E, genistein and genistein-7-O-glucoside (isoflavones) at stage M, and biochanin A (isoflavones) at N and M stages, were dramatically accumulated in MN root exudates compared to those in NF and SN, and the aboveground biomass of the plants was significantly increased (Fig. 5), suggesting these flavonoids may boost the plant’s uptake of mineral nitrogen and other nutrients, such as iron and phosphorus.
Low nitrogen level promotes the secretion of nod gene-inducing flavonoids from the legume roots. In chickpea roots inoculated with Rhizobium, a very high accumulation of the isoflavones, formononetin and biochanin A and their glycoside conjugates were found (Arfaoui et al. 2007). After inoculation with Bradyrhizobium japonicum, elevated concentrations of daidzein and genistein (isoflavones) were detected in soybean root exudate (Cooper 2004). The secretion of flavonoids varied at diverse stages of the development of the symbiotic relationship. The nod gene-inducing flavonoids, specifically luteolin (flavones), had dual actions as chemoattractants, and different flavonoids attracted different Rhizobium species (Hassan and Mathesius 2012). Kaempferol (flavonols) inhibited the polar transport of auxin in M. truncatula roots, leading to initiate the formation of nodule primordia. Meanwhile, the nod genes inducer 7,4′-dihydroxyflavone (flavones) inside the roots seemed to sustain Nod-factor synthesis in the infection thread (Zhang et al. 2009). Moreover, the levels of formononetin and ononin were systematically inhibited responding to Sinorhizobium meliloti and Nod factors, indicating that these isoflavones probably participated in the systemic modification of nodulation (Catford et al. 2006). Here, we found that the nod gene-inducing flavonoids were significantly accumulated in SN root exudates, including isoliquiritigenin (chalcones) at stage N, daidzein (isoflavones) at N and E stages, apigenin (flavones), daidzein-7-O-glucoside and genistein-7-O-glucoside (isoflavones) at stage E, genistein (isoflavones) at stage M, liquiritigenin (flavanones) at N and M stages, naringenin (flavanones) at N to M stages, formononetin-7-O-glucoside (isoflavones) at E and M stages. Additionally, flavonoids enriched significantly in the roots and root exudates in plants inoculated with rhizobia could be involved in different stages of symbiosis (Fig. 5). For instance, flavones (rhoifolin, cosmosiin and apigenin-7-O-[β-D-apiosyl-(1->2)-(6-malonyl-β-D-Glc)]) and flavonoid carbonoside (vitexin) were abundantly secreted at stage I in SN root exudates, suggesting they could be related to the recruitment or recognition of rhizobia; flavones (apigenin-7-O-[β-D-apiosyl-(1->2)-(6-malonyl-β-D-Glc)], luteoloside and cosmosiin) and flavonols (nicotiflorin, kaempferide, kaempferol and baimaside) were massively accumulated at stage N in SN roots, indicating they could be interrelated to both symbiotic infection and nodule initiation; chalcones (isoliquiritigenin and phloretin), isoflavones (3,9-dihydroxypterocarpan), flavonols (kaempferol and apigenin), flavanones (eriodictyol) and flavones (luteolin and chrysin) were largely synthesized at stage E in SN roots, demonstrating they could be associated with nodule development; chalcones (phlorizin), flavanones (liquiritigenin, naringenin, naringin and isosakuranetin), flavonols (quercetin, garbanzol and apigenin) and isoflavones (maackiain-3-O-glucoside, daidzein-7-O-glucoside, formononetin-7-O-glucoside-6″-O-malonate, formononetin, formononetin-7-O-glucoside, genistein-7-O-glucoside, glycitein-7-O-glucoside and pratensein) were abundantly accumulated at stage M in SN roots, suggesting they could be related to the maturation of nodules, the maintenance of nitrogen fixation function or systemic regulation of nodulation. Higher flavonoid levels in legume roots strongly facilitated infection via including up-regulation of nod genes and other responses in rhizobia (Liu and Murray 2016). Moreover, the expression of the nod genes in rhizobia (such as S. meliloti and B. japonicum) was suppressed by the presence of ammonium (Liu and Murray 2016). Studies on M. truncatula showed that the genes encoding chalcone-O-methyltransferase were expressed in the root hairs infected with rhizobia and in the infection zone but not in the nitrogen fixation zone of the root nodules (Chen et al. 2015). The nitrogen fixation and ammonia formation in nodules occur only in the nitrogen fixation region, so the expression of the genes involved in flavonoid synthesis may be inhibited by a high level of ammonia. Therefore, the level of available nitrogen may regulate the symbiotic nodulation between legumes and rhizobia by controlling the production of flavonoids in plants.
Flavonoid biosynthesis originates from phenylalanine. There are four pathways involved in flavonoid metabolism on the KEGG website, including flavonoid biosynthesis, flavone and flavonol biosynthesis, isoflavonoid biosynthesis and anthocyanin biosynthesis. In our study, we found that the main metabolic pathways involved in the DFs significantly enriched were the isoflavonoid biosynthesis and the flavone and flavonol biosynthesis (Fig. S2). Four key flavonoids (liquiritigenin, naringenin, kaempferol and apigenin) have been identified, and their responses to different nitrogen sources were distinct (Figs. 5, 6 and 7). Liquiritigenin, naringenin, apigenin and kaempferol were synthesized and secreted mainly responding to nitrogen deficiency and were associated with symbiotic nodulation, among which apigenin and kaempferol seemed to have a stronger response to nitrogen deficiency (Figs. 5, 6 and 7). Moreover, liquiritigenin, naringenin and kaempferol were associated with the absorption of nitrogen, which is consistent with previous findings that naringin and apigenin contributed to the mineralization of nutrients and kaempferol promoted the uptake of certain trace elements (Cesco et al. 2010; Gupta and Chakrabarty 2013; Masaoka et al. 1993).
Conclusion
In summary, our data point to a model (Fig. 8). The responses of flavonoids in the roots and root exudates of R. pseudoacacia to different nitrogen sources in the plant growth process were analyzed by metabolomic analysis. The results showed that there were significant differences in the synthesis and secretion of flavonoids in response to different nitrogen sources at various stages of plant growth. The synthesis and secretion of flavonoids were significantly increased in the plant under nitrogen deficiency conditions. Plants could respond to different sources of nitrogen by regulating the synthesis and secretion of key flavonoids. These findings provide important insights into the adaptability of legumes to different nitrogen environments and could aid in developing sustainable agricultural practices that optimize nitrogen resource utilization. Future work needs to elucidate the molecular mechanisms by which these flavonoids regulate the responses of R. pseudoacacia to different nitrogen sources and investigate the potential effects of these flavonoids on symbiotic nitrogen fixation and the growth of legume crops under varying nitrogen conditions.
Proposed mechanisms underlying how plants regulate flavonoid metabolism in response to different nitrogen sources. The big arrows with the pink gradient represent the plant roots and the light to dark color indicates the increased flavonoid synthesis and secretion at different stages of plant growth. DF, differential flavonoids. I, 1 day post-treatment (dpt). N, 3 dpt. E, 1 dpt. M, 28 dpt
Data availability
Data will be made available on request.
Abbreviations
- NF:
-
Nitrogen free
- SN:
-
Symbiotic nitrogen
- MN:
-
Mineral nitrogen
- dpt:
-
days post-treatment
- UPLC-MS/MS:
-
Ultra performance liquid chromatography tandem mass spectrometry
- PCA:
-
Principal component analysis
- HCA:
-
Hierarchical clustering analysis
- OPLS-DA:
-
Orthogonal partial least squares discriminant analysis
- FC:
-
Fold change
- VIP:
-
Variable importance for the projection
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- Chl:
-
Leaf chlorophyll content
- SL:
-
Shoot length
- SDW:
-
Shoo
t dry weight
- RL:
-
Root length
- RDW:
-
Root dry weight
- RN:
-
Root nitrogen content
- ShN:
-
Shoot nitrogen content
- SD:
-
Standard deviation
- ANOVA:
-
One-way analysis of variance
- DFs:
-
Differential flavonoids
References
Abd-Alla MH (2011) Nodulation and nitrogen fixation in interspecies grafts of soybean and common bean is controlled by isoflavonoid signal molecules translocated from shoot. Plant Soil Environ 57:453–458
Arfaoui A, El Hadrami A, Mabrouk Y, Sifi B, Boudabous A, El Hadrami I, Daayf F, Chérif M (2007) Treatment of chickpea with Rhizobium isolates enhances the expression of phenylpropanoid defense-related genes in response to infection by Fusarium oxysporum f. sp. ciceris. Plant Physiol Bioch 45:470–479
Bag S, Mondal A, Majumder A, Mondal SK, Banik A (2022) Flavonoid mediated selective cross-talk between plants and beneficial soil microbiome. Phytochem Rev 21:1739–1760
Begum AA, Leibovitch S, Migner P, Zhang F (2001) Specific flavonoids induced nod gene expression and pre-activated nod genes of Rhizobium leguminosarum increased pea (Pisum sativum L.) and lentil (Lens culinaris L.) nodulation in controlled growth chamber environments. J Exp Bot 52:1537–1543
Cao Y, Qu R, Miao Y, Tang X, Zhou Y, Wang L, Geng L (2019) Untargeted liquid chromatography coupled with mass spectrometry reveals metabolic changes in nitrogen-deficient Isatis indigotica Fortune. Phytochemistry 166:112058
Catford JG, Staehelin C, Larose G, Piché Y, Vierheilig H (2006) Systemically suppressed isoflavonoids and their stimulating effects on nodulation and mycorrhization in alfalfa split-root systems. Plant Soil 285:257–266
Cesco S, Neumann G, Tomasi N, Pinton R, Weisskopf L (2010) Release of plant-borne flavonoids into the rhizosphere and their role in plant nutrition. Plant Soil 329:1–25
Chen H, Chou M, Wang X, Liu S, Zhang F, Wei G (2013) Profiling of differentially expressed genes in roots of Robinia pseudoacacia during nodule development using suppressive subtractive hybridization. PLoS ONE 8:e63930
Chen H, Jia Y, Xu H, Wang Y, Zhou Y, Huang Z, Yang L, Li Y, Chen L, Guo J (2020) Ammonium nutrition inhibits plant growth and nitrogen uptake in citrus seedlings. Sci Hortic-Amsterdam 272:109526
Chen D, Liu C, Roy S, Cousins D, Stacey N, Murray JD (2015) Identification of a core set of rhizobial infection genes using data from single cell-types. Front Plant Sci 6:575
Chong J, Xia J (2018) MetaboAnalystR: an R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 34:4313–4314
Cooper JE (2004) Multiple responses of rhizobia to flavonoids during legume root infection. Adv Bot Res 41:1–62
Curir P, Dolci M, Galeotti F (2005) A phytoalexin-like flavonol involved in the carnation (Dianthus caryophyllus)- Fusarium oxysporum f. sp. dianthi pathosystem. J Phytopathol 153:65–67
Dai Z, Tan J, Zhou C, Yang X, Yang F, Zhang S, Sun S, Miao X, Shi Z (2019) The OsmiR396-Os GRF 8-OsF3H-flavonoid pathway mediates resistance to the brown planthopper in rice (Oryza sativa). Plant Biotechnol J 17:1657–1669
Fan M, Liu Z, Nan L, Wang E, Chen W, Lin Y, Wei G (2018) Isolation, characterization, and selection of heavy metal-resistant and plant growth-promoting endophytic bacteria from root nodules of Robinia pseudoacacia in a Pb/Zn mining area. Microbiol Res 217:51–59
García-Calderón M, Pérez-Delgado CM, Palove-Balang P, Betti M, Márquez AJ (2020) Flavonoids and isoflavonoids biosynthesis in the model legume lotus japonicus; connections to nitrogen metabolism and photorespiration. Plants 9:774
Gu Z, Eils R, Schlesner M (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849
Guenoune D, Galili S, Phillips DA, Volpin H, Chet I, Okon Y, Kapulnik Y (2001) The defense response elicited by the pathogen Rhizoctonia solani is suppressed by colonization of the AM-fungus Glomus intraradices. Plant Sci 160:925–932
Guo H, Wan X, Niu F, Sun J, Shi C, Ye JM, Zhou C (2019) Evaluation of antiviral effect and toxicity of total flavonoids extracted from Robinia pseudoacacia cv. idaho. Biomed Pharmacother 118:109335
Gupta R, Chakrabarty SK (2013) Gibberellic acid in plant: still a mystery unresolved. Plant Signal Behav 8:e25504
Hassan S, Mathesius U (2012) The role of flavonoids in root-rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. J Exp Bot 63:3429–3444
Haynes JG, Czymmek KJ, Carlson CA, Veereshlingam H, Dickstein R, Sherrier DJ (2004) Rapid analysis of legume root nodule development using confocal microscopy. New Phytol 163:661–668
He D, Singh SK, Peng L, Kaushal R, Vílchez JI, Shao C, Wu X, Zheng S, Morcillo RJ, Paré PW (2022) Flavonoid-attracted Aeromonas sp. from the Arabidopsis root microbiome enhances plant dehydration resistance. ISME J 16:2622–2632
Hoagland DR, Arnon DI (1950) The water culture method for growing plants without soil. Calif Agric Exp Stn Circ 347:2–32
Hooper AM, Tsanuo MK, Chamberlain K, Tittcomb K, Scholes J, Hassanali A, Khan ZR, Pickett JA (2010) Isoschaftoside, a C-glycosylflavonoid from Desmodium uncinatum root exudate, is an allelochemical against the development of Striga. Phytochemistry 71:904–908
Khalid M, Saeed-ur-Rahman BM, Dan-Feng H (2019) Role of flavonoids in plant interactions with the environment and against human pathogens - a review. J Integr Agr 18:211–230
Khan ZR, Midega CA, Bruce TJ, Hooper AM, Pickett JA (2010) Exploiting phytochemicals for developing a ‘push-pull’crop protection strategy for cereal farmers in Africa. J Exp Bot 61:4185–4196
Leoni F, Hazrati H, Fomsgaard IS, Moonen A, Kudsk P (2021) Determination of the effect of co-cultivation on the production and root exudation of flavonoids in four legume species using LC–MS/MS analysis. J Agr Food Chem 69:9208–9219
Li Z, Jiang H, Yan H, Jiang X, Ma Y, Qin Y (2021) Carbon and nitrogen metabolism under nitrogen variation affects flavonoid accumulation in the leaves of Coreopsis tinctoria. PeerJ 9:e12152
Li P, Li YJ, Zhang FJ, Zhang GZ, Jiang XY, Yu HM, Hou BK (2017) The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J 89:85–103
Liu C, Murray JD (2016) The role of flavonoids in nodulation host-range specificity: an update. Plants 5:33
Lv J, Dong Y, Dong K, Zhao Q, Yang Z, Chen L (2020) Intercropping with wheat suppressed Fusarium wilt in faba bean and modulated the composition of root exudates. Plant Soil 448:153–164
Masaoka Y, Kojima M, Sugihara S, Yoshihara T, Koshino M, Ichihara A (1993) Dissolution of ferric phosphate by alfalfa (Medicago sativa L.) root exudates. Plant Soil 155/156:75–78
Mathesius U (2018) Flavonoid functions in plants and their interactions with other organisms. Plants 7:30
Mohamad OA, Hao X, Xie P, Hatab S, Lin Y, Wei G (2012) Biosorption of copper (II) from aqueous solution using non-living Mesorhizobium amorphae strain CCNWGS0123. Microbes Environ 27:234–241
Nezamivand-Chegini M, Metzger S, Moghadam A, Tahmasebi A, Koprivova A, Eshghi S, Mohammadi-Dehchesmeh M, Kopriva S, Niazi A, Ebrahimie E (2022) Integration of transcriptomic and metabolomic analyses to provide insights into response mechanisms to nitrogen and phosphorus deficiencies in soybean. Plant Sci 326:111498
Okutani F, Hamamoto S, Aoki Y, Nakayasu M, Nihei N, Nishimura T, Yazaki K, Sugiyama A (2020) Rhizosphere modelling reveals spatiotemporal distribution of daidzein shaping soybean rhizosphere bacterial community. Plant Cell Environ 43:1036–1046
Parvez MM, Tomita-Yokotani K, Fujii Y, Konishi T, Iwashina T (2004) Effects of quercetin and its seven derivatives on the growth of Arabidopsis thaliana and Neurospora crassa. Biochem Syst Ecol 32:631–635
Peck MC, Fisher RF, Long SR (2006) Diverse flavonoids stimulate NodD1 binding to nod gene promoters in Sinorhizobium meliloti. J Bacteriol 188:5417–5427
Scheidemann P, Wetzel A (1997) Identification and characterization of flavonoids in the root exudate of Robinia pseudoacacia. Trees 11:316–321
Stewart AJ, Chapman W, Jenkins GI, Graham I, Martin T, Crozier A (2001) The effect of nitrogen and phosphorus deficiency on flavonol accumulation in plant tissues. Plant Cell Environ 24:1189–1197
Sugiyama A, Yamazaki Y, Yamashita K, Takahashi S, Nakayama T, Yazaki K (2016) Developmental and nutritional regulation of isoflavone secretion from soybean roots. Biosci Biotech Bioch 80:89–94
Sun X, Chen F, Yuan L, Mi G (2020) The physiological mechanism underlying root elongation in response to nitrogen deficiency in crop plants. Planta 251:1–14
Sun L, Lu Y, Yu F, Kronzucker HJ, Shi W (2016) Biological nitrification inhibition by rice root exudates and its relationship with nitrogen-use efficiency. New Phytol 212:646–656
Sun T, Zhang J, Zhang Q, Li X, Li M, Yang Y, Zhou J, Wei Q, Zhou B (2021) Integrative physiological, transcriptome, and metabolome analysis reveals the effects of nitrogen sufficiency and deficiency conditions in apple leaves and roots. Environ Exp Bot 192:104633
Tan H, Man C, Xie Y, Yan J, Chu J, Huang J (2019) A crucial role of GA-regulated flavonol biosynthesis in root growth of Arabidopsis. Mol Plant 12:521–537
Tian B, Pei Y, Huang W, Ding J, Siemann E (2021) Increasing flavonoid concentrations in root exudates enhance associations between arbuscular mycorrhizal fungi and an invasive plant. ISME J 15:1919–1930
Tyśkiewicz K, Konkol M, Kowalski R, Rój E, Warmiński K, Krzyżaniak M, Gil Ł, Stolarski MJ (2019) Characterization of bioactive compounds in the biomass of black locust, poplar and willow. Trees 33:1235–1263
Veitch NC, Elliott PC, Kite GC, Lewis GP (2010) Flavonoid glycosides of the black locust tree, Robinia pseudoacacia (Leguminosae). Phytochemistry 71:479–486
Vega-Hernández MC, Pérez-Galdona R, Dazzo FB, Jarabo-Lorenzo A, Alfayate MC, Leon-Barrios M (2001) Novel infection process in the indeterminate root nodule symbiosis between Chamaecytisus proliferus (tagasaste) and Bradyrhizobium sp. New Phytol 150:707–721
Wang L, Chen M, Lam P, Dini-Andreote F, Dai L, Wei Z (2022) Multifaceted roles of flavonoids mediating plant-microbe interactions. Microbiome 10:1–13
Wang X, Huo H, Luo Y, Liu D, Zhao L, Zong L, Chou M, Chen J, Wei G (2019) Type III secretion systems impact Mesorhizobium amorphae CCNWGS0123 compatibility with Robinia pseudoacacia. Tree Physiol 39:1533–1550
Wasson AP, Pellerone FI, Mathesius U (2006) Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18:1617–1629
Wu Q, VanEtten HD (2004) Introduction of plant and fungal genes into pea (Pisum sativum L.) hairy roots reduces their ability to produce pisatin and affects their response to a fungal pathogen. Mol Plant-Microbe Interact 17:798–804
Wuyts N, Swennen R, De Waele D (2006) Effects of plant phenylpropanoid pathway products and selected terpenoids and alkaloids on the behaviour of the plant-parasitic nematodes Radopholus similis, Pratylenchus penetrans and Meloidogyne incognita. Nematology 8: 89–101.
Xiao TT, Schilderink S, Moling S, Deinum EE, Kondorosi E, Franssen H, Kulikova O, Niebel A, Bisseling T (2014) Fate map of Medicago truncatula root nodules. Development 141:3517–3528
Yang X, Yu X, Liu Y, Shi Z, Li L, Xie S, Zhu G, Zhao P (2021) Comparative metabolomics analysis reveals the color variation between heartwood and sapwood of chinese fir (Cunninghamia lanceolata (Lamb.) Hook. Ind Crop Prod 169:113656
Yu P, He X, Baer M, Beirinckx S, Tian T, Moya YA, Zhang X, Deichmann M, Frey FP, Bresgen V (2021) Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation. Nat Plants 7:481–499
Zhalnina K, Louie KB, Hao Z, Mansoori N, Da Rocha UN, Shi S, Cho H, Karaoz U, Loqué D, Bowen BP, Zhalnina K, Louie KB, Hao Z, Mansoori N, da Rocha UN, Shi S, Cho H, Karaoz U, Loqué D, Bowen BP, Firestone MK, Northen TR, Brodie EL (2018) Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol 3:470–480
Zhang J, Subramanian S, Stacey G, Yu O (2009) Flavones and flavonols play distinct critical roles during nodulation of Medicago truncatula by Sinorhizobium meliloti. Plant J 57:171–183
Zhao X, Chen M, Li Z, Zhao Y, Yang H, Zha L, Yu C, Wu Y, Song X (2020) The response of volvariella volvacea to low-temperature stress based on metabonomics. Front Microbiol 11:1787
Zhao YH, Jia X, Wang WK, Liu T, Huang SP, Yang MY (2016) Growth under elevated air temperature alters secondary metabolites in Robinia pseudoacacia L. seedlings in Cd-and Pb-contaminated soils. Sci Total Environ 565:586–594
Zhao D, Reddy KR, Kakani VG, Reddy VR (2005) Nitrogen deficiency effects on plant growth, leaf photosynthesis, and hyperspectral reflectance properties of sorghum. Eur J Agron 22:391–403
Acknowledgements
This work was supported by Key Research & Development program of Shaanxi Province (No. 2020ZDLNY07-09), and National Natural Science Foundation of China (No. 41977052).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Responsible Editor: Ulrike Mathesius.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 34.9 MB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Y., Shen, Y., Shi, R. et al. The synthesis and secretion of key substances in the flavonoid metabolic pathway responding to different nitrogen sources during early growth stages in Robinia pseudoacacia. Plant Soil 494, 373–393 (2024). https://doi.org/10.1007/s11104-023-06286-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-023-06286-y