Abstract
Background and Aims
Leersia oryzoides, a wild relative of rice (Oryza sativa), may carry potential seed-borne bacterial endophytes which could be used to enhance growth of rice. We hypothesized that seed-associated bacteria from L. oryzoides would be compatible with rice and promote seedling growth, development, and survival.
Methods
We isolated bacteria from seed of L. oryzoides and checked compatibility with rice as well as Bermuda grass seeds for seedling growth promotion. Internal colonisation of bacteria into root cells was observed by ROS staining and microscopic observation. Growth promoting bacteria were evaluated for IAA production, phosphate solubilization and antifungal activities.
Results
Overall, ten bacteria were found to be growth promoting in rice seedlings with effects including restoration of root gravitropic response, increased root and shoot growth, and stimulation of root hair formation. All bacteria were identified by 16S rDNA sequencing. Six bacteria were found to become intracellular in root parenchyma and root hairs in rice and in Bermuda grass seedlings. Six bacteria were able to produce IAA in LB broth with highest (47.06 ± 1.99 μg ml−1) by LTE3 (Pantoea hericii). Nine isolates solubilized phosphate and inhibited at least one soil borne fungal pathogen.
Conclusions
Seed bacteria of L. oryzoides are compatible with rice. Many of these bacteria become intracellular, induce root gravitropic response, increase root and shoot growth, and stimulate root hair formation in both rice and Bermuda grass seedlings. Presence of bacteria protects seedlings from soil pathogens during seedling establishment. This research suggests that bioprospecting microbes on near relatives of rice and other crop plants may be a viable strategy to obtain microbes to improve cultivation of crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Interaction of plants with microbes is inevitable and frequent in plants growing in natural situations. These interactions between microbes and plants range from parasitism to mutualism. Some microbes are bacterial endophytes that colonize plant tissues in intracellular or intercellular spaces of roots, leaves, stems or seeds. Some endophytes do not tend to show negative effects on hosts, instead many endophytes improve the fitness of host plants (Hardoim et al. 2015; White et al. 2017). Endophytic bacteria may be acquired by plant roots through interactions with diverse microbes of the rhizosphere. Some bacteria are critical for seedling growth and survival and are vertically transmitted through seeds (Truyens et al. 2015; Verma et al. 2017; White et al. 2017). Bacterial endophytes have been reported to accelerate seed germination and seedling development in stressful and non-stressful conditions (Bent and Chanway 1998; Gond et al. 2015a, b). Endophytic microbes assist host plants directly by increasing nutrient mobilization through ‘rhizophagy’ or converting critical nutrients into usable forms (Paungfoo-Lonhienne et al. 2013; White et al. 2017), production of phytohormones (Long et al. 2008), preventing pathogen attack (Gond et al. 2015a), and increasing plant tolerance to abiotic stress, including salt, metal and heat stresses (Marulanda et al. 2010; Gond et al. 2015b).
The domestication and cultivation of crop plants may adversely affect the beneficial microbial community of plants. In one study involving the cultivation of the annual tobacco Nicotiana attenuate, Santhanam et al. (2015) showed that defensive bacterial plant microbes were progressively lost from plants after only seven years in continuous cultivation, leaving plants susceptible to a wilt disease. Reacquisition of microbes from uncultivated plants was sufficient to protect plants from disease. In another study, Irizarry and White (2017) found that the delinting process employing concentrated acids to degrade hairs on cotton seeds reduced the natural defensive seed microbes, leaving seedlings weakened and more susceptible to diseases and abiotic stress. Reacquisition of microbes from seeds of non-cultivated relatives of cotton resulted in seedlings that grew faster and were more disease and stress tolerant than untreated controls. Rice (Oryza sativa L.) is an important staple food crop with more than half of the world’s population relying on rice production for food (Molina et al. 2011). Bacterial endophytes have been reported from rice and several other crops in the recent past (Kaga et al. 2009; Hardoim et al. 2012, 2015; Gond et al. 2015a; Herrera et al. 2016). Presence of endophytic bacteria may modulate the level of phytohormones, hence affecting seedling physiology and participating in the seedling development process (Compant et al. 2005, 2010). Seed borne endophytes are thought to be specific and more closely-adapted to plants (Johnston-Monje and Raizada 2011). Rice has been in cultivation for more than 8000 years (Molina et al. 2011). It is conceivable that some of the beneficial and growth promotional microbes of ancestral rice could have been lost during domestication and the millennia of continuous cultivation, or may be lost in the future.
Leersia oryzoides (L.) Sw., also known as rice cut grass, is a wild relative of rice. This cool-season perennial native to North America is an obligate wetland grass common in agricultural drainage ditches and other flooded areas (Bouldin et al. 2004; Pierce et al. 2010). When mature, it can tolerate seasonal to permanent flooding up to 1 m in depth. It produces underground rhizomes which allow it to spread from agricultural drainage ditches into adjacent cultivated O. sativa where it becomes a problematic weed (Norsworthy et al. 2009). It can also spread by seed. We hypothesized that Leersia oryzoides seeds could carry potential endophytic bacteria which would be compatible with rice and promote seedling growth and development. In this research article we report that seed endophytes isolated from Leersia oryzoides seeds are compatible with rice and promote seedling growth, gravitropic response, and stimulate root hair formation. In this study, endophytic bacteria were also evaluated for auxin production, phosphate solubilization, and antifungal capacities. As far as we are aware this is the first report of the application of endophytic bacteria from seeds of L. oryzoides to enhance growth of rice seedlings and protect plants from pathogens.
Material and methods
Plant materials
For isolation of endophytic bacteria, Leersia oryzoides seeds were collected from plants growing near the entrance to Great Swamp National Wildlife Refuge of New Jersey, USA (40°42′45.0″N, 74°29′12.9″W). Seeds were stored in a refrigerator at 4 °C until processing. Rice (Oryza sativa L.; variety Rex) seeds were used to evaluate effects of bacteria on growth and development of seedlings. Bermuda grass (Cynodon dactylon L.) seeds were also used to screen for bacterial effects on seedling growth and development. Rex rice and Bermuda grass seeds were procured from Hancook Farm & Seed Company and stored at 4 °C in refrigerator.
Surface sterilization and isolation of bacterial endophytes
The covering lemas and paleas of Leersia oryzoides seeds were first removed by vigorous rubbing and washed with running water for 10 min. For surface sterilization, seeds were soaked in 4% NaOCl for 5 min with constant agitation on a rotary shaker. Then seeds were washed several times with sterile distilled water on a rotary shaker. Seeds were plated onto Lurea Bertani agar (LBA), yeast extract sucrose agar (YESA) and tryptic soy agar (TSA) for isolation of bacteria. Five seeds were plated onto each plate. After four to five days of incubation at 22 ± 2 °C, emerging bacterial colonies from the seeds were separated and streak plated for further purification. Cultures of isolates in LB broth containing 20% glycerol were preserved at −80 °C in a freezer in the Department of Plant Biology, Rutgers University.
Molecular identification
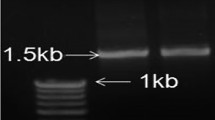
Out of 23 isolates from Leersia oryzoides seeds, the 10 most active in promotion of growth of rice seedlings were determined, then identified by 16S rDNA sequencing. For sequencing, total genomic DNA was extracted by use of a DNA extraction kit (Qiagen, USA) and the 16S rDNA sequence was amplified using universal primers 16SF (5′-AGAGTTTGATCCTGGCTCAG-3′) and 16SR (5′-CTACGGCTACCTTGTTACGA-3′). Amplified PCR products were resolved by electrophoresis in 1.5% (w/v) agarose gel stained with SYBR safe for visual examination. The PCR products were purified using a PCR purification kit (Qiagen, USA) and sent to Genewiz Inc. (South Plainfield, New Jersey) for sequencing. The sequences were BLAST searched on the NCBI GenBank database to find the closest matches.
Cleaning and inoculation of bacteria onto rice and bermuda grass seeds
Rice seed were cleaned by treating with 4% NaOCl for 1 h with constant agitation followed by 70% ethanol for 3 min. Seeds were washed several times with sterile distilled water to completely remove trace NaOCl from seeds. Bermuda grass seeds were treated with 4% NaOCl for 20 min and washed several times with sterile distilled water. Disinfected seeds of rice were inoculated with an overnight-grown suspension of 106–108 cells ml−1 of bacterial isolates including LLE3a, LLE11, LMT2b, LMT3b, LMY1a, LTE3, LTE8, LTE5a, LYE4a and LYY2b by soaking seeds for two hours in the bacterial suspensions then plating seeds on agarose plates (4–6 seeds on each plate). A total of 50 seeds were inoculated for each treatment. Rice seeds inoculated with bacteria were also placed into magenta boxes containing 15 g of sterilized potting mix in triplicate. Seedling development parameters including root and shoot lengths, gravitropic response, numbers of secondary roots and root hair formation were recorded after 7 days of incubation. Root and shoot lengths were measured for 40 seedlings from each treatment along with control. Roots of seedlings which penetrated vertically into the agar on plates were assessed to determine the % showing positive gravitropic response. Disinfected Bermuda grass seeds were also inoculated with the same bacteria and placed onto agarose plates.
ROS staining and visualization of endophytic bacteria in roots
Seven-day-old seedling roots of all the treatments (roots penetrating into agarose medium) were stained with 2.5 mM diaminobenzidine tetrachloride (DAB; Sigma- Aldrich, USA), prepared in deionised water for overnight staining. DAB-stained roots were washed with water and counter stained with aniline blue, and observed under a compound light microscope (Axioskop 20, Zeiss, Germany). DAB staining allows visualization of reactive oxygen (H2O2) produced around intra- or inter-cellular bacteria (White et al. 2014b). Roots of seven-day-old Bermuda grass seedlings inoculated with bacteria were also stained with DAB and observed under the microscope.
Root hair formation study
Seven-day-old seedling roots of rice and Bermuda grass treated with bacteria and untreated controls were observed for root hair formation. Seedlings (with and without DAB staining) with roots that penetrated into the agarose were sampled by excising two centimeters of main root, including the growing tip end. Roots were also observed microscopically for root hair development through the reverse of the plastic Petri plates.
Measurement of IAA (auxins)
The colorimetric method of Gordon and Weber (1951) was followed to assess the production of indole acetic acid (IAA) by bacteria in LB broth cultures using Salkowski reagent. For the preparation of Salkowski reagent, 1 ml of 0.5 M FeCl3 and 50 ml of distilled water were mixed with 30 ml of H2SO4 (S. Gr. 1.84). Five-days-old LB broth cultures of all bacteria were centrifuged for two min and the supernatant was collected for testing. One ml of culture supernatant was mixed with two ml of freshly prepared Salkowski reagent and the absorbance was recorded at 530 nm after 30 min of incubation. The experiment was done in triplicate. The concentration of IAA produced per ml of broth culture by bacteria was estimated by comparing absorbance with a standard curve. For making the standard curve, various concentrations of IAA (0.0, 5.0, 10.0, 25.0, 50.0, 100.0, and 200.0 μg ml−1) were prepared in LB broth media and then one ml of each concentration was mixed with two ml of Salkowski reagent. The absorbance was recorded at 530 nm after 30 min of incubation. The standard curve was obtained by plotting absorbance against the IAA concentration.
Phosphate solubilization and extracellular enzyme activity
For the phosphate solubilization assay one-day-old cultures of all the bacteria were inoculated onto Pikovskaya agar media (Pikovskaya 1948). After 5 days of incubation, clearing zones around bacterial colonies were measured. For cellulose activity, endophytic bacteria were inoculated onto yeast extract peptone agar medium composed of yeast extract 0.1 g, peptone 0 .5 g, agar 15 g, distilled water 1000 ml, pH 6, and supplemented with 0.5% Na-carboxymethyl cellulose (CMC). After 5 days of incubation, the plates were flooded with 0.2% aqueous solution of congo red for 15 min and de-stained with 1 M NaCl for 15 min. For pectinase activity, the endophytic bacteria were inoculated onto the pectin agar plates prepared by dissolving 5.0 g yeast extract, 5.0 g pectin, 15 g agar in 1000 ml water. Plates were incubated for 5–7 days at laboratory ambient temperature (≈ 22 °C). Plates were flooded with freshly-prepared 1% aqueous solution of CTAB for 10 min. Clear zones around colonies for both enzymes were measured.
Antifungal activities on plates
Endophytic bacteria were screened against three soil borne fungal pathogens including Fusarium oxysporum, Curvularia sp. and Alternaria sp. for their antifungal activities. Experiments were done with the dual culture technique on PDA plates and the percentage inhibition of the growth of the pathogen was calculated using the formula: % inhibition of radial growth = R1-R2/ R1 × 100 (Whipps 1997). In this formula R1 is the radial distance grown by the pathogen on the control plate and R2 is the distance grown on a line between the inoculation of the pathogen and the antagonist on the test plate.
Assessment of in vivo antagonism to Fusarium oxysporum
Disinfected Rex rice, Poa and Bermuda grass seeds were treated with endophytic bacteria, incubated at lab ambient temperature overnight, and then inoculated with the fungal pathogen Fusarium oxysporum (104–106 cells ml−1) along with the non-bacterial-treated control. After four to five days of incubation seedling roots were observed microscopically for Fusarium oxysporum infection within root tissues.
Statistical analysis
The SPSS-16 program was used for ANOVA followed by post hoc Duncan analysis at 0.05 level of probability to compare significance differences in root and shoot lengths among the treatments. We used non-transformed data for analysis. Means are presented with standard errors.
Results
Isolation and identification
Twenty-three bacterial isolates were recovered from seeds of Leersia oryzoides and preserved at −80 °C. All the isolates were screened for growth promotion capacity in rice seedlings. Isolates (10) showing growth promotion in rice were identified through 16S rDNA sequencing and BLAST searches of sequences in the NCBI database. Isolates, their GenBank accession numbers, and their closest matches are given in Table 1. Three bacteria were identified as species of Pantoea (LLE3a- P. agglomerans, LTE3- P. hericii and LYY2b– Pantoea vagans); four were identified as species of Pseudomonas (LLE11- Pseudomonas sp., LMT3b- P. parafulva, LMY1a- P. baetica and LYE4a- P. oryzyhabitans); LMT2b was identified as Microbacterium sp.; LTE8 was identified as Paenibacillus sp. and LTE5a was identified as Chryseobacterium sp. with 99 or 100% identity (Table 1).
Effect of inoculation of bacteria on rice seedling development
Out of 23 isolates recovered from Leersia oryzoides seeds, ten isolates showed positive growth promotion activity and effects on rice seedling development; thirteen isolates were either ineffective or inhibitory to rice seedling growth. Bacterial endophytes promoted increases in root and shoot lengths in rice seedlings (Fig. 1). Treatments of LLE3a, LMT2b, LMT3b, LTE3, LTE5a, LYE4a and LYY2b significantly increased root length (P ≤ 0.05). Inoculation with LLE3a, LMT2b, LTE3, LYE4a and LYY2b significantly increased shoot length (P ≤ 0.05) (Fig. 2). In comparison to root and shoot lengths (2.18 ± 0.16, 2.03 ± 0.14 cm) in controls, seedlings treated with LTE3 (Pantoea hericii) showed maximum root and shoot lengths (4.16 ± 0.15, 2.89 ± 0.17 cm) followed by LYY2b (Pantoea vagans), LYE4a (Pseudomonas oryzihabitans) and LLE3a (Pantoea agglomerans). Inoculation with bacteria onto seeds promoted positive gravitropism (i.e., seedling roots grew downward into agarose medium); LMT2b and LYY2b (70%), LTE3 (60%), LYE4a (75%), LMT3b and LTE8 (50%), and LLE3a, LLE11 and LTE5a (45%) in comparison to controls (20%) (Table 2). Except for LTE8 and LMT3b, all bacteria significantly promoted root hair formation in treated seedling roots of rice in comparison to controls (without bacteria). Seeds treated with Microbacterium sp. (LMT2b), Pantoea hericii (LTE3), Pseudomonas oryzihabitans (LYE4a) and Pantoea vagans (LYY2b) showed abundant production of long root hairs (Fig. 3, Table 2). These bacteria also promoted seedling development including increasing root and shoot lengths and root hair formation in Bermuda grass (Fig. 5). Root apical meristems appeared to be in better condition in seedlings inoculated with bacteria than controls free of bacteria in both rice and Bermuda grass seedlings.
Effect of inoculation of bacteria onto rice seedlings showing root and shoot lengths after 7 days of incubation on 0.7% agarose plate: where (a) shows that the root length of control is significantly less than bacterial treatments LLE3a, LMT2b, LMT3b, LTE3, LTE5a, LYE4a and LYY2b but not significantly different with LLE11, LMY1a and LTE8 (P ≤ 0.05, N = 40); (b) shows that the shoot length of the control is significantly less than treatments with bacteria LLE3a, LMT2b, LTE3, LYE4a and LYY2b but not significantly different with LLE11, LMT3b, LMY1a, LTE8 and LTE5a (P ≤ 0.05, N = 40). Different letters show significant differences among the means of treatments
Effect of endophytic bacteria on root hair development in rice seedlings: seven-day-old seedling roots were stained with DAB overnight: where (a) and (b) are controls (without bacteria) with no or very few root hairs; (c), (d), (e), (f) (g), (h) and (i) are inoculated with bacteria; LTE5a (e), LMTb (f) and LLE3a (h), were not observed to be intracellular, while LTE3 (c), LYY2b (d), LMTb (g), LYE4a (i) were confirmed to be intracellular
ROS staining and visualization of bacteria within cells of rice and Bermuda grass
Except LLE11, LTE8 and LTE5a, all treatment showed dark brown staining with DAB in comparison to the light stain seen in controls without bacteria (Table 2, Fig. 3). In control sedlings, no intercellular or intracellular bacteria were seen, although intercellular fungal mycelium was sometimes observed (Fig. 4). Out of 10 growth promoting bacteria, six, i.e. LMT2b (Microbacterium sp.), LMY1a (Pseudomonas baetica), LTE3 (Pantoea hericii), LTE8 (Paenibacillus sp.), LYE4a (Pseudomonas oryzihabitans) and LYY2b (Pantoea vagans), were confirmed to be endophytic and intracellular through microscopic examination. In ROS staining of rice seedling roots, L-form (cell wall deficient) bacteria were observed in root parenchyma and root hairs (Fig. 4). L-form endophytic bacteria were also seen in Bermuda grass root parenchyma and root hairs after ROS staining (Fig. 5). Strain LLE3a (Pantoea agglomerans) was observed on surfaces of rice roots but could not be confirmed to be intracellular in rice; LLE3a was, however, observed to be intracellular in Bermuda grass roots (Fig. 5).
Microscopic visualisation of endophytic bacteria inside root parenchyma and root hairs of rice seedlings after staining with DAB overnight: where (a), (b) and (c) are controls (without bacteria), showing cells that are free of internal H2O2-staining structures, however fungal mycelium found in (c) (arrow); Internal L-form bacteria (arrows) stain red to dark brown in root parenchyma (d), and root hairs (e) and (f) where seedlings were inoculated with LTE3, and root parenchyma (g) and root hairs (h) where seedlings were inoculated with LYY2b, while in root hairs (i) seedlings were inoculated with LMT2b
Microscopic observation of root hairs and bacterial endophytes inside root parenchyma and root hairs of Bermuda grass seedlings after staining with DAB overnight (seven-day-old seedlings): where in the bacteria-free control (a) and (b) no or very small root hair initials are seen and cells are free of internal reactive oxygen staining; in (c) and (d), where seedlings were inoculated with LLE3a, (e) and (f) inoculated with LYE4a, and (g) and (h) inoculated with LTE3, very long and abundant root hairs (arrows) with L-form bacteria within root cells (arrows) observed; (i) and (j) inoculated with LTE3 show L-form bacteria within root hair cells (arrows)
IAA (auxin) production, phosphate solubilization and enzyme activity
Out of ten bacteria evaluated for IAA production, six including LLE3a (Pantoea agglomerans), LLE11 (Pseudomonas sp.), LTE3 (Pantoea hericii), LTE5a (Chryseobacterium sp.), LYE4a (Pseudomonas oryzihabitans) and LYY2b (Pantoea vagans) showed production of auxin in LB broth medium (Table 3). LTE3 produced the highest levels of auxin (47.06 ± 1.99 μg ml−1), followed by LYY2b (23.63 ± 1.19 μg ml−1) and LLE3a (13.26 ± 0.88 μg ml−1). Except for LTE5a (Chryseobacterium sp.), all isolates showed phosphate solubilisation activity. Strains LLE3a, LMT2b, LMY1a, LTE3, and LYY2b were found most active in phosphate solubilization (Table 3). Five isolates, including LLE3a (Pantoea agglomerans), LMT2b (Microbacterium sp.), LTE3 (Pantoea hericii), LYE4a (Pseudomonas oryzihabitans) and LYY2b (Pantoea vagans), showed both extracellular cellulase and pectinase activity. However isolate LMY1a showed only pectinase activity (Table 3).
Antifungal activity
In co-culture experiments seven bacterial isolates were found to inhibit all three fungal pathogens tested. Inhibition of the fungi Fusarium oxysporum, Curvularia sp. and Alternaria sp. by the most inhibitory isolates was as follows: LLE3a-Pantoea agglomerans (29.25, 54.9, 40% inhibition, respectively); LLE11- Pseudomonas sp. (23.89, 32.35, 39.39%, respectively); LTE3- Pantoea hericii (30.13, 39.78, 26.67%, respectively), LTE5a- Chryseobacterium sp. (38.57, 56.57, 40%, respectively), and LYY2b- Pantoea vagans (29.27, 42.71, 19.05%, respectively) (Table 4). Surface sterilized rice and Bermuda grass seeds inoculated with bacterial endophyte LTE3 (Pantoea hericii) showed less colonization by Fusarium oxysporum than non-inoculated controls (Fig. 6a-h). Microscopic visualization of roots of rice seedlings showed that roots of seedlings inoculated with bacteria had significantly less infection by the Fusarium pathogen, compared to the non-bacterial controls (Fig. 6i-l).
Protection from the soil borne fungal pathogen Fusarium oxysporum by pre-treatment with endophytic bacterium Pantoea hericii (LTE3): Images (a), (b) and (c) are from controls of Poa, rice and Bermuda, respectively, inoculated only with the fungus, and (d), (e), (f) were pre-treated with LTE3 and then fungus; image (g) is from a control rice seedling (without bacteria) inoculated with fungus (agarose plate), image (h) was from seedlings treated with LTE3 and then the fungus (agarose plate); (i) and (j) show intercellular hyphal infection colonization in root parenchyma of seedlings treated only with the fungus, whereas in bacterium-treated (k) and (l) no fungal infection was seen, rather intracellular L-form bacteria are readily observed (arrows)
Discussion
Seed associated endophytic bacteria have been proposed to be tightly associated with plant growth and development from the beginning of germination (Johnston-Monje and Raizada 2011). Of twenty-three bacterial isolates from Leersia oryzoides seeds, ten (43.5%) were found to promote growth and development of rice seedlings. Leersia oryzoides is a wild relative of rice (Vaughan 1994; Ge et al. 2004) that grows naturally in swamp areas of North America. We hypothesized that L. oryzoides could host rice-compatible seed-borne bacterial endophytes. All 10 of the isolates were identified by 16S rDNA sequencing. Recovered bacterial endophytes mostly included Pseudomonads (4) and Pantoea spp. (3) (Table 1). LMT2b (Microbacterium sp.) and LTE5a (Chryseobacterium sp.) have not commonly been reported previously as endophytes.
Endophytism and compatibility
Endophytes are present in all plants and their presence may have important consequences ecologically for host plants which could be applied in agriculture (Rosenblueth and Martinez-Romero 2006; White et al. 2014a). In the present study we found six bacteria that we could confirm became intracellular in plant root cells (root hairs and/or root parenchyma cells) of rice and Bermuda grass seedlings after ROS staining. DAB stains H2O2 produced around bacteria inside cells due to defensive production of reactive oxygen by plant cells (White et al. 2014b). Some bacteria were found to be endophytic and intracellular in Bermuda grass but we could not determine their intracellular presence in rice. For example, LLE3a was found on the surface of root hairs in rice, but was observed to become intracellular as well as epiphytic of root hairs and root parenchyma in Bermuda grass seedlings (Fig. 5). This could be due to a compatibility issue where the bacterium did not enter rice cells, or perhaps this bacterium became intracellular but did not elicit a defensive response in rice and H2O2 was not produced. The mechanisms of entry of bacteria into cells are still not well understood. It has been hypothesized that openings in roots where branch roots emerge and damaged root hairs or wounds may be important sites of entry (Sørensen and Sessitsch 2006). Paungfoo-Lonhienne et al. (2013) proposed that bacteria may be actively taken into cells in a process that may be similar to phagocytosis. It is also possible that some bacteria actively invade root cells as endoparasites using their enzymatic capacities to degrade cell wall barriers to entry. In this respect, five endophytic bacteria in the present study show production of extracellular cellulase and pectinase, and they all were found to be endophytic either to rice or Bermuda grass seedlings. Production of hydrolytic enzymes may help in intracellular colonisation and development of endophytism. Variable sizes and shapes of bacteria were seen in root hairs and root meristems. This is due to bacterial loss of cell walls and formation of L-forms as bacteria enter plant cells (Walker et al. 2002; White et al. 2017). Although difficult to confirm, we hypothesize that bacteria lose their cell walls and enter plant cells at the root tip meristem where cell walls are thin. Symbiosis of plants with L-form of bacteria is non-pathogenic and frequently their intracellular or intercellular presence protects plants from diseases (Amijee et al. 1992; Walker et al. 2002). Amijee and collaborators have shown that L-forms of Bacillus subtilis protect Chinese cabbage seedlings from fungal pathogens during seed germination. Reports of large numbers of endophytic bacteria in plant tissues could be explained by their presence as non-pathogenic L-forms in planta (Allan et al. 2009). As plant cells differentiate many of the intracellular bacteria are degraded. The process of degradation of microbial endophytes within roots has been termed ‘rhizophagy’ (Paungfoo-Lonhienne et al. 2013; White et al. 2017). Through the process of rhizophagy, microbes that have both a free-living and endophytic phase may aid plants by scavenging organic or inorganic nutrients in the soil, then entering into plant tissues where they are degraded and nutrients are transferred to plants (Beltran-Garcia et al. 2014; White et al. 2015). We have also observed that some bacteria that enter roots are able to exit root epidermal cells during root hair formation where bacteria in hairs leave the hair at the elongating hair tip where the cell wall is thin (White et al. 2017).
Seedling development
Positive root gravitropic response, where roots grow downward, is an important developmental behavior during seedling establishment. We found that all the endophytic bacteria promoted positive gravitropic response in rice seedlings and further stimulated root growth and increased root architecture (i.e., more branch roots) in comparison to bacterial-free controls where roots lacked gravitropic response and reduced root growth and branching (Table 2, Figs. 1 and 2). Enhancing root architectural development during seedling establishment is controlled by the auxin/cytokinin ratio (Aloni et al. 2006), and microbial associations during seedling development may modulate hormone levels and hence improve proper development of roots (Ortíz-Castro et al. 2009). Four endophytic bacteria, LTE3 (Pantoea hericii), LYY2b (Pantoea vagans), LYE4a (Pseudomonas oryzihabitans) and LLE3a (Pantoea agglomerans) significantly (p ≤ 0.05) accelerated both root and shoot length growth in rice seedlings. Most of the isolates were also found to produce auxins and possess phosphate solubilization ability. Isolates LTE3 (Pantoea hericii) showed the highest production of auxin and also promoted the greatest root and shoot growth, followed by LYY2b (Pantoea vagans). Production of auxin was seen to roughly correlate with the level of seedling growth. During seedling development plants may interact with seed microbes, and microbes may release phytohormones and other molecule which may directly or indirectly modulate seedling morphogenesis (Long et al. 2008). Phosphorous is one of the major limiting nutrients for plant development due to its unavailability in many soils. Although it is present abundantly in many soils, it is often in an insoluble form (Miller et al. 2010). Phosphate solubilization capacity of bacteria, particularly those that cycle between a free-living soil phase and an endophytic phase, could be one of the important ways that endophytic microbes promote growth in plants (Oteino et al. 2015).
Root hair formation
Root hair development is important in enabling an increased soil-plant interface. Root hair formation increases root surface area and hence increases nutrient and water uptake by plants (Datta et al. 2011; Mercado-Blanco and Prieto 2012). Long and abundant root hairs have become recognised as an important trait for agricultural crops, increasing the nutrient acquisition capacity of plants. It has been reported that root hairs improve the uptake of immobile nutrients including phosphate and potassium (Datta et al. 2011). In the present study we found that inoculation of endophytic bacteria onto seeds induced the formation of long and abundant root hairs in rice and Bermuda grass seedlings (Table 2, Figs. 3–5). It is known that root hair development, density and length is influenced by soil nutrients and genetic factors (Hogh-Jensen and Pedersen 2003; Datta et al. 2011; Nestler et al. 2016). Previous research (Datta et al. 2011) suggests that in most of angiosperms, root hairs develop in the differentiation zone by epidermal cell extension. The pattern of root hair formation on plants is also different from species to species. For example, in rice root hairs develop in a random pattern, but in Aradidopsis root hairs are developed in files of root hairs and non-hair cells (Datta et al. 2011; Salazar-Henao et al. 2016). The role of root hairs in the development of efficient nitrogen-fixing nodules in legumes through symbiosis with rhizobia has been well studied (Kijne 1992; Mercado-Blanco and Prieto 2012). Literature also suggests that presence of root hairs is crucial for colonisation of roots by rhizospheric bacteria (Mercado-Blanco and Prieto 2012). A few studies on endophytic bacteria have reported that root hairs play key role in development of endophytism in plants with bacteria entering roots through root hairs (Oteino et al. 2015; Prieto et al. 2011). However, our research suggests that endophytic bacteria enter into root cells at the root apical meristem, and then induce root hair formation in the root epidermal layer (Verma et al. 2017; White et al. 2017). Root hair formation could be triggered by production of IAA (auxin) by these intracellular bacteria, effectively modulating root hair development (Verma et al. 2017). Presence of intracellular bacteria in roots was seen to correlate with higher levels of ROS compared to non-inoculated controls. Several studies report that auxin stimulates ROS production, root cell elongation and promotes seedling development (Morre et al. 1995; Krishnamurthy and Rathinasabapathi 2013; Ivanchenko et al. 2013). Here we suggest that endophytic bacterium-produced IAA directly, or with ROS, may be modulating root hair development. In Arabidopsis roots, ROS was found to regulate plant cell expansion and root hair development in growing regions of roots (Foreman et al. 2003). Regardless of the mechanism, it is apparent that endophytic bacteria are promoting root hair development in rice seedlings.
Antifungal activity against soil borne fungal pathogens
Except for LTE8 (Paenibacillus sp.), all endophytes inhibited at least two fungal pathogens tested in co-culture experiments (Table 4). Pantoea agglomerans (LLE3a), Pseudomonas sp. (LLE11), Pantoea hericii (LTE3), Chryseobacterium sp. (LTE5a), and Pantoea vagans (LYY2b) were found to be potent growth inhibitors of Fusarium oxysporum, Curvularia sp. and Alternaria sp. In examination of in vivo Fusarium oxysporum infection in seedling roots with and without Pantoea hericii (LTE3) we observed that treatment of surface sterilized seeds with Pantoea hericii (LTE3) significantly suppressed F. oxysporum colonisation on Poa, rice and Bermuda grass seedlings (Fig. 6a-h). In contrast, we observed intercellular and intracellular fungal infection in root parenchyma and root hairs of seedlings that had not been treated with bacteria (Fig. 6i-j). Very few or no hyphae were found in seedlings inoculated with Pantoea hericii (LTE3) (Fig. 6k-l). Endophytic bacteria may suppress the growth of soil borne fungal pathogens in several ways, including antibiosis, competition, or indirectly by inducing defense gene expression in host plants (Prieto et al. 2011). Recently, an endophytic Pantoea ananatis was reported to produce an antifungal indole-derivative compound (Aman and Rai 2016). Endophytic pseudomonads have been reported to produce antifungal metabolites, including pyrrolnitrin, phenazines and pyoluteorin (Compant et al. 2005). Bacterial endophytes compete with pathogens for space and nutrient availability. Endophytic bacteria may produce antifungal lipopeptides and also induce the up-regulation of defense genes in host plants (Gond et al. 2015a). With the present data we cannot identify which of these mechanisms may explain how these endophytes are suppressing soil borne pathogens. However more production of ROS in roots with endophyte could be an indication that host genes are being affected by the endophytes. Direct involvement of ROS is also known in defense against pathogens (Lamb and Dixon 1997; Huckelhoven and Kogel 2003). Further, ROS induces defense-related genes in plants (Levine et al. 1994; Torres et al. 2006). Two endophytic bacteria, Corynebacterium avescens and Bacillus pumilus from rice seeds were shown to be growth promoting and also compete with pathogen Azospirillum brasilense in the rhizosphere (Bacilio-JimeÂnez et al. 2001). Pseudomonas sp., a seed borne endophyte of Phragmites australis, was reported to suppress the pathogen Fusarium oxysporum (White et al. 2017). Prieto et al. (2011) found that internal colonisation of root hairs in olive roots by Pseudomonas spp. was associated with bio-control against Verticillium wilt.
Finally, we conclude that seed-associated bacteria from the wild relative of rice, i.e. Leersia oryzoides, are compatible with rice seeds and promote growth and root hair formation during seedling establishment. These bacteria also defend plants from fungal pathogens. Our findings indicate that endophytic bacteria from wild relatives of crops could be sources of microbes to develop consortia to enhance crop development and as biocontrol agents for sustainable crop production.
References
Allan EJ, Hoischen C, Gumpert J (2009) Bacterial L-forms. Adv Appl Microbiol Elsevier Inc 68:1–39. doi:10.1016/S0065-2164(09)01201-5
Aloni R, Aloni E, Langhans M, Ullrich CI (2006) Role of cytokinin and auxin in shaping root architecture: regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann Bot 97:883–893
Aman M, Rai RV (2016) Antifungal activity of novel indole derivative from endophytic bacteria Pantoea ananatis 4G-9 against Mycosphaerella musicola. Biocontrol Sci Tech 26(4):476–491
Amijee F, Allan EJ, Waterhouse RN, Glover LA, Paton AM (1992) Nonpathogenic association of L-form bacteria (Pseudomonas syringae pv. phaseolicola) with bean plants (Phaseolus vulgaris L.) and its potential for biocontrol of halo blight. Biocontrol Sci Tech 2:203–214
Bacilio-JimeÂnez M, Aguilar-Flores S, del Valle MV, PeÂrez A, Zepeda A, Zenteno E (2001) Endophytic bacteria in rice seeds inhibit early colonization of roots by Azospirillum brasilense. Soil Biol Biochem 33:167–172
Beltran-Garcia M, White JF, Prado FM, Prieto KR, Yamaguchi LF, Torres MS, Kato MJ, Medeiros MHG, Di Mascio P (2014) Nitrogen acquisition in Agave tequilana from degradation of endophytic bacteria. Sci Rep 4:6938. doi:10.1038/srep06938
Bent E, Chanway CP (1998) The growth-promoting effects of a bacterial endophyte on lodgepole pine are partially inhibited by the presence of other rhizobacteria. Can J Microbiol 44:980–988
Bouldin JL, Farris JL, Moore MT, Cooper CM (2004) Vegetative and structural characteristics of agricultural drainages in the Mississippi Delta landscapes. Environ Pollut 132:403–411
Compant S, Duffy B, Nowak J, Clément C, Barka EA (2005) Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl Environ Microbiol 71:4951–4959
Compant S, Clément C, Sessitsch A (2010) Plant growth promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42:669–678
Datta S, Kim CM, Pernas M, Pires ND, Proust H, Tam T, Vijayakumar P, Dolan L (2011) Root hairs: development, growth and evolution at the plant-soil interface. Plant Soil 346:1–14
Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S et al (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422:442–446
Ge S, Guo Y, Zhu Q (2004) Molecular phylogeny and divergence of the rice tribe oryzeae with special reference to the origin of the genus Oryza. In: Toriyama K, Heong KL, Hardy B (eds) Rice is life: scientific perspectives for the 21st century. Proceeding of the world rice research conference, IRRI and Japan international research Center for Agriculture Science, pp 40–43
Gond SK, Bergen MS, Torres MS, White JF (2015a) Endophytic Bacillus spp. produce antifungal lipopeptides and induce host defence gene expression in maize. Microbiol Res 172:79–87
Gond SK, Torres MS, Bergen MS, Helse Z, JFJr W (2015b) Induction of salt tolerance and up-regulation of aquaporin genes in tropical corn by rhizobacterium Pantoea agglomerans. Lett Appl Microbiol 60:392–399
Gordon SA, Weber RP (1951) Colorimetric estimation of indole acetic acid. Plant Physiol 26(1):192–195
Hardoim PR, Hardoim CCP, van Overbeek LS, van Elsas JD (2012) Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS One 7(2). doi:10.1371/journal.pone.0030438
Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, Döring M, Sessitsch A (2015) The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev 79:293–320
Herrera SD, Grossi C, Zawoznik M, Groppa MD (2016) Wheat seeds harbour bacterial endophytes with potential as plant growth promoters and biocontrol agents of Fusarium graminearum. Microbiol Res 186–187:37–43
Hogh-Jensen H, Pedersen M (2003) Morphological plasticity by crop plants and their potassium use efficiency. J Plant Nutr 26:969–984
Huckelhoven R, Kogel KH (2003) Reactive oxygen intermediates in plant microbe interactions: who is who in powdery mildew resistance? Planta 216:891–902
Irizarry I, White JF (2017) Application of bacteria from non-cultivated plants to promote growth, alter root architecture and alleviate salt stress of cotton. J Appl Microbiol. doi:10.1111/jam.13414
Ivanchenko MG, Desireeden O, Monhausen GB, Dubrovsky JG, Bednarova A, Krishnan N (2013) Auxin increases the hydrogen peroxide (H2O2) concentration in tomato (Solanum lycopersicum) root tips while inhibiting root growth. Ann Bot. doi:10.1093/aob/mct181 www.aob.oxfordjournals.org
Johnston-Monje D, Raizada MN (2011) Conservation and diversity of seed associated endophyes in Zea across boundaries of evolution, ethnography and ecology. PLoS One 6(6). doi:10.1371/journal.pone.0020396
Kaga H, Mano H, Tanaka F, Watanabe A, Kaneko S, Morisaki H (2009) Rice seeds as sources of endophytic bacteria. Microbes Environ 24(2):154–162
Kijne JW (1992) The Rhizobium infection process. In: Stacey G, Burris RH, Evans HJ (eds) Biological nitrogen fixation. Chapman and Hall, New York, pp 349–398
Krishnamurthy A, Rathinasabapathi B (2013) Oxidative stress tolerance in plants: novel interplay between auxin and reactive oxygen species signaling. Plant Signal Behav. doi:10.4161/psb.25761
Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48:251–275
Levine A, Tenhaken R, Dixon R, Lamb CJ (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79:583–593
Long HH, Schmidt DD, Baldwin IT (2008) Native bacterial endophytes promote host growth in a species-specific manner; phytohormone manipulations do not result in common growth responses. PLoS One 3(7):e2702. doi:10.1371/journal.pone.0002702
Marulanda A, Azcon R, Chaumont F, Ruiz-Lozano JM, Aroca R (2010) Regulation of plasma membrane aquaporins by inoculation with a Bacillus megaterium strain in maize (Zea mays L.) plants under unstressed and salt-stressed conditions. Planta 232:533–543
Mercado-Blanco J, Prieto P (2012) Bacterial endophytes and root hairs. Plant Soil 361:301–306
Miller SH, Browne P, Prigent-Cambaret C, Combes-Meynet E, Morrissey JP, O’Gara F (2010) Biochemical and genomic comparison of inorganic phosphate solubilisation in Pseudomonas species. Environ Microbiol Rep 2:403–411
Molina J, Sikora M, Garud N, Flowers J, Rubinstein S, Reynolds A, Huang P, Jackson S, Schaal BA, Bustamante CD, Boyko AR, Purugganan MD (2011) Molecular evidence for a single evolutionary origin of domesticated rice. Proc Natl Acad Sci 108(20):8351. doi:10.1073/pnas.1104686108
Morre DJ, Brightman AO, Hidalgo A, Navas P (1995) Selective inhibition of auxin-stimulated NADH oxidase activity and elongation growth of soybean hypocotyls by thiol reagents. Plant Physiol 107:1285–1291
Nestler J, Keyes SD, Wissuwa M (2016) Root hair formation in rice (Oryza sativa L.) differs between root types and is altered in artificial growth conditions. J Exp Bot 67:3699–3708
Norsworthy JK, Scott RC, Smith KL, Still J, Meier J (2009) Herbicide options for rice cutgrass (Leersia oryzoides) control. Weed Technol 23:1–5
Ortíz-Castro R, Contreras-Cornejo HA, Macías-Rodríguez L, López-Bucio J (2009) The role of microbial signals in plant growth and development. Plant Signal Behav 4(8):701–712
Oteino N, Lally RD, Kiwanuka S, Lloyd A, Ryan D, Germaine KJ, Dowling DN (2015) Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front Microbiol 6:745. doi:10.3389/fmicb.2015.00745
Paungfoo-Lonhienne C, Schmidt S, Webb R, Lonhienne T (2013) Rhizophagy- a new dimension of plant-microbe interactions, in de Briujn FJ (ed) Molecular microbial ecology of the rhizosphere. Wiley-Blackwell. Pub John Wiley & Sons, Inc
Pierce SC, Moore MT, Larsen D, Pezeshki SR (2010) Macronutrient (N, P, K) and redoximorphic metal (Fe, Mn) allocation in Leersia oryzoides (rice cutgrass) grown under different flood regimes. Water Air Soil Pollut 207:73–84
Pikovskaya RI (1948) Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Microbiology 17:362–370
Prieto P, Schilirò E, Maldonado-González M, Valderrama R, Barroso-Albarracín JB, Mercado-Blanco J (2011) Root hairs play a key role in the endophytic colonization of olive roots by Pseudomonas spp. with biocontrol activity. Microb Ecol 62:435–445
Rosenblueth M, Martinez-Romero E (2006) Bacterial endophytes and their interactions with hosts. Mol Plant-Microbe Interact 19:827–837
Salazar-Henao JE, Vélez-Bermúdez IC, Schmidt W (2016) The regulation and plasticity of root hair patterning and morphogenesis. Development 143:1848–1858
Santhanam R, Luu VT, Weinhold A, Goldberg J, Oh Y, Baldwin JT (2015) Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. PNAS 112(36):E5013–E5020. doi:10.1073/pnas.1505765112
Sørensen J, Sessitsch A (2006) Plant-associated bacteria lifestyle and molecular interactions. In: van Elsas JD, Jansson JK, Trevors JT (eds) Modern soil microbiology, 2nd edn. CRC press, pp 211–236
Torres MA, Jones JDG, Dangl JL (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141:373–378
Truyens S, Weyens N, Cuypers A, Vangronsveld J (2015) Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ Microbiol Rep 7(1):40–50
Vaughan DA (1994) The relationship between the genus Oryza and other grasses. In: The wild relative of rice: a genetic resources hand book. IRRI, Manila, pp 3–6
Verma SK, Kingsley K, Irizarry I, Bergen M, Kharwar RN, White JF (2017) Seed vectored endophytic bacteria modulate development of rice seedlings. J Appl Microbiol 22:1680–1691
Walker R, Ferguson CMJ, Booth NA, Allan EJ (2002) The symbiosis of Bacillus subtilis L-forms with Chinese cabbage seedlings inhibits conidial germination of Botrytis cinerea. Lett Appl Microbiol 34:42–45
Whipps JM (1997) Developments in the biological control of soil-borne plant pathogens. Adv Bot Res 26:1–134
White JF, Torres MS, Sullivan RF, Jabbour RE, Chen Q, Tadych M, et al (2014a) Occurrence of Bacillus amyloliquefaciens as a systemic endophyte of vanilla orchids. Microsc Res Tech doi:10.1002/jemt.22410
White JF Jr, Torres MS, Somu MP, Johnson H, Irizarry I, Chen Q, Zhang N, Walsh E, Tadych M, Bergen M (2014b) Hydrogen peroxide staining to visualize bacterial infections of seedling root cells. Microsc Res Tech 77:566–573
White JF, Chen Q, Torres MS, Mattera R, Irizarry I, Tadych M Bergen M (2015) Collaboration between grass seedlings and rhizobacteria to scavenge organic nitrogen in soils. AoB Plants 7. doi:10.1093/aobpla/plu093
White JF, Kingsley KI, Kowalski KP, et al (2017) Disease protection and allelopathic interactions of seed-transmitted endophytic Pseudomonads of invasive seed grass (Phragmites australis). Plant Soil doi:10.1007/s11104-016-3169-6
Acknowledgements
The authors are thankful to the Department of Plant Biology, Rutgers University, NJ for use of research facilities. SKV is thankful to UGC, India for providing a Raman Post Doctoral fellowship (No.-F 5-11/2016 IC) for the year (2015-16) to research in USA. SKV and RNK thank the Head and Coordinator CAS, FIST of Botany, B.H.U., Varanasi, India for providing facilities and leave to pursue research on endophytes. The authors are also thankful for support from the John E. and Christina C. Craighead Foundation, USDA-NIFA Multistate Project W3147, the Rutgers Turf Grass Research Center, and the New Jersey Agricultural Experiment Station.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible editor: Stéphane Compant.
Rights and permissions
About this article
Cite this article
Verma, S.K., Kingsley, K., Bergen, M. et al. Bacterial endophytes from rice cut grass (Leersia oryzoides L.) increase growth, promote root gravitropic response, stimulate root hair formation, and protect rice seedlings from disease. Plant Soil 422, 223–238 (2018). https://doi.org/10.1007/s11104-017-3339-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-017-3339-1