Abstract
Background and aims
High-throughput 454 pyrosequencing was applied to investigate differences in bacterial and fungal communities between replant and closely situated control non-replant (fallow) soils.
Methods
The V1-V3 region of the bacterial 16S rRNA gene and the ITS1 region of fungi from the different soils were sequenced using 454 pyrosequencing (Titanium chemistry), and data were analysed using the MOTHUR pipeline.
Results
The bacterial phyla Proteobacteria, Actinobacteria and Acidobacteria dominated in both fallow and replant apple orchard soils, and community composition at both phylum and genus level did not significantly differ according to NP-MANOVA. The fungal phyla Ascomycota, Zygomycota and Basidiomycota were dominant, and communities also did not differ in composition at either phylum or genus level. High positive Pearson correlations with plant growth in a plant growth assay performed with apple rootstocks plantlets were detected for the bacterial genera Gp16 and Solirubrobacter (r: >0.82) and fungal genera Scutellinia, Penicillium, Lecythophora and Paecilomyces (r: >0.65). Strong negative correlations with plant growth were detected for the bacterial genera Chitinophaga and Hyphomicrobium (r: <−0.78) and the fungal genera Acremonium, Fusarium and Cylindrocarpon (r: <−0.81).
Conclusions
Study findings are in part consistent with those of previous research, but also highlight associations between apple plants and certain microbial genera. The functional role of these genera in affecting soil health and fertility should be further investigated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Apple replant disease (ARD) is common in all major apple growing areas in the world. Most disease symptoms are evident shortly (1–3 months) after the replanting of apple trees on a site where apple trees were growing previously (Mazzola and Manici 2012), although the disease can affect apple yield and quality during the whole orchard life. The etiology of ARD is still not well understood, despite innumerous studies that have been conducted on the topic over the past few decades. In fact, problems in re-establishing old orchard sites have been documented for more than 200 years (Mai and Abawi 1981). The disease has been attributed to various abiotic factors, including pH highs and lows, phytotoxins, poor soil nutrition/structure, heavy metal contamination and cold or drought stress, however, the majority of evidence indicates that the disease is primarily a result of biotic factors (Mazzola 1998). Studies using soil fumigation (Browne et al. 2006; Mai and Abawi 1981; Spath et al. 2015; Yim et al. 2013), soil pasteurisation and soil sterilisation (Jaffee 1982; Manici et al. 2013; Yim et al. 2013) showed significantly increased growth of apple trees in fumigated and pasteurised soils when compared to untreated soils. Most likely, abiotic factors influence the symptoms of ARD, but are not the main cause of the disease.
Soil-borne microorganisms are considered to be an important component of soil function, being involved in the release of soil mineral nutrients, plant disease resistance, and plant diseases (Zhang et al. 2013). Fungi of the genera Cylindrocarpon and Rhizoctonia, as well as the oomycetes Phytophthora and Pythium are considered to be essential in the etiology of ARD, and these pathogens have been found frequently at different replant disease sites (Kelderer et al. 2012; Tewoldemedhin et al. 2011). Several Fusarium species have also been reported to be associated with ARD, although the role of the genus as a pathogen is controversial (Tewoldemedhin et al. 2011). The severity of ARD is also considered to be related to the presence of nematodes, such as Pratylenchus penetrans (Mazzola 1998).
The role of bacteria in ARD has been considerably less investigated than the role of fungi. Nonetheless, bacteria belonging to the Actinomycetes, as well as to the genera Bacillus and Pseudomonas have been implicated in ARD (Mazzola et al. 2002; Mazzola and Manici 2012; Utkhede et al. 1992). Certain species of Streptomyces, members of the Actinobacteria, have been reported to alleviate Rhizoctonia root infections in apple (Zhao et al. 2009). Similarly, different Pseudomonas species have been shown to be important in both causing and suppressing ARD (Mazzola et al. 2002). According to numerous authors however, the role of bacteria in ARD is debatable (Dullahide et al. 1994; Mazzola 1998; Tewoldemedhin et al. 2011).
The plant microbiome can be considered an extension of the host genome, as almost all tissues in plants host diverse microbial communities (Turner et al. 2013). As such, the plant microbiome serves as a key determinant of plant health and productivity, and manipulation of the microbiome has the potential to reduce the incidence of plant disease, thus resulting in higher yields. Recent studies have highlighted the ability of plant-associated microbiomes to influence plant traits including disease resistance, growth and abiotic stress tolerance (Mendes et al. 2011; Panke-Buisse et al. 2015). Despite its complexity and dynamism, the plant microbiome should be considered increasingly more in the future, when interpreting experimental data (Turner et al. 2013).
The recent development of ‘next-generation sequencing methods’ such as 454 pyrosequencing, has allowed the microbial communities in different environments to be efficiently sequenced and identified. Pyrosequencing techniques are capable of providing a more comprehensive picture of all microbes present in a sample, including low-abundance species (La Duc et al. 2012). The technique has been used in numerous studies to investigate bacterial communities in soils (Uhlik et al. 2013; Xiong et al. 2014; Yang et al. 2012; Zhang et al. 2013), and recently, a study focusing on bacterial communities using high-throughput sequencing to investigating ARD soils in China was published (Sun et al. 2014a). However, to our knowledge, no studies exist in which both the bacterial and fungal communities of soils affected by ARD have been investigated by high-throughput sequencing.
The aim of this study was to elucidate both bacterial and fungal populations in the rhizosphere most highly correlated with plant growth reduction, the main indicator of the multifactorial phenomena of ARD (Mazzola and Manici 2012; Scotto La Massese et al. 1988). Findings of a previous transnational survey on nine multi-generation apple orchards selected from specialised apple growing areas in Central Europe (Italy, Germany and Austria) showed significantly higher plant growth in soils from driving lanes or fallow plots close to the orchard (fallow) than in replanted rows (replant) in a plant growth assay (Manici et al. 2013). The highest growth in almost all orchards was observed in soils after disinfection by irradiation. A subset of soils from that study encompassing three sets of soils with a trend of higher plant growth in the fallow soils than in the replant soils was selected for the present study (significantly higher growth in soils from Germany and Austria, but not in soil from Italy). A deep-sequencing approach was employed targeting the V1-V3 region of the bacterial 16S rRNA gene and the ITS1 region of the fungi contained in fallow and replant rhizospheric soil samples from each of three orchards after a plant growth bioassay performed in a previous survey. The hypothesis was, that given the primary role of biotic components on ARD (highlighted in the previous survey by the comparison of plant growth in original soil samples with sterilised soils), the differences in plant growth between replant and fallow soils could be attributed to differences in the microbial communities in the soils.
Material and methods
Sample selection
The study was performed on rhizosphere soil samples collected at the end of a bioassay test which was performed in a previous study for evaluating soil health in nine orchards of Central Europe (Manici et al. 2013). The rhizospheric soil samples for the current study were taken from the bioassay, in which rooted cuttings of clonal M9 rootstocks were grown in plastic pots for 85 days on soil samples taken from replant and fallow sites from three of the nine multi-generation apple orchards under investigation (Nachtweih- Rhineland Palatinate, Germany; 50.625204 N 6.96336 E; Haidegg - Styria, Austria, 47.578064 N 13.45415 E; and Egma/Neustift- South Tyrol, Italy, 46.311590 N, 11.272514 E). The three sampling sites were chosen based on their similar plant growth responses, whereby, higher plant growth was observed in fallow soils compared with the paired replant soils. Rhizosphere soil samples were obtained from soil adhering to plantlet roots after the plastic pot was removed from the clod of soil and plants were gently shaken. Then, rhizosphere soils from each replicate were gently taken by hand and mixed to obtain a total of three samples representative of each fallow or replant soil from the three orchards.
DNA extraction
DNA extraction was conducted from 0.25 g amounts of −20 °C stored soils, using the NucleoSpin®Soil extraction kit for DNA, RNA and protein purification (Macherey-Nagel, Düren, Germany) according to the instructions provided by the manufacturer. The SL1 buffer provided was used during the extraction process.
Analyses of bacterial and fungal rRNA-ITS regions by fingerprinting (ARISA)
Bacterial community structure was determined by amplification of the 16S-23S rRNA gene ITS region from extracted DNA using the bacteria-specific Joe-labelled forward primer bRISAfor (5′-TGCGGCTGGATCCCCTCCTT-3′) and bRISArev reverse primer (5′-CCGGGTTTCCCCATTCGG-3′) (Hartmann et al. 2005; Kovacs et al. 2010). Fungal communities were characterized by amplification of ITS genes using the fungal-specific Joe-labelled forward primer F-ITS1 (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and F-ITS2 reverse primer (5′-TCCTCCGCTTATTGATATGC-3′) according to the conditions described elsewhere (Ranjard et al. 2000). Amplification was performed in volumes of 25 μL under previously described conditions (Chemidlin Prévost-Bouré et al. 2014; Hartmann et al. 2005). Thermal cycling comprised 30 cycles of denaturation at 94 °C for 1 min, annealing at 53 °C for 1 min and extension at 72 °C for 1 min, followed by a final extension at 72 °C for 10 min. PCR products (3 μl) were mixed with 0.5 μL internal-lane DNA standard (Genescan 500 ROX, Applied Biosystems Inc.) and 10 μL deionized formamide, before denaturation for 2 min at 95 °C and being placed on ice. Separation, detection and basic GeneScan analysis of fluorescently labelled 16S rRNA gene and ITS products were performed on an automated ABI PRISM 3100 DNA sequence analyzer (Applied Biosystems Inc., Foster City, CA). For each treatment, three replicates were analyzed. In addition, a mixture of DNA was prepared from 3 μl aliquots of each DNA extract from replicate soils to identify the potential mismatch between representative DNA mixtures used in subsequent deep sequencing.
Resulting fingerprints were analyzed in R according to procedures described recently (Abdo et al. 2006; Schutte et al. 2008). Fragments were expressed as a relative proportion of the total signal in each fingerprint. The significance of groupings by soil type and treatment was tested using NP-MANOVA (P < 0.05), whereas distances between pairs of replant-fallow soils for bacterial and fungal datasets were tested with ANOSIM in the PAST program software (Hammer et al. 2001).
PCR amplification and 454 pyrosequencing
PCR amplification, and high-throughput sequencing was performed at GATC Biotech (Konstanz, Germany). Because of the high degree of similarity among replicate soil samples observed using ARISA analysis, total community DNA for 454 pyrosequencing was extracted from pooled triplicate soil samples. The V1-V3 region of the 16S rRNA gene of bacteria was amplified using the primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 534R (5′-ATTACCGCGGCTGCTGG-3′), generating a 507 bp product (Methe et al. 2012). The ITS1 region of fungi was amplified using the primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′), generating a product varying from 300 to 500 bp in length (Buee et al. 2009). Each of the primers was synthesised together with a sequencing adaptor (A adapter 5′- CCATCTCATCCCTGCGTGTCTCCGACTCAG-3′, B adapter 5′- CCTATCCCCTGTGTGCCTTGGCAGTCTCAG-3′) and the forward primer was in addition modified with a MID tag, so that multiple samples could be pooled together and sequenced on the same half plate. For the V1-V3 sequencing, the FLX A adapter was fused with the 534R primer, and the FLX B adapter was fused with the 27F primer. For ITS sequencing, the FLX A adapter was fused with the ITS1F primer, and the FLX B adapter was fused with the ITS2 primer. Amplicons were uni-directionally sequenced from the FLX A adapter site with a 454 Life Sciences Genome Sequencer GS FLX instrument (454 Life Sciences, Brandford, CT, USA) using Titanium chemistry.
Bacterial 16S rRNA gene and fungal ITS sequences were analysed using the MOTHUR v.1.33.3 software pipeline (64 bit executionable) according to Standard Operating Procedures for 454 (Schloss et al. 2009). Quality data files were initially denoised in order to remove sequences that contained sequencing errors. Sequences were removed that had > 2 differences with the primer region, > 1 difference with the barcode, had a length of < 200 bp (bacteria) or 150 bp (fungi), or had homopolymers > 8 nt in length. Unique bacterial sequences were aligned against the SILVA rRNA gene database (Quast et al. 2013) and trimmed such that only overlapping sequence was considered. Fungal sequences were pairwise compared to the recently established unified fungal UNITE ITS database (Kõljalg et al. 2013). Chimeras were removed using the chimera.uchime command. Pre-clustering was conducted at 2 %, and rarefaction curves were generated using a 97 % identity cut-off. Operational taxonomic units (OTUs) were binned at 97 % identity. The number of sequences included in analyses for fungal and bacterial datasets (29,748 and 11,678, respectively) was based on the lowest number of sequences found in all samples. Three diversity estimates (InvSimpson diversity, Shannon diversity and Chao richness estimate) and Good’s coverage were calculated for bacteria and fungi, using MOTHUR.
Data analysis
Plant growth assay data obtained in a previous study (Manici et al. 2013) from three replicates per soil sample (three orchards, two positions within the orchard) were re-subjected to a two-way ANOVA for position within the orchard (replant and fallow) and orchard location (Germany, Austria and Italy) using Statgraphics centurion software (2005 STATPOINT Inc. Virginia, USA). Multivariate analyses and comparison of diversity were performed using the PAST program software for data analysis in paleoecology (Hammer et al. 2001).
Data matrices of rhizospheric bacterial and fungal phylum and genus abundance were ln (x + 1) transformed and subjected to one-way NP-MANOVA for position within orchards (fallow and replant). Principal component analysis (PCA) was inferred from physical and chemical parameters of fallow and replant soils of the three orchards selected for this study. To estimate diversity, data were ln (x + 1) transformed, pooled and compared with the Shannon diversity T test and diversity profiling using PAST Software ver. 3.0 was conducted (Hammer et al. 2001).
As plant growth is the main indicator of soil health and soil suppressiveness (Nielsen and Winding 2002), Pearson’s correlation was calculated (using Microsoft Excel) between the abundance of fungal and bacterial genera and plant growth obtained in the same soil samples (number of paired counts: 6). Genera present in all samples, with an abundance of > 10 in at least one sample, and with correlations r < −0.4 or r > 0.4 were included in results tables.
Accession numbers
The sequences obtained in this study have been submitted to the NCBI Sequence Read Archive (SRA) under the Bioproject SUB801699.
Results
Soil physical-chemical parameters and plant growth data analysis
Sandy clay soils from Nachtweih (Austria), loam sandy soils from Egma (Italy) and clay loam soils from Nachtweih (Germany) were included in the study. Soil parameters were found to differ at the three orchards and no significant correlations could be made between fallow and replant soils with the various physical-chemical parameters and soil biological variables (Manici et al. 2013). PCA of the soils is shown in Fig. ESM1. Fallow and replant soils differed significantly (P < 0.001) in terms of plant growth (Table 1). In contrast, orchard location did not lead to significantly different plant growth. There was no significant interaction between the two analyzed variables. Based on these findings, studies focused on differences between microbial communities in fallow and replant soils.
ITS fingerprinting
NM-MDS Bray-Curtis similarity index 3D ordinations were most successful in representing sample data with lowest stress values (stressbacteria = 0.15; stressfungi = 0.14). Analysis of bacterial and fungal fingerprints in the span of 50 to 650 bp showed that samples grouped according to soil sample origin (Egma, Haidegg or Nachtweih) and treatment (fallow or replant) in an NM-MDS analysis (Fig. 1). The replicates of each soil type were found to group together. NP-MANOVA showed that all six groups for bacteria and fungi differed significantly (P < 0.001; total sum of squares = 7.961 and 7.518; within group sum of squares = 1.656 and 1.549, respectively).
Closer examination of distribution of groups and calculation of distance between pairs of replant-fallow soils for bacterial and fungal datasets, respectively, also showed that the NM-MDS Bray-Curtis similarities between Egma replant-fallow soils were significantly higher (P < 0.001; 0.18 ± 0.10 and 0.24 ± 0.05, respectively) than for the Haidegg replant-fallow soils (0.06 ± 0.03 and 0.13 ± 0.08, respectively) and Nachtweih replant-fallow soils (0.02 ± 0.04 and 0.11 ± 0.08, respectively), as calculated by ANOSIM. Replant soils were also found to cluster together.
Pyrosequencing-derived bacterial diversity
A total of 256,749 sequence reads for bacteria were obtained from the 1/8th region of the Picotitre plate using a GX FLX sequencer. After denoising and quality control processing, 85,812 bacterial sequences were included in analyses. OTU assignment conducted at the 97 % sequence similarity level resulted in 28,337 OTUs. The total number of reads and OTUs did not differ greatly between the different soil treatments (Table 2). After quality processing, the lowest number of sequences found in a sample was 11,678, and to compare all soil samples without bias, downstream analyses were conducted on the first 11,678 sequences in all soils. Average read length was 213 bp. In total, 437 bacterial genera were detected in all soils.
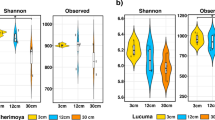
High parametric and non-parametric diversity indices were recorded for all soils in the study (Table 2). Shannon, Invsimpson (both parametric) and Chao (non-parametric estimator) diversity indices showed that the Egma fallow soil had the highest community diversity. The higher variability seen among the Egma fallow replicates in ITS fingerprinting likely contributed to this higher diversity. Coverage values between 0.63 and 0.68 were obtained for all soils, indicating between 63 and 68 % species detection rates. Results of rarefaction analysis indicate that sequence coverage was not complete in any of the 6 libraries generated, and that deeper sequencing would likely identify additional OTUs (Fig. 2a).
All of the sequences obtained from the six soils were classified to 25 different bacterial phyla, using the MOTHUR pipeline analysis, although of the 85,812 sequences, 13,361 sequences were not able to be classified. The communities in all soils were dominated by Proteobacteria, Actinobacteria and Acidobacteria, which comprised between 69 and 74 % of all sequences. There were only minor differences between the bacterial communities of the soils at the phylum level (Fig. 3a), and fallow and replant soils were not found to differ significantly according to NP-MANOVA analyses. The remaining phyla represented were present at an abundance of < 4 % of all sequences in soils. Principal components analysis (PCA) of phylum level diversity indicated a separate clustering of fallow and replant soils (Fig. 4a). The replant soils all clustered within the X+ Y- quadrant, while the fallow soils grouped in the X- Y+ quadrant. The first two axes explained 95 % of the variance.
The three dominant phyla were further investigated at the order level (Fig. ESM2), and no significant differences were observed between fallow and replant soils. According to PCA, unclassified orders and the order Actinomycetales contributed the most to the differentiation of replant and fallow soils within the phylum Actinobacteria (data not shown). For the phylum Proteobacteria, unclassified orders and the order Pseudomonadales had the highest contribution on differentiation (data not shown), while for the Acidobacteria, the orders Gp6 and Gp4 contributed the most to the differentiation of replant and fallow soils (data not shown).
According to the bacterial genus composition, the diversity in the replant (375 genera) and fallow (357 genera) soils differed significantly (Shannon T test, P < 0.001). There was higher genus diversity in replant soils than in fallow soils (Shannon H: 5.55 and 5.47, respectively). The diversity profile (not shown) showed that the two communities differed both in richness and in evenness. The finding that orchard location did not exert a significant influence on plant growth initiated an investigation of correlation determination between the abundance of genera and plant growth in the three fallow soils and in the three replant soils (Table 3). High correlations between plant growth and the genera Gp16 and Solirubrobacter (r: 0.87 and 0.82, respectively) were detected, indicating these genera to positively influence plant growth. The most negative correlations between plant growth and genus abundance were recorded for Chitinophaga (r: −0.82), Hypomicrobium (r: −0.78), Nitrospira (r: −0.77), Devosia (r: −0.72) and Sphingomonas (r: −0.71).
Pyrosequencing-derived fungal diversity
A total of 239,409 sequence reads for fungi were obtained from the 1/8th region of the Picotitre plate using a 454 genome sequencer. After quality control processing and denoising, 202,041 fungal sequences were included in analyses. Table 2 shows the number of OTUs (97 % sequence similarity level) in each soil. Downstream analyses were conducted on 29,748 sequences in each soil, which was the smallest number of sequences in any of the six soils. Average read length was the same as for bacteria, but for fungal ITS datasets, sequences > 150 bp in length were also included in the analysis. In total, 331 fungal genera were detected in all soils. Figure 2b shows the fungal rarefaction curves in each of the six soils. In contrast to the results of the bacterial sequencing, results indicate that sequence coverage was closer to completion in all libraries generated.
At all three sites investigated, > 99 % of sequences from both fallow and replant soils were comprised of fungi belonging to the phyla Ascomycota, Zygomycota and Basidiomycota (Fig. 3b). In total, fungi from seven phyla were detected, and there were no significant differences between fallow and replant soils at the phylum level. In all soils with the exception of the Haidegg and Nachtweih fallow soils, Ascomycota were present in the highest numbers. A higher proportion of Zygomycota were however detected in both the Haidegg and Nachtweih fallow soils, compared with the replant soils collected at the same sites. No differences were observed between the Egma fallow and replant soils. The PCA of fungal communities in soil samples at the phylum level indicated less distinct clustering of the replant and fallow soils (Fig. 4b), with the Egma fallow and replant soils grouping closely together. Almost all the variance was explained by the two axes (98.3 %).
A comparison of fungal communities in fallow and replant soils at the family level (abundance >5 %) was conducted (Fig. ESM3), and analysis indicated no significant differences in the soils. The families Mortierellaceae and Nectriaceae were found to exert the most impact in PCA (data not shown).
Fungal diversity of fallow and replant soils (266 and 262 genera, respectively) did not differ according to the Shannon T test. This finding was supported by the graphic diversity profile (not shown).
Fungal community composition of apple orchard fallow and replant soils did not significantly differ at the genus level. Higher numbers of the pathogenic genera Acremonium, Cylindrocarpon and Fusarium were detected in replant soils, and plant growth was highly and negatively correlated with these genera (r: −0.84, −0.83, −0.81; Table 4). Negative correlations with plant growth were, in addition, recorded for Lophiostoma (r: −0.75), Exophiala (r: −0.71) and Schizothecium (r: −0.64). Fungal genera detected at relative high abundance and negatively correlated with plant growth included Cryptococcus (RA: 3.94 %, r: −0.58), Humicola (RA: 3.15 %, r: −0.46), Gibberella (RA: 2.13 %, r: −0.43 %) and Gibellulopsis (RA: 1.7 %, r: −0.33). Low numbers of the genera Armillaria (r: −0.28) were detected in all soils in this study (RA: <0.01 %).
High positive correlations of plant growth with the genera Scutellinia (r: 0.72), Penicillium (r: 0.68), Lecythophora (r: 0.67) and Paecilomyces (r: 0.65) were recorded. The most abundant fungal genus detected in all soils was Mortierella, and higher numbers of this genus were found in all fallow soils than their respective paired replant soil (r: 0.63).
Discussion
Fallow and replant soils from 3 apple orchards
Physical-chemical data from fallow and replant soils at the three orchards investigated showed that soils clustered together depending on site, rather than soil position within the orchard (Fig. ESM1). Also, Egma fallow and replant soils grouped more closely together than the fallow and replant soils from Haidegg and Nachtweih. These results were consistent with the findings of the plant growth tests, where the difference in shoot length between fallow and replant soils was not significant in the Egma soils, but was in the Haidegg and Nachtweih soils (Manici et al. 2013).
ARISA analysis of ITS regions showed all six soils to differ significantly for both bacterial and fungal communities, supporting the findings of the PCA of physical-chemical data. The Egma soils were more similar to each other than the Haidegg and Nachtweih soils, again providing supporting evidence for the findings of physical-chemical data and plant growth tests (Manici et al. 2013) and indicating that the more similar microbial communities in the Egma fallow and replant soils attributed to the reduced effect of decline between the two soils. The DNA replicates from most soil types grouped closely together (Fig. 1), although there was some heterogeneity within the Egma and Haidegg fallow soil samples in the bacterial analysis and in the Haidegg replant soil in the fungal analysis, resulting in the replicates grouping less tightly. As the fourth ITS sample representing a pooled DNA extract was correctly placed in close proximity of the three ITS soil replicates for all soils, the pooled DNA extracts were used for pyrosequencing targeting the 16S rRNA and ITS genes.
Bacterial analyses
The calculated sequence coverage values for bacteria (63–68 %) were lower than those found by others (Sun et al. 2014b; Uhlik et al. 2013; Zhang et al. 2013). The lower coverage values are reiterated in the rarefaction curves, which indicate that deeper sequencing would most likely have identified greater diversity. Nonetheless, the high Shannon indexes obtained for all soils indicate that significant diversity was captured using this approach.
Fallow and replant soils were not found to differ significantly from each other at the phylum and genus levels, indicating that the microbial composition did not largely vary between the replanted rows and row strips in apple orchards. These findings are consistent with those observed for root endophytic fungal communities in fallow and replant soils whereby most species were detected in both soil types, although differences in species abundance existed (Manici et al. 2013). The most frequently occurring phyla detected in this study were Acidobacteria, Actinobacteria and Proteobacteria, supporting the findings of others (St. Laurent et al. 2008; Sun et al. 2014b). Correlations of bacterial genus abundance in soils with plant growth were conducted in order to identify the bacterial genera positively and negatively associated to plant growth, the main indicator of soil health and of replant disease (Mazzola and Manici 2012). Negative correlations between the abundance of the dominant genera Pseudomonas, Sphingomonas and Lysobacter and plant growth were found (Table 3) although the correlation for Pseudomonas was weak (r: −0.03, not shown). In a separate study investigating replant disease, it was reported that Lysobacter and Pseudomonas both increased in abundance at the replant site (Sun et al. 2014a). These authors speculated that the abundance of Lysobacter in replanted soils was linked with its ability to prolifically produce novel antibiotics in response to high levels of pathogenic fungi in the soils. Pseudomonas species have been reported to both cause and suppress ARD in the northwest USA (Mazzola et al. 2002). The poor correlation obtained in this study indicates that most likely, Pseudomonas did not play a significant role in the decreased soil fertility of replant soils investigated. Despite a null correlation with plant growth in this study, Sphingomonas has been largely reported in the literature as a plant growth promoter (Maheshwari 2011) and active in the biocontrol of pathogenic bacteria (Innerebner et al. 2011). This may suggest that various species play different roles in different environments.
No literature concerning a negative impact on plant growth of other genera negatively correlated with plant growth in this study and listed in Table 3 could be found. In contrast, Hyphomicrobium, Nitrosospira, Rhizobium and Gemmatimonas are genera known to be involved in resource acquisition in different plant processes. The higher occurrence in replant soils of these bacteria with known positive impacts on soil fertility may possibly have been due to cropping practices applied to replanted soils, which aim to maintain high fertility in planted areas within the orchards.
The phylum Acidobacteria has commonly been reported as dominant in soils (Gottel et al. 2011; St. Laurent et al. 2008; Zhang et al. 2013), supporting the findings of this study. The genus Gp16 was found to be highly positively correlated with plant growth. No evidence relating Gp16 to plant disease or disease suppression exists in the literature, however, bacteria in the phylum Acidobacteria were reported present at higher frequencies in the rhizosphere of healthy plants than in diseased plants (Yin et al. 2013). The beneficial effect on plant growth in fallow soils from Gp16 most likely resulted from plant growth promotion by the bacteria, and contributed to increased soil fertility.
In all soils, with the exception of the Haidegg fallow soil, Actinobacteria were found to be the second most dominant phylum of bacteria. At the order level, Actinomycetales, Solirubrobacterales and Thermoleophilales were found in fallow soils in higher numbers than in their respective replant soil (Fig. ESM2). At the genus level, unclassified sequences were again found to dominate (78.2 % in all soil samples). A high positive correlation between the genus Solirubrobacter and plant growth was found. This finding is in contrast to research with peach replant disease, where a high negative correlation (r: −0.774) was reported for Solirubrobacter sp. and fresh peach shoot weights (Yang et al. 2012). It would appear that bacteria of the genus Solirubrobacter interacted differently in apple soils, and have a beneficial effect on apple plants.
Bacteria of the phyla Chloroflexi, Bacterioides Planctomycetes, Verrucomicrobia, Firmicutes and Gemmatimonadetes were present at levels below 4 % in all soils. Nonetheless, a high negative correlation between the genus Chitinophaga (member of the Bacteroides phylum) and plant growth was detected. A similar finding was reported in a study investigating the role of bacterial communities in suppression of Rhizoctonia solani bare patch disease of wheat, whereby high numbers of Chitinophaga were detected in the rhizosphere of diseased wheat plants (Yin et al. 2013). Interestingly, the genera listed in Table 3 with high correlations to plant growth (r > 0.4 or < −0.4) account for a total of 9.0 % of the total number of sequences obtained by pyrosequencing targeting bacteria. This low proportion is mainly due to a large component of unidentified bacteria at the genus level, which accounted for 64 % of the total sequences. The paucity of literature linking the bacterial genera observed with higher positive and negative correlations to plant growth may be explained by their minor incidence in the soil samples. Indeed, these low proportions make an inference of the specific functional roles of these genera difficult.
Fungal and oomycete analyses
Fungal coverage values of 0.99 were obtained for all soils by pyrosequencing, confirming that the diversity in the soils was adequately investigated. The Egma soils were very similar. These findings support those of the plant growth assay, where only minimal differences between the Egma fallow and replant soils were recorded, and lead to the conclusion that plant health in replanted apple orchards is closely linked to soil fungal communities.
The finding that the fungal community composition was not significantly different at the phylum and genus levels initiated a deeper investigation into correlations of fungal genera in soils with plant growth. The genera listed in Table 4 accounted for 70.7 % of all fungal sequences detected, indicating a high representation of the genera most significantly correlated with plant growth. As was the case for bacteria, almost all fungal genera were found in both replant and fallow soils, and few genera were only found in one soil type. The strongest negative correlation with plant growth (r: −0.84) was found for the genus Acremonium, a genus reported to cause root rot or replant disease in asparagus (Blok and Bollen 1995) and in muskmelon (Garcia-Jimenez et al. 1994). The genus has also been detected in soils suffering from peach replant disease (Yang et al. 2012). The role of the pathogen in decreased soil fertility in the soils under investigation is unknown, but the low relative abundance of the genus (0.67 %) would indicate it not to be a major player.
Cylindrocarpon is the only root inhabiting fungus for which evidence of a direct role in ARD in previous studies has been shown (Manici et al. 2013; Tewoldemedhin et al. 2011), and a strong negative correlation between Cylindrocarpon and plant growth was found in this study (r: −0.83). These findings suggest a link between Cylindrocarpon (RA: 1.41 %) and reduced soil fertility, and support recent research whereby endophyte populations of Cylindrocarpon were, along with Pythium species, the main pathogens found in ARD soils at nine sites in Germany, Austria and Italy (Manici et al. 2013). Several other genera detected in this study including Ilyonectria, Thelonectria, Neonectria and Haematonectria, genera which fall into the group conventionally called Cylindrocarpon-like fungi (Chaverri et al. 2011), were also found to be negatively correlated to plant growth and confirm the overall negative impact of this group of fungi on plant growth in apple orchards.
Fusarium is typically considered a member of the root rot complex of apple because it is the most frequently occurring genus among root-colonising fungi. However, Fusarium was shown to exhibit only weak pathogenicity as a root endophyte in several previous studies (Manici et al. 2003; Tewoldemedhin et al. 2011). Negative correlations of Fusarium with plant growth were observed in this study (r: −0.81), suggesting that Fusarium species inhabiting the rhizophere can have a relevant negative role on plant growth. It is possible that the contrasting results are due to different Fusarium populations in the plant roots and in the rhizosphere soils.
Genera with negative correlations with plant growth, but relevant due to high abundances in the soils include Exophiala, Giberella and Cryptococcus. The genus Exophiala includes black yeasts, species of which have not been reported previously as plant pathogens. Similarly, no reports on plant pathogenicity of Cryptococcus, an encapsulated yeast, could be found, despite evidence that members of the genus are found predominantly in soils and rotting vegetation (Di Menna 1954). In contrast, evidence exists that Giberella is involved with plant disease (Maxin et al. 2012; Sanzani et al. 2013), and the finding of this genus in higher numbers in the replant soils could indicate a role of this fungus in ARD. The genus Gibberella includes the perfect stage of several Fusarium species, and the negative correlation of this genus with plant growth further supports the overall pathogenic role of Fusarium sp. in replanted orchards.
The genera Mortierella, Tetracladium and Penicillium were the most abundant fungal genera found to be highly positively correlated with plant growth (RA of 34.4 %, 12.1 % and 0.67 %, respectively; Table 3). This may be due to the impact of natural vegetation in fallow soils. Species of these genera have been suspected to be causal agents of replant disease (Catska et al. 1988; Mazzola and Manici 2012; Utkhede et al. 1992), although evidence also indicates that certain species can have a commensal or mutualistic relationship with apple plants (Vega et al. 2010). Penicillium is known to be capable of the production of numerous biologically active compounds, and to act as a bacterial antagonist and plant growth promoter (Khan et al. 2008; Nicoletti and de Stefano 2012). The positive correlations between Penicillium and Tetracladium and plant growth suggests that the species present in the soils most likely were not pathogenic, but rather acted in a beneficial way and increased soil fertility. High positive correlations with plant growth were demonstrated for the fungal genera Lecythophora and Paecilomyces. Both genera have been reported to live as saprophytes in soil (Domsch et al. 2007; Mazzola 1999). Species of Lecythophora were reported to produce antifungal metabolites which are able to inhibit the growth of pathogenic fungi (Chakravarty and Hiratsuka 1994), supporting a beneficial role of this genus in soils. Paecilomyces was also isolated from apple tree roots in a previous study (Manici et al. 2013) but has never been linked to ARD. In this study, Paecilomyces was found in higher numbers in fallow soils than replant soils. Paecilomyces has been reported to actively produce the phytohormones gibberellins and indole acetic acid, both plant growth promoters (Khan et al. 2012). This production would help to explain the high association between the presence of the genus and apple seedling plant growth.
Scutellinia is a genus of cup-fungi belonging to the family Pyronemataceae. The high association between this genus and plant growth indicates a possible beneficial role of this genus in reducing replant disease in soils. No evidence of any involvement of this genus in replant diseases could however be found in the literature.
Species of the genera Pythium, Phytophthora and Rhizoctonia, despite varying in frequency, have been routinely isolated from ARD plant roots and soils (Mazzola 1998, 1999; Mazzola and Manici 2012). In the transnational study of soils from Austria, Germany and Italy, Pythium species were identified as one of the main pathogenic components of ARD (Manici et al. 2013). Neither Pythium nor Phytophthora were however detected in this study, because the ITS primers used do not target oomycetes. The primer pair used has been reported to have a narrower target spectrum and/or lower PCR efficiencies than other primer pairs, and is known to be unable to multiply the ITS1 region of a large number of fungi (Op De Beeck et al. 2014). In addition, the amplification of DNA from basidiomycetes has been reported to not occur as efficiently as for other fungi, and such biases may also have contributed to the lack of Rhizoctonia sequences detected.
Conclusions
The results of this study support the findings of previous research, indicating that ARD is a complicated disease, for which most likely, various biotic factors are responsible (Mazzola 1998; Tewoldemedhin et al. 2011). Deep-throughput sequencing of fallow and replant soils revealed that the bacterial and fungal community composition did not differ significantly at either the phylum or genus level. However, correlations of bacterial and fungal genus abundance with plant growth revealed co-existence in the rhizosphere of organisms with both high negative and positive correlations. The strong associations of the genera Acremonium, Cylindrocarpon and Fusarium with replant soils support previous findings whereby these genera have already been associated with replant diseases. In contrast, the associations with replant disease found for Chitinophaga, Hyphomicrobium and Nitrosospira, bacterial genera not known to be associated with ARD, open several new options related to these newly discovered involvements in ARD. At the same time, the bacterial (Gp16 and Solirubrobacter) and fungal genera (Penicillium, Paecilomyces) which were positively associated with plant growth, deserve further investigation into their potential beneficial functional role. In conclusion, the findings of this study investigating both bacterial and fungal communities in parallel, demonstrate the advantages of deep-throughput sequencing in helping to elucidate the biotic components of disease complexes which affect replanted apple orchards. Future analyses should however aim for species level identification, because numerous genera contain species characterised by beneficial, neutral and pathogenic relationships with plant hosts. Indeed, this new research approach indicates a need for further study of certain bacteria and fungi which may potentially be exploited for the biological control of ARD as well as for maintaining biological fertility of soils in permanent crops, which are often affected by fertility decline.
References
Abdo Z, Schuette UM, Bent SJ, Williams CJ, Forney LJ, Joyce P (2006) Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ Microbiol 8:929–938
Blok WJ, Bollen GJ (1995) Fungi on roots and stem bases of asparagus in the Netherlands: species and pathogenicity. Eur J Plant Pathol 101:15–24
Browne GT, Connell JH, Schneider SM (2006) Almond replant disease and its management with alternative pre-plant soil fumigation treatments and rootstocks. Plant Dis 90:869–876
Buee M, Reich M, Murat C, Morin E, Nilsson RH, Uroz S, Martin F (2009) 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol 184:449–456
Catska V, Vancura V, Prikryl Z, Hudska G (1988) Artificial induction of the apple replant problem by Penicillium claviforme inoculation. Plant Soil 107:127–136
Chakravarty PH, Hiratsuka Y (1994) Evaluation of Lecythophora hoffmannii as a potential biological control agent against a blue stain fungus on Populus tremuloides. J Plant Dis Protect 101:74–79
Chaverri P, Salgado C, Hirooka Y, Rossman AY, Samuels GJ (2011) Delimitation of Neonectria and Cylindrocarpon (Nectriaceae, Hypocreales, Ascomycota) and related genera with Cylindrocarpon-like anamorphs. Stud Mycol 68:57–78
Chemidlin Prévost-Bouré N, Dequiedt S, Thioulouse J, Lelièvre M, Saby NPA, Jolivet C, Arrouays D, Plassart P, Lemanceau P, Ranjard L (2014) Similar processes but different environmental filters for soil bacterial and fungal community composition turnover on a broad spatial scale. PLoS ONE 9:e111667
Di Menna ME (1954) Cryptococcus terreus n.sp., from soil in New Zealand. J Gen Microbiol 11:195–197
Domsch KH, Gams W, Anderson T-H (2007) Compendium of soil fungi. APS, New York
Dullahide S, Stirling G, Nikulin A, Stirling A (1994) The role of nematodes, fungi, bacteria, and abiotic factors in the etiology of apple replant problems in the Granite Belt of Queensland. Aust J Exp Agric 34:1177–1182
Garcia-Jimenez J, Velazquez M, Jorda C, Alfaro-Garcia A (1994) Acremonium species as the causal agent of muskmelon collapse in Spain. Plant Dis 78:416–419
Gottel NR, Castro HF, Kerley M, Yang Z, Pelletier DA, Podar M, Karpinets T, Uberbacher E, Tuskan GA, Vilgalys R, Doktycz MJ, Schadt CW (2011) Distinct microbial communities within the endosphere and rhizosphere of populus deltoides roots across contrasting soil types. Appl Environ Microbiol 77:5934–5944
Hammer O, Harper D, Ryan P (2001) PAST: paleontological statistics software package for evolution and data analysis. Palaeontol Electron 4:1–9
Hartmann M, Frey B, Kolliker R, Widmer F (2005) Semi-automated genetic analyses of soil microbial communities: comparison of T-RFLP and RISA based on descriptive and discriminative statistical approaches. J Microbiol Methods 61:349–360
Innerebner G, Knief C, Vorholt JA (2011) Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl Environ Microbiol 77:3202–3210
Jaffee BA (1982) Fungi associated with roots of apple seedlings grown in soil from an apple replant site. Plant Dis 66:942–944
Kelderer MM, Manici L, Caputo F, Thalheimer M (2012) Planting in the ‘inter-row’ to overcome replant disease in apple orchards: a study on the effectiveness of the practice based on microbial indicators. Plant Soil 357:381–393
Khan SA, Hamayun M, Yoon H, Kim HY, Suh SJ, Hwang SK, Kim JM, Lee IJ, Choo YS, Yoon UH, Kong WS, Lee BM, Kim JG (2008) Plant growth promotion and Penicillium citrinum. BMC Microbiol 8:231
Khan A, Hamayun M, Kang S-M, Kim Y-H, Jung H-Y, Lee J-H, Lee I-J (2012) Endophytic fungal association via gibberellins and indole acetic acid can improve plant growth under abiotic stress: an example of Paecilomyces formosus LHL10. BMC Microbiol 12:3
Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson K-H (2013) Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277
Kovacs A, Yacoby K, Gophna U (2010) A systematic assessment of automated ribosomal intergenic spacer analysis (ARISA) as a tool for estimating bacterial richness. Res Microbiol 161:192–197
La Duc MT, Vaishampayan P, Nilsson H, Torok T, Venkateswaran K (2012) Pyrosequencing-derived bacterial, archaeal, and fungal diversity of spacecraft hardware destined for mars. Appl Environ Microbiol 78:5912–5922
Maheshwari DK (2011) Bacteria in Agrobiology: Plant Growth Responses. Springer
Mai WF, Abawi GS (1981) Controlling replant diseases of pome and stone fruits in Northeastern United States by preplant fumigation. Plant Dis 65:859–864
Manici L, Ciavatta C, Kelderer M, Erschbaumer G (2003) Replant problems in South Tyrol: role of fungal pathogens and microbial population in conventional and organic apple orchards. Plant Soil 256:315–324
Manici LM, Kelderer M, Franke-Whittle IH, Rühmer T, Baab G, Nicoletti F, Caputo F, Topp A, Insam H, Naef A (2013) Relationship between root-endophytic microbial communities and replant disease in specialized apple growing areas in Europe. Appl Soil Ecol 72:207–214
Maxin P, Weber RWS, Pedersen HL, Williams M (2012) Control of a wide range of storage rots in naturally infected apples by hot-water dipping and rinsing. Postharvest Biol Technol 70:25–31
Mazzola M (1998) Elucidation of the microbial complex having a causal role in the development of apple replant disease in washington. Phytopathology 88:930–938
Mazzola M (1999) Transformation of soil microbial community structure and Rhizoctonia-suppressive potential in response to apple roots. Phytopathology 89:920–927
Mazzola M, Manici LM (2012) Apple replant disease: Role of microbial ecology in cause and control. Annu Rev Phytopathol 50:45–65
Mazzola M, Granatstein DM, Elfving DC, Mullinix K, Gu YH (2002) Cultural management of microbial community structure to enhance growth of apple in replant soils. Phytopathology 92:1363–1366
Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Andersen GL, Bakker PAHM, Raaijmakers JM (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100
Methe B, Nelson K, Pop M (2012) A framework for human microbiome research. Nature 486:215–221
Nicoletti R, de Stefano M (2012) Penicillium restrictum as an antagonistc of plant pathogenic fungi. In Dynamic Biochemistry, Process Biotechnology and Molecular Biology. Global Science books
Nielsen MN, Winding, A (2002) Microorganisms as indicators of soil health. National Environmental Research Institute
Op De Beeck M, Lievens B, Busschaert P, Declerck S, Vangronsveld J, Colpaert JV (2014) Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS One 9:e97629
Panke-Buisse K, Poole AC, Goodrich JK, Ley RE, Kao-Kniffin J (2015) Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J 9:980–989
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596
Ranjard L, Poly F, Nazaret S (2000) Monitoring complex bacterial communities using culture-independent molecular techniques: application to soil environment. Res Microbiol 151:167–177
Sanzani SM, Cariddi C, Roccotelli A, Garganese F, Fallanaj F, Ippolito A (2013) First report of Gibberella avenacea causing wet apple core rot in Italy. J Plant Pathol 95:217
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schutte UM, Abdo Z, Bent SJ, Shyu C, Williams CJ, Pierson JD, Forney LJ (2008) Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl Microbiol Biotechnol 80:365–380
Scotto La Massese C, Minot JC, Voisin R, Palmieri M (1988) Value of a biological test for estimating the influence of soil type, previous crop, and soil sterilization on the growth of peach and apple. Acta Horticult 233:53–59
Spath M, Insam H, Peintner U, Kelderer M, Kuhnert-Finkernagel R, Franke-Whittle IH (2015) Linking soil biotic and abiotic factors to apple replant disease: a greenhouse approach. J Phytopathol 163:287–299
St. Laurent A, Merwin I, Thies J (2008) Long-term orchard groundcover management systems affect soil microbial communities and apple replant disease severity. Plant Soil 304:209–225
Sun J, Zhang Q, Zhou J, Wei Q (2014a) Illumina amplicon sequencing of 16S rRNA tag reveals bacterial community development in the rhizosphere of apple nurseries at a replant disease site and a new planting site. PLoS One 9, e111744
Sun J, Zhang Q, Zhou J, Wei Q (2014b) Pyrosequencing technology reveals the impact of different manure doses on the bacterial community in apple rhizosphere soil. Appl Soil Ecol 78:28–36
Tewoldemedhin YT, Mazzola M, Labuschagne I, McLeod A (2011) A multi-phasic approach reveals that apple replant disease is caused by multiple biological agents, with some agents acting synergistically. Soil Biol Biochem 43:1917–1927
Turner T, James E, Poole P (2013) The plant microbiome. Genome Biol 14:209
Uhlik O, Musilova L, Ridl J, Hroudova M, Vlcek C, Koubek J, Holeckova M, Mackova M, Macek T (2013) Plant secondary metabolite-induced shifts in bacterial community structure and degradative ability in contaminated soil. Appl Microbiol Biotechnol 97:9245–9256
Utkhede RS, Vrain TC, Yorston JM (1992) Effects of nematodes, fungi and bacteria on the growth of young apple trees grown in apple replant disease soil. Plant Soil 139:1–6
Vega FE, Simpkins A, Aime MC, Posada F, Peterson SW, Rehner SA, Infante F, Castillo A, Arnold AE (2010) Fungal endophyte diversity in coffee plants from Colombia, Hawaii, Mexico and Puerto Rico. Fungal Ecol 3:122–138
Xiong W, Zhao Q, Zhao J, Xun W, Li R, Zhang R et al (2014) Different continuous cropping spans significantly affect microbial community membership and structure in a vanilla-grown soil as revealed by deep pyrosequencing. Microb Ecol. doi:10.1007/s00248-014-0516-0
Yang J-I, Ruegger PM, McKenry MV, Becker JO, Borneman J (2012) Correlations between root-associated microorganisms and peach replant disease symptoms in a California soil. PLoS One 7:e46420
Yim B, Smalla K, Winkelmann T (2013) Evaluation of apple replant problems based on different soil disinfection treatments—links to soil microbial community structure? Plant Soil 366:617–631
Yin C, Hulbert SH, Schroeder KL, Mavrodi O, Mavrodi D, Dhingra A, Schillinger WF, Paulitz TC (2013) Role of bacterial communities in the natural suppression of Rhizoctonia solani bare patch disease of wheat (Triticum aestivum L.). Appl Environ Microbiol 79:7428–7438
Zhang Q, Sun J, Liu S, Wei Q (2013) Manure refinement affects apple rhizosphere bacterial community structure: a study in sandy soil. PLoS One 8:e76937
Zhao X, Tewoldemedhin Y, Mcleod A, Mazzola M (2009) Multiple personalities of Streptomyces spp. isolated from the rhizosphere of apple cultivated in brassica seed meal amended soils. Phytopathology 99:S150–S160
Acknowledgments
Financial support for the BIO-INCROP project was provided by the CORE Organic II Funding Body, partners of the FP7 ERA-Net project, CORE Organic II (Coordination of European Transnational Research in Organic Food and Farming systems, project no. 249667). Ljubica Begovic is gratefully acknowledged for her assistance.
Compliance with Ethical Standards
In the work conducted, there were no potential conflicts of interest. In addition, no humans or animals were used in the study. The content and authorship of the submitted manuscript has been approved by all authors, and all of the reported work is original.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Stéphane Compant .
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. ESM1
Principal component analysis of physical-chemical characteristics, microbial biomass and basal respiration in fallow and replant soil samples. Abbreviations: EF (Egma fallow), ER (Egma replant), HF (Haidegg fallow), HR (Haidegg replant), NF (Nachtweih fallow) and NR (Nachtweih replant). (DOCX 2330 kb)
Fig. ESM2
Comparison of bacterial communities in fallow and replant soils at the order level. A) Proteobacteria. B) Acidobacteria. and C) Actinobacteria. Only orders present at an abundance >1 % are shown. Abbreviations: EF (Egma fallow), ER (Egma replant), HF (Haidegg fallow), HR (Haidegg replant), NF (Nachtweih fallow) and NR (Nachtweih replant). (DOCX 93 kb)
Fig. ESM3
Comparison of fungal communities in fallow and replant soils at the family level. Only families present at an abundance >5 % are shown. Abbreviations: EF (Egma fallow), ER (Egma replant), HF (Haidegg fallow), HR (Haidegg replant), NF (Nachtweih fallow) and NR (Nachtweih replant). (DOCX 43 kb)
Rights and permissions
About this article
Cite this article
Franke-Whittle, I.H., Manici, L.M., Insam, H. et al. Rhizosphere bacteria and fungi associated with plant growth in soils of three replanted apple orchards. Plant Soil 395, 317–333 (2015). https://doi.org/10.1007/s11104-015-2562-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-015-2562-x