Abstract
ELO-like elongase is a condensing enzyme elongating long chain fatty acids in eukaryotes. Eranthis hyemalis ELO-like elongase (EhELO1) is the first higher plant ELO-type elongase that is highly active in elongating a wide range of polyunsaturated fatty acids (PUFAs) and some monounsaturated fatty acids (MUFAs). This study attempted using domain swapping and site-directed mutagenesis of EhELO1 and EhELO2, a close homologue of EhELO1 but with no apparent elongase activity, to elucidate the structural determinants critical for catalytic activity and substrate specificity. Domain swapping analysis of the two showed that subdomain B in the C-terminal half of EhELO1 is essential for MUFA elongation while subdomain C in the C-terminal half of EhELO1 is essential for both PUFA and MUFA elongations, implying these regions are critical in defining the architecture of the substrate tunnel for substrate specificity. Site-directed mutagenesis showed that the glycine at position 220 in the subdomain C plays a key role in differentiating the function of the two elongases. In addition, valine at 161 and cysteine at 165 in subdomain A also play critical roles in defining the architecture of the deep substrate tunnel, thereby contributing significantly to the acceptance of, and interaction with primer substrates.
Key Message
Structural and functional analysis of the first higher plant ELO-type elongase. Structural factors critical for catalytic activity and substrate specificity determined.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biosynthesis of many nutritionally important very long chain PUFAs such as docosahexaenoic acid (DHA, 22:6n-3) and arachidonic acid (ARA, 20:4n-6) in mammals and some eukaryotic microorganisms involves an elongation process of cyclic four sequential catalytic reactions: condensation, keto-reduction, dehydration, and enoyl-reduction, by adding a two-carbon unit to long chain PUFAs. The first step of the four reactions in a cycle is catalyzed by a condensing enzyme (3-ketoacyl-CoA synthase, also known as elongase) with high substrate specificity, as compared to the other three enzymes catalyzing subsequent steps, in terms of the chain length and the number of double bonds. Thus, it is the key enzyme to initiate the process for fatty acid elongation (Leonard et al. 2004; Jakobsson et al. 2006; Qiu et al. 2020).
Elongase is a special type of condensing enzymes catalyzing Claisen reactions using long chain acyl-CoA as a primer and malonyl-CoA as an extender in the biosynthesis of very long chain fatty acids (Heath and Rock 2002). Two types of elongases have been found in eukaryotes that do not share the similarity of sequence, structure, and catalytic mechanism. The FAE type of elongases occurs in plants mainly for saturated fatty acid and MUFA elongations (James Jr. et al. 1995; Haslam and Kunst 2013). It has one or two transmembrane α-helices in the N-terminus of sequences anchored to endoplasmic reticulum membrane with a large soluble catalytic domain facing towards the cytosol where a Cys-His-Asn catalytic triad is used for the condensation reaction (Ghanevati and Jaworski 2002; Haslam and Kunst 2013; Blacklock 2021). On the other hand, the ELO type of elongases occurs in higher animals and some eukaryotic microorganisms mainly for PUFA elongation; but it can elongate saturated and monounsaturated fatty acids in some eukaryotic species where no FAE type of elongases exists (Toke and Martin 1996; Leonard et al. 2004). The ELO-like elongases mainly for PUFA elongation were initially identified in fungus Mortierella alpina and worm Caenorhabditis elegans (Beaudoin et al. 2000; Parker-Barnes et al. 2000). Afterwards, many elongases for PUFA elongation have been identified in algae (Meyer et al. 2004; Pereira et al. 2004), fish (Agaba et al. 2005) and mammals (Jakobsson et al. 2006; Logan et al. 2014). In general, this type of elongases possesses six or seven transmembrane α-helices stretched to the whole sequence to form a transmembrane helix barrel surrounding a substrate tunnel facing towards the cytosol where conserved histidines along with other residues are used for generating a hydrogen-bonding network to facilitate the condensation reaction (Nie et al. 2021).
Interestingly, a functional ELO-like elongase (Eranthis hyemalis ELO-like elongase 1 or EhELO1) was recently found in winter aconite (Eranthis hyemalis) where it is involved in the biosynthesis of two unusual very long chain PUFAs docosadienoic acid (DDA, 22:2n-6) and docosatrienoic acid (DTA, 22:3n-3) (Aitzetmüller 1996; Meesapyodsuk et al. 2018). EhELO1 is the first ELO-type elongase identified in higher plants and possesses the elongation activity on a wide range of PUFAs as well as some MUFAs. Meanwhile, RNA sequencing analysis of winter aconite also identified a close homologue coding for Eranthis hyemalis ELO-like elongase 2 (EhELO2), but with no apparent fatty acid elongase activity. This study aimed at using domain swapping and site-directed mutagenesis of the two highly homologous sequences to elucidate the structural factors determining the catalytic activity and substrate specificity of the plant ELO-type elongase.
Materials and methods
Materials
Linoleic acid, alpha-linolenic acid, and standard fatty acids were purchased from Nu-Chek Prep, Inc. (Elysian, MN, USA). HP Taq DNA polymerase, deoxynucleoside triphosphate, nucleic acid purification kits and DNA extraction kits were acquired from Bio Basic Inc. (Markham, ON, Canada). Q5 high-fidelity DNA polymerase was obtained from New England Biolabs (Ipswich, MA, USA). Intermediate vector pGEM-T, HPLC and GC grade solvents, yeast and Escherichia coli media were either purchased from VWR (Edmonton, AB, Canada) or Fisher Scientific (Ottawa, ON, Canada). Restriction enzymes, DNA ligase, and S.c. EasyComp Transformation kits were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Oligo primers were synthesized from Sigma-Aldrich (Oakville, ON, Canada).
Construction of EhELO1 and EhELO2 chimeras and site-directed mutants
The full-length cDNA of EhELO2 (accession number, AXS59120) was cloned by RT-PCR using total RNA prepared from E. hyemalis developing seeds as a template using primers EhELO2-F and EhELO2-R (Table S1). Amplified full-length EhELO2 was cloned into an intermediated pGEM-T cloning vector and subsequently subcloned into a yeast expression vector, pHVX2 generating EhELO2/pHVX2. To create both chimeras and site directed mutant genes, the overlap extension PCR technique was adopted (Heckman and Pease 2007; Meesapyodsuk and Qiu 2014). Two overlapping fragments of each construct were amplified by PCR with primers (Table S1) and proofreading enzyme Q5 polymerase using EhELO1/pHVX2 (Meesapyodsuk et al. 2018), EhELO2/pHVX2 or corresponding chimeric gene as templates. The fragments were gel purified, mixed, and used as the template for amplifying the full-length chimeric and site directed mutagenesis genes.
Yeast expression
All mutant constructs were subcloned into pHVX2 (Volschenk et al. 1997) under the control of the constitutive promoter PGK1 and transformed into yeast Saccharomyces cerevisiae, INVSc1 using S.c. EasyComp transformation kit. Transformants were selected on a synthetic dropout medium (SD-Leu) containing 2% (w/v) glucose, 0.17% (w/v) yeast nitrogen base, 0.5% (w/v) ammonium sulfate, and 0.06% (w/v) dropout mix without leucine. To assess the elongation activity, recombinant yeast cells were grown overnight in 5 mL of SD-Leu medium at 30 °C. Yeast cells were then harvested and washed with sterile water and resuspended in a 5-mL of the same medium supplemented with or without 0.25 mM linoleic and 0.25 mM alpha-linolenic acids in the presence of 0.1% Tergitol (Nonidet P-40, Sigma). Cultures were incubated at 20 °C with shaking for 2–4 days. The cells were then harvested and washed once with 1% Tergitol and once with sterile water. Yeast pellets were used for fatty acid analysis.
Fatty acid analysis
Yeast total fatty acids were directly transmethylated to fatty acid methyl esters (FAMEs) using 2 mL of 1% (v/v) H2SO4 in methanol. The yeast samples were incubated at 80 °C for 2 h and then cooled on ice. The FAMEs were extracted by adding 1 mL of 0.9% NaCl and 2 mL of hexane. After centrifugation at 2500 rpm for 5 min, the upper layer containing FAMEs was transferred into a new tube, dried under nitrogen gas, and dissolved with 200 µL of hexane. Total FAME samples were analyzed on an Agilent 6890N gas chromatography equipped with flame ionization detection and a DB-23 column (30 M × 0.25 mm with 0.25-µm film thickness, J&W Scientific). The temperature program was maintained at 160 °C for 1 min, then increased to 240 °C at a rate of 4 °C per min, and held for 10 min. Elongation efficiencies (%) of exogenously fed fatty acid substrates in yeast transformants were calculated by the formula as [(product) × 100/(substrate + product)] using values corresponding to the peak area of FAMEs.
Topology prediction and structural modeling
The transmembrane protein topology of EhELO1 and EhELO2 was predicted using an online web server of TOPCONs (Tsirigos et al. 2015) with five of the topology predictors (Octopus, Philius, PolyPhobius, Scampi, and Spoctopus). Three-dimensional structure was de novo modeled by Alphafold (Mirdita et al. 2022) and presented using ChimeraX (Pettersen et al. 2021).
Results
Sequence and functional comparison of two ELO-like elongases EhELO1 and EhELO2 in winter aconite
Two cDNAs coding for ELO-type 3-ketoacyl-CoA synthase or elongase (EhELO1 and EhELO2) with similar length were identified by searching a transcriptome of winter aconite developing seeds (SRA accession number: SRP149907) using ELO-type elongases from fungi and animals as queries (Oh et al. 1997; Parker-Barnes et al. 2000; Agaba et al. 2005). EhELO1 and EhELO2 shared about 68% of amino acid identity and 80% of sequence similarity with conserved substitutions. Further sequence analysis showed that both EhELO1 and EhELO2 possessed seven transmembrane helices and four conserved motifs KxxExxDT, FLHxxHH, HxxxYxYY and TxxQxxQ in four transmembrane helices (III, IV, V, VI), which are typical features of ELO-type elongases (Leonard et al. 2004) (Fig. 1). However, functional analysis of the two elongase genes in yeast Saccharomyces cerevisiae showed that EhELO1 was highly active for elongating a wide range of 18 ℃ and 20 ℃ PUFAs such as 18:2–9,12 and 20:2–11,14, 18:3–9,12,15 and 20:3–11,14,17, as well as some 18–24 ℃ monounsaturated fatty acids (MUFAs) such as 18:1–11 and 20:1–13, while EhELO2 did not exhibit any elongase activity on all substrates tested including saturated, monounsaturated and polyunsaturated fatty acids (Fig. 2) (Meesapyodsuk et al. 2018). The structure similarity and functional difference of the two highly homologous sequences offered an opportunity to investigate the structural factors for catalytic activity and substrate specificity.
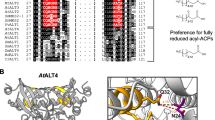
Comparison of two ELO-type elongase sequences EhELO1 and EhELO2 from winter aconite. Amino acids with black background are identical. Transmembrane helices were denoted by lines on top of sequences with arrows towards the cytosolic side. Four conserved motifs in four transmembrane helices (III, IV, V, VI) were indicated by red star. Letter A, B, C, D on top of sequence indicate the start sites of four subdomains in the C-terminal domain. Two cysteine residues forming a disulfide in the loops of transmembrane helices II and III, and of VI and VII at the lumen side of endoplasmic reticulum were highlighted by green color. Mutagenized amino acids were highlighted by red color
Elongation activity of two large domain-swapping chimeras of EhELO1 and EhELO2 in yeast. A Fatty acid analysis of the two chimeras in yeast without exogenous fatty acid feeding. Detected elongation products of 18:1–11 were 20:1–13, 22:1–15 and 26:1–19. B Fatty acid analysis of the two chimeras in yeast with exogenous feeding of linoleic acid (LA, 18:2–9,12) and α-linolenic acid (ALA, 18:3–9,12,15). Elongated products of 18:2–9,12 were 20:2–11,14 and 22:2–13,16 (DDA) while Elongated products of 18:3–9,12,15 were 20:3–11,14,17 and 22:3–13,16,19 (DTA)
Effects of domain swapping between EhELO1 and EhELO2
To identify structural determinants for the functional difference in the two highly homologous sequences, EhELO1 and EhELO2 were first dissected in the middle of two sequences located between transmembrane helices III and VI into two large half domains, which were then swapped with each other, giving rise to two chimeras: chimera 1 and chimera 2 (Fig. 3). Chimera 1 was comprised of the N-terminal domain of EhELO1 and the C-terminal domain of EhELO2 while chimera 2 was made of the N-terminal domain of EhELO2 and the C-terminal domain of EhELO1. Functional analysis of the two chimeras in yeast showed that without exogenous feeding of PUFAs, chimera 2, like wild type EhELO1, could effectively elongate the endogenous 18:1–11 to 20:1–13 and further to 26:1–19, while chimera 1, like wild type EhELO2, could not elongate MUFAs (Fig. 2A). And with exogenous feeding of two PUFAs, chimera 2, also similar to wild type EhELO1, could effectively elongate 18:2–9,12 and 18:3–9,12,15 to 20:2–11,14 and 20:3–11,14,17, and further to 22:2–13,16 (DDA) and 22:3–13,16,19 (DTA), respectively, while chimera 1, similar to wild type EhELO2, did not exhibit any elongase activity on the two PUFAs (Fig. 2B). In addition, chimera 2, also like wild type EhELO1, possessed the higher elongation activity towards ω-3 PUFAs over ω-6 PUFAs. These results clearly indicate that the C-terminal half of EhELO1 is critical for the elongase activity on both PUFAs and MUFAs.
Effects of C-terminal subdomain swapping of EhELO1 and EhELO2. A Schematic Illustration of subdomain swapping of chimeras. Black box: EhELO1. Grey box: EhELO2. B Elongation activity of chimeras on MUFAs and PUFAs was represented by the category relative to the native EhELO1: + , 4 − 10%; + + , 11 − 50%; + + + , 51 − 100%; ND not detected
Afterwards, the C-terminal half domain of EhELO1 was further dissected at the loops of four transmembrane helices into four subdomains: A (p141-p172), B (p173-p207), C (p208-p234), D (p235-p275), and chimeras with different combinations of these subdomains were constructed for functional analysis in yeast (Fig. 3). Results showed that chimera 3 comprising the N-terminal domain and the C-terminal subdomains A and B of EhELO2 and the C-terminal subdomains C and D of EhELO1 retained considerable PUFA elongation activity, but lost MUFA elongation activity. This result indicates that the C-terminal subdomains A and B of EhELO1 are essential for MUFA elongation, while the C-terminal subdomains C and D of EhELO1 are required for PUFA elongation.
Chimera 4 comprising the N-terminal domain and the C-terminal subdomains C and D of EhELO2 and the C-terminal subdomains A and B of EhELO1 lost the elongation activity of both MUFAs and PUFAs. This result indicates that the C-terminal subdomains C and D of EhELO1 are essential for PUFA elongation, but they are also required for MUFA elongation. Without subdomains C and D, subdomains A and B are not adequate for MUFA elongation.
Chimera 5 comprising the N-terminal domain and the C-terminal subdomain D of EhELO2, and the C-terminal subdomains A, B and C of EhELO1 retained substantial PUFA and MUFA elongation activity. However, it exhibited higher elongation activity towards ω-6 PUFAs over ω-3 PUFAs. This result indicates that C-terminal subdomain C, but not subdomain D, of EhELO1 is essential for PUFA and MUFA elongations.
Chimera 6 comprising the N-terminal domain and the C-terminal subdomains B, C and D of EhELO2, and the C-terminal subdomain A of EhELO1 lost both PUFA and MUFA elongation activity. As compared with that of chimera 5, this result indicates the C-terminal subdomains B and C of EhELO1 are essential for the elongation of PUFAs and MUFAs.
Chimera 7 comprising the N-terminal domain and the C-terminal subdomains A, B and D of EhELO2, and the C-terminal subdomain C of EhELO1 did not exhibit any MUFA elongation activity, but still retained some residual PUFA elongation activity. This result confirms that subdomain B is essential for MUFA elongation and subdomain C is essential for PUFA elongation.
Chimera 8 comprising the N-terminal domain and the C-terminal subdomains B and D of EhELO2, and the C-terminal subdomains A and C of EhELO1 produced the similar result as chimera 7. This result indicates that subdomain A does not enhance the capacity of subdomain C for PUFA elongation. As compared to that of chimera 5, this result also confirms subdomain B, not subdomain A, is essential for MUFA elongation.
Chimera 9 comprising the N-terminal domain and the C-terminal subdomains A and D of EhELO2, and the C-terminal subdomains B and C of EhELO1 effectively elongated both PUFAs and MUFAs. However, it also exhibited higher elongation activity to ω-6 PUFAs over ω-3 PUFAs. This result reaffirms that C-terminal subdomains B and C of EhELO1 are critical for the elongation of MUFAs and PUFAs, and both are essential for MUFA elongation while only subdomain C is essential for PUFA elongation.
Effect of site-directed mutagenesis of amino acids in the subdomain C of EhELO1
As the C-terminal subdomain C of EhELO1 is most critical for both MUFA and PUFA elongations relative to that of EhELO2, we then looked into the residues in the subdomain, particularly those in the transmembrane helix VI that differentiate the two elongases. Site-directed mutagenesis of these residues in EhELO1 by switching them to corresponding counterparts in EhELO2 showed that there were little effects on the elongase activity in substitutions of F212C, W218V, L221F and T225G, as these mutants still retained comparable elongation efficiencies on both MUFAs such as 18:1–11 and PUFAs including ω-6 PUFAs such as 18:2–9,12 and ω-3 PUFAs such as 18:3–9,12,15. However, dramatic change was observed in the substitution mutant G220S where the activity of both MUFA elongation and PUFA elongation was completely lost (Fig. 4, Table 1).
Effects of site-directed metagenesis of the residue at position 220 in EhELO1 and EhELO2. A Fatty acid analysis of wild type of EhELO1 and substitution of the glycine in EhELO1 with serine (G220S) fed with LA and ALA. B Fatty acid analysis of wild type of EhELO2 and substitution of the serine in EhELO2 with glycine (S220G) fed with LA and ALA
Effect of site-directed mutagenesis in EhELO2
To confirm the amino acid at 220 in subdomain C plays a key role in differentiating the function of the two highly homologous elongases, the serine residue at the position in EhELO2 was substituted with its counterpart glycine in EhELO1 (S220G). The mutagenesis showed that the substitution restored the elongation activity of EhELO2 on PUFAs where the elongation efficiencies of ω-6 PUFAs and ω-3 PUFAs reached about 7% and 10%, respectively (Fig. 4, Table 1). However, substitutions of the serine in EhELO2 with a small hydrophobic amino acid valine (S220V) or a similar hydroxy amino acid threonine (S220T) did not exhibit any elongation activity. This result indicates glycine at the position is absolutely required for the elongation activity.
Effect of site-directed mutagenesis of amino acids in the other regions of EhELO1
Next, we investigated the residues that were highly conserved in the other regions than subdomain C of EhELO1 that might be important for the elongase activity. Two strictly conserved cysteine residues were found at positions 108 and 237 located in the loops of transmembrane helices II and III, and transmembrane helices VI and VII. Changes of the cysteine residue at 108 to alanine or glutamine (C108A or C108Q) resulted in complete loss of elongase activity on both MUFAs and PUFAs (Table 1). This result indicates that the cysteine is essential and might be involved in the formation of a disulfide bond with that at 237, which was believed to be required for maintaining the conformation and stability of ELO-type elongases (Nie et al. 2021).
In addition, a systematic tryptophan scanning mutagenesis (Wojciechowski et al. 2015) was employed to introduce tryptophan residues at positions near the catalytic histidine sites of the helices on the rationale that a large and hydrophobic tryptophan would be tolerated at positions facing the hydrophobic interior of the membrane lipids, but not at positions facing the substrate tunnel. The mutagenesis showed that changes of valine at 161 (V161W), methionine at 164 (M164W) and cysteine at 165 to tryptophan (C165W) in the transmembrane helix IV of subdomain A resulted in the total loss of elongase activity on both MUFAs and PUFAs. Changes of threonine at 162 (T162W) lost MUFA elongation activity, but still retained some residual activity on PUFA elongation. On the other hand, changes of threonine at 159 (T159W) and isoleucine at 163 (I163W) to tryptophan retained substantial elongase activity on both MUFAs and PUFAs (Table 1). This result indicates valine at 161, methionine at 164 and cysteine at 165, but not threonine at 159 and isoleucine at 163, might be critically involved in defining the structure of the substrate tunnel for catalytic activity.
Discussion
Identification of two highly homologous ELO-like elongases EhELO1 and EhELO2 with essentially different activity provides a good opportunity to investigate the structural determinants for the catalytic activity and substrate specificity of the elongase using domain swapping and site-directed mutagenesis. For domain swapping analysis, we first dissected the two sequences carefully in the middle of sequences guided by the topological structure into two halves, then swapped them with each other to give two chimeras. Functional analysis of the two chimeras pinpointed the C-terminal half of EhELO1 for the critical role in both PUFA and MUFA elongations. Afterwards, the C-terminal half domain of EhELO1 and EhELO2 was further dissected into four subdomains guided by the topological structure. Functional analysis of subdomain-swapped chimeras showed that essentiality of subdomain C and B of EhELO1 for PUFA and/or MUFA elongation. Subdomain D of EhELO1 is not required for elongation of either MUFAs or PUFAs; however, it might have a role in assisting subdomain C in elongating ω-3 PUFAs, as both chimeras 5 and 9 without this subdomain exhibits the higher elongation activity to ω-6 PUFAs over ω-3 PUFAs, which is opposite to the activity of the native EhELO1. Interestingly, previous reports also noted that these regions of ELO-type elongases could affect the specificity of acyl-CoA substrates and length of final products (Denic and Weissman 2007; Vrinten et al. 2010).
After revealing the critical role of C-terminal subdomain C of EhELO1 for elongating both MUFAs and PUFAs, we then utilized site-directed mutagenesis to interrogate the residues in the subdomain that could differentiate the two elongases. Functional analysis of reciprocal switch mutants pinpointed the critical role of the glycine at 220 in the subdomain C of EhELO1 for the functionality of the two elongases, as substitution of it to its counterpart in EhELO2 (G220S) lost all elongation activity and substitution of the serine at the same position in EhELO2 to glycine (S220G) restores the elongation activity of EhELO2 towards both ω-6 PUFAs and ω-3 PUFAs. It is interesting to note that the glycine residue at the position was found to be highly conserved in a group of ELO elongases from microalgae with Δ9 PUFA elongation activity (Figure S1) (Qi et al. 2002; Petrie et al. 2010; Sayanova et al. 2011; Li et al. 2011).
The three-dimensional structure of a human ELO elongase has been recently resolved (Nie et al. 2021). It possesses a narrow substrate tunnel formed by two units of three transmembrane α-helices (II–IV and V–VII). Structural modeling of the plant ELO-type elongases showed that the glycine at 220 in transmembrane helix VI of EhELO1 faces the middle substrate tunnel; thus, would play a critical role in defining the architecture of the substrate tunnel for substrate acceptance and interaction (Fig. 5). With the small size, it could widen the pocket, and with the flexibility, it could help accommodate different sizes and shapes of fatty acyl chains. Therefore, the change of this residue to serine in EhELO1 might have disrupted the structure of the substrate tunnel.
Predicted three-dimensional structure of EhELO1. Transmembrane helices were labeled by I to VII. Two cysteine residues (108, 237) in forming a disulfide bond, critical residues facing the substrate channel (Gly at 220, Val at 161, Met at 164 and Cys at 165) were shown in red stick. Electron density map of 3-keto eicosanoyl-CoA in the substrate tunnel was shown in blue mesh
According to the modeled three-dimensional structure of EhELO1, the substrate tunnel is mainly defined by directed amino acids on transmembrane helices IV, VI, and VII. Besides the glycine at 220 in transmembrane helix VI, valine at 161, methionine at 164, and cysteine at 165 in transmembrane helix IV also face the middle substrate tunnel. However, threonine at 159 and isoleucine at 163 in transmembrane helix IV face the opposite side of the tunnel (Fig. 5). Changing those facing the substrate tunnel to tryptophan resulted in the total loss of the elongase activity on both PUFAs and MUFAs while changing those facing the opposite side to tryptophan still retained substantial elongation activity on both MUFAs and PUFAs. This implies that valine at 161, methionine at 164, and cysteine at 165 in transmembrane helix IV also play vital roles in defining the architecture of the substrate tunnel due to the critical positions. Substitution of these amino acids with tryptophan, an amino acid with a large and aromatic side chain might have disrupted the structure probably by closing off the tunnel. On the other hand, substitution of threonine at 159 and isoleucine at 163 with tryptophan could be tolerant as they face the interior of the membrane lipids and do not contribute significantly to the formation of the substrate tunnel for substrate acceptance and interaction.
In summary, this study has determined the structural factors critical for catalytic activity and substrate specificity of the first higher plant ELO-type elongase using domain swapping and site-directed mutagenesis. The information gained is useful for designing this type of enzymes to produce specific very long chain PUFAs for nutraceutical uses.
Data availability
Enquiries about data availability should be directed to the authors.
Abbreviations
- EhELO1:
-
Eranthis hyemalis Fatty acid elongase 1
- EhELO2:
-
Eranthis hyemalis Fatty acid elongase 2
- FAMEs:
-
Fatty acid methyl esters
- MUFAs:
-
Monounsaturated fatty acids
- PUFAs:
-
Polyunsaturated fatty acids
References
Agaba MK, Tocher DR, Zheng X, Dickson CA, Dick JR, Teale AJ (2005) Cloning and functional characterisation of polyunsaturated fatty acid elongases of marine and freshwater teleost fish. Comp Biochem Physiol B Biochem Mol Biol 142:342–352. https://doi.org/10.1016/j.cbpb.2005.08.005
Aitzetmüller K (1996) An unusual fatty acid pattern in Eranthis seed oil. Lipids 31:201–205. https://doi.org/10.1007/BF02522621
Beaudoin F, Michaelson LV, Hey SJ, Lewis MJ, Shewry PR, Sayanova O, Napier JA (2000) Heterologous reconstitution in yeast of the polyunsaturated fatty acid biosynthetic pathway. Proc Natl Acad Sci U S A 97:6421–6426. https://doi.org/10.1073/pnas.110140197
Blacklock BJ (2021) Fatty acid elongation by ELOVL condensing enzymes depends on a histidine nucleophile. Nat Struct Mol Biol 28:462–464. https://doi.org/10.1038/s41594-021-00609-2
Denic V, Weissman JS (2007) A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 130:663–677. https://doi.org/10.1016/j.cell.2007.06.031
Ghanevati M, Jaworski JG (2002) Engineering and mechanistic studies of the Arabidopsis FAE1 beta-ketoacyl-CoA synthase, FAE1 KCS. Eur J Biochem 269:3531–3539. https://doi.org/10.1046/j.1432-1033.2002.03039.x
Haslam TM, Kunst L (2013) Extending the story of very-long-chain fatty acid elongation. Plant Sci 210:93–107. https://doi.org/10.1016/j.plantsci.2013.05.008
Heath RJ, Rock CO (2002) The Claisen condensation in biology. Nat Prod Rep 19:581–596. https://doi.org/10.1039/b110221b
Heckman KL, Pease LR (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2:924–932. https://doi.org/10.1038/nprot.2007.132
Jakobsson A, Westerberg R, Jacobsson A (2006) Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog Lipid Res 45:237–249. https://doi.org/10.1016/j.plipres.2006.01.004
James DW Jr, Lim E, Keller J, Plooy I, Ralston E, Dooner HK (1995) Directed tagging of the arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell 7:309–319
Leonard AE, Pereira SL, Sprecher H, Huang Y-S (2004) Elongation of long-chain fatty acids. Prog Lipid Res 43:36–54. https://doi.org/10.1016/S0163-7827(03)00040-7
Li M, Ou X, Yang X, Guo D, Qian X, Xing L, Li M (2011) Isolation of a novel C18-Δ9 polyunsaturated fatty acid specific elongase gene from DHA-producing Isochrysis galbana H29 and its use for the reconstitution of the alternative Δ8 pathway in Saccharomyces cerevisiae. Biotechnol Lett 33:1823–1830. https://doi.org/10.1007/s10529-011-0626-4
Logan S, Agbaga M-P, Chan MD, Brush RS, Anderson RE (2014) Endoplasmic reticulum microenvironment and conserved histidines govern ELOVL4 fatty acid elongase activity. J Lipid Res 55:698–708. https://doi.org/10.1194/jlr.M045443
Meesapyodsuk D, Qiu X (2014) Structure determinants for the substrate specificity of acyl-CoA Δ9 desaturases from a marine copepod. ACS Chem Biol 9:922–934. https://doi.org/10.1021/cb400675d
Meesapyodsuk D, Ye S, Chen Y, Chen Y, Chapman RG, Qiu X (2018) An engineered oilseed crop produces oil enriched in two very long chain polyunsaturated fatty acids with potential health-promoting properties. Metab Eng 49:192–200. https://doi.org/10.1016/j.ymben.2018.08.009
Meyer A, Kirsch H, Domergue F, Abbadi A, Sperling P, Bauer J, Cirpus P, Zank TK, Moreau H, Roscoe TJ, Zähringer U, Heinz E (2004) Novel fatty acid elongases and their use for the reconstitution of docosahexaenoic acid biosynthesis. J Lipid Res 45:1899–1909. https://doi.org/10.1194/jlr.M400181-JLR200
Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M (2022) ColabFold: making protein folding accessible to all. Nat Methods 19:679–682. https://doi.org/10.1038/s41592-022-01488-1
Nie L, Pascoa TC, Pike ACW, Bushell SR, Quigley A, Ruda GF, Chu A, Cole V, Speedman D, Moreira T, Shrestha L, Mukhopadhyay SMM, Burgess-Brown NA, Love JD, Brennan PE, Carpenter EP (2021) The structural basis of fatty acid elongation by the ELOVL elongases. Nat Struct Mol Biol 28:512–520. https://doi.org/10.1038/s41594-021-00605-6
Oh CS, Toke DA, Mandala S, Martin CE (1997) ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem 272:17376–17384. https://doi.org/10.1074/jbc.272.28.17376
Parker-Barnes JM, Das T, Bobik E, Leonard AE, Thurmond JM, Chaung LT, Huang YS, Mukerji P (2000) Identification and characterization of an enzyme involved in the elongation of n-6 and n-3 polyunsaturated fatty acids. Proc Natl Acad Sci U S A 97:8284–8289. https://doi.org/10.1073/pnas.97.15.8284
Pereira SL, Leonard AE, Huang Y-S, Chuang L-T, Mukerji P (2004) Identification of two novel microalgal enzymes involved in the conversion of the omega3-fatty acid, eicosapentaenoic acid, into docosahexaenoic acid. Biochem J 384:357–366. https://doi.org/10.1042/BJ20040970
Petrie JR, Mackenzie AM, Shrestha P, Liu Q, Frampton DF, Robert SS, Singh SP (2010) Isolation of three novel long-chain polyunsaturated fatty acid Δ9-elongases and the transgenic assembly of the entire pavlova salina docosahexaenoic acid pathway in nicotiana benthamiana1. J Phycol 46:917–925. https://doi.org/10.1111/j.1529-8817.2010.00870.x
Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE (2021) UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci 30:70–82. https://doi.org/10.1002/pro.3943
Qi B, Beaudoin F, Fraser T, Stobart AK, Napier JA, Lazarus CM (2002) Identification of a cDNA encoding a novel C18-Delta(9) polyunsaturated fatty acid-specific elongating activity from the docosahexaenoic acid (DHA)-producing microalga, Isochrysis galbana. FEBS Lett 510:159–165. https://doi.org/10.1016/s0014-5793(01)03247-1
Qiu X, Xie X, Meesapyodsuk D (2020) Molecular mechanisms for biosynthesis and assembly of nutritionally important very long chain polyunsaturated fatty acids in microorganisms. Prog Lipid Res 79:101047. https://doi.org/10.1016/j.plipres.2020.101047
Sayanova O, Haslam RP, Calerón MV, López NR, Worthy C, Rooks P, Allen MJ, Napier JA (2011) Identification and functional characterisation of genes encoding the omega-3 polyunsaturated fatty acid biosynthetic pathway from the coccolithophore Emiliania huxleyi. Phytochemistry 72:594–600. https://doi.org/10.1016/j.phytochem.2011.01.022
Toke DA, Martin CE (1996) Isolation and characterization of a gene affecting fatty acid elongation in Saccharomyces cerevisiae. J Biol Chem 271:18413–18422. https://doi.org/10.1074/jbc.271.31.18413
Tsirigos KD, Peters C, Shu N, Käll L, Elofsson A (2015) The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. https://doi.org/10.1093/nar/gkv485
Volschenk H, Viljoen M, Grobler J, Petzold B, Bauer F, Subden RE, Young RA, Lonvaud A, Denayrolles M, Van Vuuren HJJ (1997) Engineering pathways for malate degradation in saccharomyces cerevisiae. Nat Biotechnol 15:253–257. https://doi.org/10.1038/nbt0397-253
Vrinten PL, Hoffman T, Bauer J, Qiu X (2010) Specific protein regions influence substrate specificity and product length in polyunsaturated fatty acid condensing enzymes. Biochemistry 49:3879–3886. https://doi.org/10.1021/bi902028w
Wojciechowski D, Fischer M, Fahlke C (2015) Tryptophan scanning mutagenesis identifies the molecular determinants of distinct barttin functions. J Biol Chem 290:18732–18743. https://doi.org/10.1074/jbc.M114.625376
Acknowledgements
This is National Research Council of Canada publication number 58456.
Funding
The authors have not disclosed any funding.
Author information
Authors and Affiliations
Contributions
DM and XQ designed the experiments. DM performed the experiments on domain swapping and site-directed mutagenesis as well as functional analysis of all mutants. KS conducted the three-dimensional structure modelling. DM and XQ analyzed the data and wrote the paper. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
We declare there is no conflict of interest associated with this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Meesapyodsuk, D., Sun, K. & Qiu, X. Structural and functional analysis of plant ELO-like elongase for fatty acid elongation. Plant Mol Biol 114, 90 (2024). https://doi.org/10.1007/s11103-024-01490-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11103-024-01490-5