Abstract
Phenylpropanoids, a class of specialized metabolites, play crucial roles in plant growth and stress adaptation and include diverse phenolic compounds such as flavonoids. Phenylalanine ammonia-lyase (PAL) and chalcone synthase (CHS) are essential enzymes functioning at the entry points of general phenylpropanoid biosynthesis and flavonoid biosynthesis, respectively. In Arabidopsis, PAL and CHS are turned over through ubiquitination-dependent proteasomal degradation. Specific kelch domain-containing F-Box (KFB) proteins as components of ubiquitin E3 ligase directly interact with PAL or CHS, leading to polyubiquitinated PAL and CHS, which in turn influences phenylpropanoid and flavonoid production. Although phenylpropanoids are vital for tomato nutritional value and stress responses, the post-translational regulation of PAL and CHS in tomato remains unknown. We identified 31 putative KFB-encoding genes in the tomato genome. Our homology analysis and phylogenetic study predicted four PAL-interacting SlKFBs, while SlKFB18 was identified as the sole candidate for the CHS-interacting KFB. Consistent with their homolog function, the predicted four PAL-interacting SlKFBs function in PAL degradation. Surprisingly, SlKFB18 did not interact with tomato CHS and the overexpression or knocking out of SlKFB18 did not affect phenylpropanoid contents in tomato transgenic lines, suggesting its irreverence with flavonoid metabolism. Our study successfully discovered the post-translational regulatory machinery of PALs in tomato while highlighting the limitation of relying solely on a homology-based approach to predict interacting partners of F-box proteins.
Key message
Despite its highest sequence homology with Arabidopsis CHS-interacting KFB among 31 tomato KFBs, SlKFB18 does not function in CHS degradation, while predicted PAL-interacting SlKFBs function in PAL degradation in tomato.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phenylpropanoids are a group of specialized metabolites, encompassing flavonoids, condensed tannins, hydroxycinnamoyl compounds, volatile phenylpropanoids, and monolignols (Deng and Lu 2017; Dong and Lin 2021; Garibay‑Hernández et al. 2021). They are ubiquitously present in the plant kingdom (Liu et al. 2015; Garibay‑Hernández et al. 2021) and play vital roles in plant survival. Monolignols, for instance, serve as building blocks of lignin, providing rigidity and hydrophobicity to vascular bundles (Muro‑Villanueva et al. 2019). Flavonoids protect plants from various stresses (Agati et al. 2020; Shomali et al. 2022) and regulate plant growth and development by modulating auxin transport and scavenging reactive oxygen species (Brown et al. 2001; Yin et al. 2014; Muhlemann et al. 2018; Tan et al. 2019; Chapman and Muday 2021; Teale et al. 2021). Kaempferol, a flavonol aglycone, serves as a precursor for ubiquinone, an essential respiratory cofactor. (Soubeyrand et al. 2018, 2021; Fernández‑Del-Río et al. 2020; Berger et al. 2022). Moreover, numerous phenylpropanoids, especially flavonoids, exhibit beneficial properties for human health, such as anti-cancer, anti-diabetes, and antioxidant activities (Wedick et al. 2012; Tu et al. 2017; Bondonno et al. 2019; Wen et al. 2021; Prasanna and Upadhyay 2021; Micek et al. 2021; Xian et al. 2021; Slika et al. 2022). Therefore, understanding phenylpropanoid biosynthesis and its regulation is imperative for engineering enhanced phenylpropanoid production in crops (Sun et al. 2020; Chen et al. 2021).
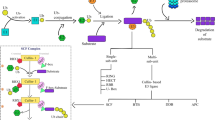
Phenylpropanoid biosynthesis starts with the deamination of phenylalanine to produce cinnamic acid by phenylalanine ammonia-lyase (PAL) (Zhang and Liu 2015). Subsequent hydroxylation and ligation reactions produce p-coumaroyl-CoA, a precursor for hydroxycinnamoyl compounds like monolignols and flavonoids (Vogt 2010) (Fig. 1a). The first enzyme directing flux from the production of hydroxycinnamoyl compounds to flavonoid biosynthesis is chalcone synthase (CHS) that produces naringenin chalcone using p-coumaroyl-CoA and malonyl-CoA (Fig. 1a) (Grotewold 2006; Saito et al. 2013). Then, sequential reactions of isomerases, hydroxylases, and reductases generate basic structures of flavonoid skeletons such as flavonol aglycones and anthocyanidins (Wen et al. 2020).
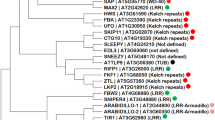
Proteolytic regulation steps in the phenylpropanoid pathway in Arabidopsis and a phylogenetic tree of tomato KFBs. a Chalcone synthase (CHS) and phenylalanine ammonia-lyase (PAL) are regulated through ubiquitin-mediated proteolysis in Arabidopsis. AtKFBCHS and AtKFBPAL are kelch-domain containing F-box proteins responsible for ubiquitination of CHS and PAL, respectively. b Phylogenetic analysis with 31 putative SlKFBs retrieved from the tomato genome and 9 characterized KFBs from other plant species identified two sub clades including AtKFBCHS and AtKFBPAL. A phylogenetic tree was constructed using Maximum Likelihood method with 1000 bootstrap samples and the JTT model. The tree was constructed with the full-length protein sequences of 31 SlKFBs from the tomato reference genome (SL4.0 build; ITAG4.0 annotation) (Tomato Genome Consortium 2012) and 9 KFBs from Arabidopsis, rice, grape, and muskmelon (marked with open circle) known for their role in regulating phenylpropanoid metabolism. Branch lengths are drawn to scale, with the scale bar indicating the number of amino acid substitutions per site. The four AtKFBPALs (AtKFB1, 20, 39, and 50) are clustered in Clade 1, and the KFBs known to regulate flavonoids are in Clade 2. SlACIF1 serves as an outgroup
The phenylpropanoid pathway is regulated through intricate mechanisms, including feed-forward, feed-back, transcriptional, post-transcriptional, and post-translational regulations (Yin et al. 2012; Xu et al. 2015; Shin et al. 2015; Verweij et al. 2016; Zhang et al. 2017; Ohno et al. 2018; Wang et al. 2020). Recent studies have revealed that PAL and CHS activities are regulated post-translationally through ubiquitin-dependent proteasomal degradation (Zhang et al. 2013, 2017; Gu et al. 2019; Mao et al. 2022; Zhao et al. 2023). Ubiquitination is a protein modification that adds the small regulatory protein called ubiquitin (Ub) to lysine residues of substrate proteins and poly-ubiquitinated proteins are subsequently degraded by the 26S proteasome (Hristova et al. 2020). Ubiquitination requires ubiquitin-activating enzyme (E1), ubiquitin conjugating enzyme (E2), and ubiquitin ligase (E3). Ubiquitin is activated by the E1 enzyme, and then transferred to E2. The E3 complex then adds ubiquitin from E2 to target proteins. The E3 ubiquitin ligase includes E2 binding protein, scaffold protein, adaptor protein, and substrate binding protein such as F-box proteins (Gray and Estelle 2000). Several Kelch domain-containing F-box proteins (KFBs) were identified as subunits of E3 ligase functioning in the ubiquitination of PAL and CHS, the two vital enzymes functioning at the entry points of the general phenylpropanoid pathway and flavonoid pathway, respectively (Fig. 1a) (Zhang and Liu 2015; Zhang et al. 2017). In Arabidopsis, four KFBs (KFB1, KFB20, KFB39, and KFB50) function in the PAL ubiquitination and overexpression of these KFBs significantly reduces phenylpropanoid contents (Zhang et al. 2013, 2017). AtKFBCHS, FvKFB1, and VviKFB7 directly interact with CHS in Arabidopsis, strawberry, and grape, respectively, which leads to the degradation of CHS (Zhang et al. 2017; Mao et al. 2022; Zhao et al. 2023). In rice (Oryza sativa), ibf1 mutant having a defective KFB (IBF1) contains increased flavonoid contents, and muskmelon (Cucumis melo) cultivars with elevated CmKFB expression have decreased flavonoid contents (Shao et al. 2012; Feder et al. 2015). Although the interacting partners of IBF1 and CmKFB remain unknown, these findings imply a role of IBF1 and CmKFB in flavonoid metabolism, either directly or indirectly. Notably, tomato leaves expressing CmKFB contain reduced levels of flavonoids, suggesting that tomato flavonoid metabolism is likely regulated through KFB-mediated ubiquitination and degradation (Feder et al. 2015).
Tomato is one of the most widely consumed vegetables globally, serving as an excellent source of beneficial phytonutrients, including phenylpropanoids (Chandra et al. 2012; Anwar et al. 2019). Tomato accumulates various phenylpropanoids including flavonoids, caffeic acid derivatives, stilbenes, coumarins, monolignols, aurones (Zhang et al. 2015b). Despite advances in our understanding of the phenylpropanoid metabolism in tomato (Zhang et al. 2015b; Tohge et al. 2017; Rosa‑Martínez et al. 2023), the post-translational regulation of phenylpropanoid metabolism in tomato remains unknown. In this study, we aimed to identify tomato KFBs (SlKFBs) involved in phenylpropanoid metabolism in tomato (Solanum lycopersicum). Our homology study identified 31 genes encoding putative KFB in the tomato genome, and we investigated their functions in phenylpropanoid metabolism.
Materials and methods
Genetic material and plant growth conditions
Micro-Tom was obtained from the tomato genetics resource center located at the University of California, Davis, led by C.M. Rick. To conduct the BiFC analysis, we used Nicotiana benthamiana. Tomato and tobacco plants were grown under controlled conditions of 22 °C ± 1 °C with a 16 h day and 8 h photoperiod.
Retrieval of KFBs from tomato genome
Kelch-domain containing F-box proteins (KFBs) were identified in the tomato genome (SL4.0 build; ITAG4.0 annotation) (Tomato Genome Consortium 2012) using the Jackhmmer program (version 2.41.2) within HmmerWeb (https://www.ebi.ac.uk/Tools/hmmer/) (Fernandez‑Pozo et al. 2015; Potter et al. 2018). A Hidden Markov Model (HMM) profile was constructed by initially querying the sequence of Kelch domain and F-box domain in the Arabidopsis KFBCHS (AT1G23390). This model was then employed to iteratively search the tomato protein database, which includes UniProtKB and SwissProt. The search was conducted separately for proteins containing either the Kelch domain or F-box domain until no additional proteins were added to the retrieved protein lists. Proteins containing both the F-box and Kelch domain were selected by cross-referencing the protein lists obtained with the HMM profile of each domain separately. In total, this method identified 31 KFBs in the tomato genome.
Gene matrix construction
To generate the amino acid sequence identity matrix, we utilized Clustal Omega (version: Clustal2.1). We used the default configurations for the analysis.
Yeast two hybrid (Y2H) assay
The coding sequences (CDS) of seven SlKFBs (Solyc01g005970, Solyc03g120320, Solyc03g120330, Solyc05g005150, Solyc06g066770, Solyc06g083550, Solyc10g080610), AtKFBCHS (At1g23390), CmKFB (XP008446188), SlCHS1 (Solyc09g091510), SlCHS2 (Solyc05g053550), SlCHI (Solyc05g010320), SlF3H (Solyc02g083860), SlPAL5 (M83314), and AtCHS (At5g13930) with the appropriate restriction enzyme site at the end of the CDS were synthesized from Twist Bioscience (CA, USA). Arabidopsis PAL1, PAL2, PAL3, and PAL4 were cloned from vectors purchased from ABRC (stock number: pDEST-DB004H07 for AtPAL1, pDEST-DB030E01 for AtPAL2, CIW05433 for AtPAL3, and pDEST-DB101F06 for AtPAL4) using primer numbers 32–39 (Supplementary Table S1). The CDS of KFBs were subcloned into the pGADT7 vector (catalog number: 630442, Takara Bio, Otsu, Japan), while the CDS of CHS, PAL, CHI, F3H were subcloned into the pGBKT7 vector (catalog number: 630443, Takara Bio, Otsu, Japan). The empty pGADT7 and pGBKT7 vectors were used as negative controls. The constructed vectors were then co-transformed into the yeast strain Y2HGold (catalog number: 630489, Takara Bio, Otsu, Japan) using the lithium acetate-mediated transformation method (Gietz and Woods 2002). To screen the transformed yeast strains, we used SD media (5 g of ammonium sulfate, 3.4 g of yeast nitrogen base without amino acids, 20 g of d-glucose, 20 g of agar per liter) supplemented with dropout amino acids, excluding leucine and tryptophan. We used two SD media, dropout-SD and dropout-SD with Aureobasidin A (AbA), to assess protein–protein interactions. AUR1-C, a mutated version of the AUR1 reporter gene in this Y2H system, allows yeast to survive on media containing AbA. AbA inhibits the wild-type AUR1 protein, which is lethal to yeast, but AUR1-C provides resistance. Dropout-SD lacked leucine, tryptophan, histidine, and adenine, while dropout-SD with AbA was the same as dropout-SD but included AbA for strong selectivity.
Bimolecular fluorescence complementation (BiFC) assay
Two different BIFC systems were used in this study. For the BiFC with PAL, we adopted a vector system from (Han et al. 2022). PCR amplification was performed using specific primers (Supplementary Table S1). The PCR products of KFBs (full length or truncated proteins lacking F-box domain) were cloned into the pUC19/Vc-C vector (Addgene #183,158), while SlPAL5 was cloned into the pYL322d1/Vn-C vector (Addgene #183,154). The pUC19/Vc-c constructs were then linearized with the AscI enzyme, and the pYL322d1/Vn-C constructs were linearized with AscI and SbfI enzymes. Additionally, a linearized DNA fragment containing the mCherry marker fused with a nuclear localization sequence (NLS) was obtained from pUC19/NLS-mChe (Addgene #183,162) using the AscI enzyme. The three DNA fragments were assembled into the plant binary vector, pYL1300UaUf (Addgene #183,173) using the NEBuilder® HiFi DNA Assembly Cloning Kit (catalog number: E5520S, NEB, Ipswich, MA, USA). The protocol for the BIFC with CHS was adopted from a previous report with slight modifications (Nakabayashi et al. 2015). The F-box domains of SlKFB18 and AtKFBCHS were removed to avoid substrate degradation. The truncated KFBs (lacking F-box domain) and the full-length of CHSs were amplified with the attB sequence at the end of CDS by PCR with specific primers listed in Supplementary Table S1. The PCR products were subsequently inserted into the pCC1155 (Zhang et al. 2020) for gateway cloning using BP Clonase™ II Enzyme Mix (catalog number: 11789020; Thermo Fisher Scientific, Waltham, MA, USA). The pCC1155 with KFB constructs were subcloned into the pBatTL-B-sYFPn vector, and the pCC1155 with CHS constructs were subcloned into pBatTL-B-sYFPc using LR Clonase™ II Enzyme Mix (catalog number: 11791; Thermo Fisher Scientific, Waltham, MA, USA). Empty pBatTL-B-sYFPn and pBatTL-B-sYFPc were utilized as negative controls.
All BiFC constructs were transformed into A. tumefaciens strain GV3101 and co-infiltrated into 4 weeks-old Nicotiana benthamiana leaves. The YFP, Venus, and mCherry fluorescence signals were detected using a confocal scanning microscope (Olympus IX81‐DSU) 48 h after infiltration.
Generation of transgenic lines
To generate Arabidopsis transgenic lines overexpressing SlKFB13 and SlKFB14, coding sequences of SlKFB13 and SlKFB14 were synthesized by Twist Bioscience (San Francisco, CA, USA). These sequences were subsequently cloned into the pCC1155 entry vector using the BP Clonase™ II enzyme mix (catalog no. 11789020; Thermo Fisher Scientific, Waltham, MA, USA), and then were recombined into the pCC0995 destination vector via the LR Clonase™ II enzyme mix (catalog no. 11791; Thermo Fisher Scientific, Waltham, MA, USA). These constructs were then introduced into wild-type Arabidopsis plants (Col-0) using the Agrobacterium tumefaciens-mediated floral dip method, as described by (Zhang et al. 2006). The resulting T1 generation seedlings were initially selected on soil with 0.2% (w/v) BASTA (glufosinate ammonium), and surviving plants were subsequently transplanted to fresh soil for further assays. To determine the expression levels of SlKFBs in the transgenic Arabidopsis lines, RT-PCR was performed using specific primers No. 24 and 25 for SlKFB13, No. 26 and 27 for SlKFB14, and No. 28 and 29 for AtTUB3 as an internal control.
To generate tomato transgenic lines overexpressing SlKFB18 and CRISPR lines,
the gRNA-containing (5'-GCTTCAACAAGCCGAAGCCG-3') pHSE401 CRISPR vectors to target the upstream region of the SlKFB18 gene and the pGWB502 overexpression vectors harboring the SlKFB18 CDS were introduced into the A. tumefaciens GV3101 using the heat shock method. The tissue culture-mediated tomato transformation was conducted using the previously described method with slight modifications (Gupta and Van Eck 2016). Cotyledons from 10 days-old seedlings were excised and incubated on filter paper on preculture media plates containing 4.3 g of Murashige and Skoog (MS) salt, 100 mg of myoinositol, 1 ml of modified Nitsch vitamins stock(composed of 10 mg of glycine, 50 mg of nicotinic acid, 2.5 mg of pyridoxine HCl, 2.5 mg of thiamine HCl, 2.5 g of folic acid, and 0.2 mg of d-biotin in 10 ml), 20 g sucrose, 5.2 g of TC gel, and 2 ml of 1 mg/ml trans-zeatin stock per litter, with the pH adjusted to 6. The cotyledons were incubated under a 16 h photoperiod for 24 h. Agrobacterium cells carrying the vector were incubated overnight and subsequently harvested via centrifugation. The pellet of Agrobacterium cells was re-suspended in a buffer containing 4.3 g of MS salts, 100 mg of myoinositol, 0.4 mg of thiamine HCl, and 20 g of sucrose per liter, with the pH adjusted to 5.8. The cotyledons were then immersed in the Agrobacterium suspension for 5 min and placed back onto preculture media for co-culture in the dark at 22 °C for 48 h. The cotyledons were then transferred to callus induction media, which contained 3 ml of 100 mg/ml timentin stock and 3 mg/l hygromycin per liter and were incubated for 2 weeks. Once small calli had formed, the cotyledons with the calli were transferred to a shoot induction media. The concentration of trans-zeatin in shoot induction media was reduced to 50% of the amount in the callus induction media, while all other ingredients remained the same. The calli were transferred to fresh shoot induction media every 2 weeks until shoot formation was observed. Once the shoots reached a minimum length of 2 cm with at least one node, they were excised and planted in a root regeneration media composed of 4.3 g of MS salt, 1 ml of modified nitch vitamins stock, 30 g of sucrose, 8 g of Difco Bacto Agar, 2 ml of 100 mg/ml timentin stock, 3 mg of hygromycin, and 1 mg of indole acetic acid per liter with the pH adjusted to 6. When the roots were sufficiently formed, the plantlets were moved to soil and grown until seed maturation. The CRISPR knock-out lines were isolated via sequencing of the target genes. To check the genotype of modified region of SlKFB18, the primer set, No. 30 and 31, was used (Supplementary Table S1). Transgenic plants overexpressing SlKFB18 were isolated based on their resistance to hygromycin. The expression of SlKFB18 was then confirmed by qRT-PCR using the primer set of No. 22 and 23. SlACTIN2 expression was used as an internal control (Supplementary Table S1).
PAL activity
The PAL activity was measured by a modified method of the procedure described in (Kim et al. 2015). Total proteins were extracted from the leaves of 3 weeks-old Arabidopsis, which were pulverized in a Benchmark BeadBlaster 24 homogenizer (Benchmark Scientific, NJ). The powdered tissue was then incubated in extraction buffer comprising 0.1 M Tris–HCl (pH 8.3), 10% glycerol, and 5 mM dithiothreitol (DTT) for one hour and then crude proteins were collected after centrifugation. Protein concentrations in the extracts were quantified via the Bradford assay, employing the Bradford Reagent (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s protocol. The enzyme reaction of PAL was started by adding 150 µl of protein extract with 400 µl of a reaction buffer that contained 5 mM L-phenylalanine, and the mixture was incubated at 37 °C for 90 min. The reaction was terminated by adding 40 µl of 30% (v/v) acetic acid. The product of the enzyme reaction was then extracted with ethyl acetate, the volume of which was 600 µl, and subsequently concentrated using an Eppendorf Vacufuge Plus (Eppendorf, Hamburg, Germany). The dried extract was then redissolved in 100 µl of 50% methanol and 10 µl of extract was analyzed using HPLC with a solvent B (100% acetonitrile) gradient in solvent A (0.1% formic acid in water). The gradient starting from 12 to 30% of solvent B over 2.6 min, increasing from 30 to 95% in the next 4 min, and holding at 95% for an additional 3 min. The flow rate was set at 0.7 ml/min, and the column temperature was maintained at 40 °C. The PAL reaction product, trans-cinnamic acid, was quantified by measuring the peak area at 270 nm and comparing it to a calibrated curve of a standard trans-cinnamic acid solution (Sigma-Aldrich, St. Louis, MO, USA).
RNA extraction and gene expression analysis
Leaf samples from 4 weeks old tomato plants were collected and immediately frozen in liquid nitrogen. The samples were then homogenized using a Benchmark BeadBlaster 24 homogenizer (Benchmark Scientific, Edison, NJ, USA) with 500 µl of 1.25 mm Zirconia oxide beads (A Norstone Company, Bridgeport, PA, USA) to obtain a complete ground sample. Total RNA was extracted from the samples using Trizol reagent (Life Technologies Inc., Gaithersburg, MD, USA) according to the manufacturer's instructions. For cDNA synthesis, 2 µg of total RNA and a reverse transcription kit (catalog number: 4368814; Thermo Fisher Scientific, Waltham, MA, USA) were used. Quantitative real-time Reverse Transcription PCR (qRT-PCR) and RT-PCR were performed using PCR kits (catalog number: FERK1071; Thermo Fisher Scientific, Waltham, MA, USA, and catalog number: K1081; Thermo Fisher Scientific, Waltham, MA, USA) with 1 µl of cDNA. AtTUB3 (AT5G62700) and SlACTIN2 (Solyc11g005330) were used as internal controls for gene expression analysis in Arabidopsis and tomato, respectively. Specific forward and reverse primers, as listed in Supplementary Table S1, were used for PCR.
Phenylpropanoid quantification
The first three true leaves from 3 weeks old tomato plants and 3rd and 4th rosette leaves of 3 weeks old Arabidopsis were used for metabolite analysis using 50% (v/v) methanol at 65 °C for 1 h. The extracts were then subjected to analysis using an UltiMate 3000 HPLC system (ThermoFisher Scientific, Waltham, MA, USA) equipped with an Acclaim™ 120 C18 column (75 mm × 3 mm; 2.2 μm) coupled with a C18 guard column (10 mm × 3 mm; 5 μm) (ThermoFisher Scientific, Waltham, MA, USA). Metabolites from Arabidopsis samples were separated using a mobile phase composed of solvent A (0.1% formic acid (v/v) in water) and solvent B (100% acetonitrile), with a gradient of 5% to 14% for 2.2 min, followed by 14% to 18% (v/v) solvent B for 9 min, and finally 18% to 95% solvent B for 3.5 min. Three kaempferol glycosides were identified by comparing the HPLC profiles of wild type, ugt78d1, and ugt78d2 mutants, following previous studies (Yin et al. 2012, 2014). The levels of kaempferol glycosides were compared based on their HPLC peak areas. Sinapate esters, including sinapoylmalate and sinapoylglucose, were identified based on their retention times and UV spectra, as determined in previous studies (Kim et al. 2015, 2020; Li et al. 2015; Perez et al. 2021; Shin et al. 2023). Sinapoylmalate was quantified using sinapic acid as its standard (catalog number: D7927; Sigma-Aldrich, St. Louis, MO, USA).
Metabolites from the tomato samples were separated with a mobile phase consisting of solvent A (0.1% formic acid (v/v) in water) and solvent B (100% acetonitrile) with a gradient of 3% to 18% for 17 min, followed by 18% to 50% (v/v) solvent B for 6 min. The contents of rutin and chlorogenic acid were quantified based on peak area and a standard curve of rutin (catalog number: R5143; Sigma-Aldrich, St. Louis, MO, USA) and chlorogenic acid (catalog number: C0181; TCI America, Portland, OR, USA).
Results
putative SlKFBs were identified in the tomato genome
From the tomato genome (SL4.0 Assembly), we identified a total of 31 genes encoding proteins with both F-box and Kelch domains, which we considered to be putative KFB homologs in tomato. We designated them as SlKFB1 to SlKFB31 based on their positions within the tomato chromosomes (Supplementary Table S2). To identify SlKFB candidates targeting CHS and PAL, we conducted a comparative analysis with the amino acid sequences of tomato KFBs and previously characterized KFBs from Arabidopsis, rice, grape, and muskmelon that were reported to regulate phenylpropanoid production (Shao et al. 2012; Zhang et al. 2013, 2015a, 2017; Feder et al. 2015; Zhao et al. 2023) (Fig. 1b). In the phylogenetic tree, four PAL-interacting KFBs from Arabidopsis (AtKFB01, AtKFB20, AtKFB39, and AtKFB50) were clustered in clade 1, while clade 2 included KFBs functioning in flavonoid metabolism, including Arabidopsis CHS-interacting KFB (AtKFBCHS) (Fig. 1b).
Four tomato KFBs, SlKFB13 (Solyc03g120320), SlKFB14 (Solyc03g120330), SlKFB21 (Solyc06g066770), and SlKFB29 (Solyc10g080610), were clustered with PAL-interacting KFBs in clade 1 (Zhang et al. 2013, 2015a) (Fig. 1b). SlKFB13, SlKFB14, and SlKFB21 showed 45 to 50% amino acid sequence identities with AtKFB1 and AtKFB20, while SlKFB29 showed 36% and 37% amino acid sequence identities with AtKFB1 and AtKFB20, respectively (Supplementary Fig. S1). All four SlKFBs displayed sequence identities ranging from 30 to 36% with AtKFB39 and AtKFB50. SlKFB28 (Solyc09g066210), the closest KFB to those in clade 1, showed only 24% to 27% sequence identities with AtKFB1, 20, 39, and 50 (Fig. 1b, Supplementary Fig. S1). Thus, SlKFB13, SlKFB14, SlKFB21, and SlKFB29 in clade 1 were selected as PAL-interacting SlKFB candidates.
SlKFB18 was the only SlKFB in clade 2 having AtKFBCHS, IBF1, VviKFB7, and CmKFB, the KFBs functioning in flavonoid metabolism (Fig. 1b). SlKFB22 (Solyc06g083550) was SlKFB close to clade 2, but SlKFB22 showed 20% to 24% sequence identities with AtKFBCHS, IBF1, CmKFB, and VviKFB7 while SlKFB18 exhibited over 40% sequence identity with them (Supplementary Fig. S1). Thus, SlKFB18 was selected as a CHS-targeting KFB candidate.
SlKFB13, SlKFB14, SlKFB21, and SlKFB29 regulate PAL stability
In Arabidopsis, four Phenylalanine Ammonia-Lyase (PAL) enzymes, AtPAL1, AtPAL2, AtPAL3, and AtPAL4, function redundantly (Rohde et al. 2004; Huang et al. 2010) and four KFBs, AtKFB1, AtKFB20, AtKFB39, and AtKFB50, participate redundantly in the ubiquitination of all four AtPALs (Zhang et al. 2013, 2015a). We identified six PAL homologs (Solyc05g056170, Solyc10g086180, Solyc09g007890, Solyc09g007910, Solyc09g007900, and Solyc09g007920) in the tomato genome (SL4.0 build; ITAG4.0 annotation) (Tomato Genome Consortium 2012) that showed over 70% sequence identities with Arabidopsis PALs (Supplementary Fig. S2). Solyc10g086180, Solyc09g007890, Solyc09g007910, Solyc09g007900, and Solyc09g007920 were shown to be upregulated in AtMYB12 overexpression tomato, along with other flavonoid biosynthesis enzymes (Zhang et al. 2015b). Additionally, SlPAL5 (M83314.1) has been reported as a tomato PAL in previous studies (Guo and Wang 2009; Løvdal et al. 2010). Interestingly, all seven SlPALs are more closely related to AtPAL1 and AtPAL2 than to AtPAL3 and AtPAL4 in the phylogenetic tree (Supplementary Fig. S2). As SlPAL5 has been previously characterized (Guo and Wang 2009; Løvdal et al. 2010), we decided to use SlPAL5 to identify PAL-interacting SlKFBs in tomato.
In our Y2H analysis, we included four SlKFBs, SlKFB13 (Solyc03g120320), SlKFB14 (Solyc03g120330), SlKFB21 (Solyc06g066770), and SlKFB29 (Solyc10g080610) from clade 1 (Fig. 1b). Among them, SlKFB14, SlKFB21 and SlKFB29 interacted with SlPAL5 (Fig. 2). Notably, SlKFB13 (Solyc03g120320) did not interact with SlPAL5 in our Y2H assay, despite its high sequence similarity with SlKFBs in clade 1 (Fig. 1b, 2; Supplementary Fig. S1).
Three SlKFBs interacted with SlPAL5 in the Y2H assay SlKFBs were fused with the activation domain (AD), and SlPAL5 was fused with the binding domain (BD). SD media excluding leu and trp were used to check the introduction of AD and BD vectors in the yeast. SD media excluding leu, trp, his, and ade were used to assess protein interactions. SD media excluding leu, trp, his, and ade along with Aureobasidin A (AbA) were used to increase stringency of interactions. SlKFB14, 21, and 29 in clade 1 showed interaction with SlPAL5, while SlKFB13 in the same clade and SlKFB18 in clade 2 did not. The pair of AD-T and DB-P53 was used as a positive control, and the pair of AD and BD vector without insert DNA was used as a negative control
To test if the interaction of SlKFB14/21/29 with SlPAL5 affects protein stability, we conducted BiFC experiments using intact SlKFBs and truncated SlKFBs (SlKFB (∆)) with the F-box domain removed (Fig. 3). In this test, we included a nuclear-localized mCherry cassette to evaluate proper transformation (Fig. 3a). All tested samples showed mCherry signals in nucleus, indicating properly expressed transgenes. The detection of the Venus signal was evident in leaves that were infiltrated with SlKFB14, SlKFB21, and SlKFB29 lacking their F-box domains, along with SlPAL5 (Fig. 3b). However, this signal was not observed with intact SlKFBs (Fig. 3b). This result suggests that SlPAL5, upon interacting with SlKFBs, undergoes degradation through a mechanism requiring the F-box domain, likely involving ubiquitination-mediated degradation.
BiFC assay for the interactions of SlKFBs with SlPAL5. a The illustration of the BiFC vector used in this assay. P35s (35S promoter), mChe (mCherry CDS), NLS (Nuclear localization signal sequence), Tnos (Nos terminator), Vn (Venus N-terminal fragment), Vc (Venus C-terminal fragment), PAL (Phenylalanine ammonia-lyase CDS), and KFB (Full length or partial CDS of Kelch domain-containing F-box). b SlKFB14/21/29 interacting with SlPAL5 exhibited fluorescence when their F-box domain was removed, but intact SlKFBs co-infiltrated with SlPAL5 did not show fluorescence. The BiFC was conducted using intact SlKFBs, and truncated SlKFBs that lack the F-box domains (SlKFB14 (∆F), SlKFB21 (∆F), and SlKFB29 (∆F) that first 46, 51 and 49 amino acids were removed, respectively). Images captured under identical exposure conditions depict bright field, mCherry (captured using Wide‐band Green‐Red excitation light with a center wavelength of 593 nm, and a TRITC filter), Venus (captured using blue excitation light with a center wavelength of 470 nm and a FITC filter) and merged two-channel views. More than five infiltrated leaves were examined, and similar images were obtained
In Arabidopsis, all four KFBs redundantly interact with four PALs (AtPAL1 ~ AtPAL4) (Zhang et al. 2013, 2015a). As there are at least six additional PAL-encoding genes in the tomato genome, SlKFB14/21/29 may interact with other SlPALs in addition to SlPAL5. Similarly, SlKFB13 may interact with other PAL proteins, although it did not interact with SlPAL5. To test if SlKFB13 functions in PAL degradation, we took advantage of Arabidopsis system. Arabidopsis rosette leaves accumulate sinapoylmalate, a blue fluorescent phenylpropanoid (Ruegger and Chapple 2001), causing Arabidopsis leaves to emit a bluish color under UV light. Conversely, Arabidopsis plants with reduced sinapoylmalate would appear red under UV light due to chlorophyll auto-fluorescence (Ruegger and Chapple 2001). As PAL activity is necessary for the production of phenylpropanoids, including sinapoylmalate, accelerated PAL degradation can be detected with leaf UV fluorescence. Given that tomato PALs showed over 80% sequence identities with Arabidopsis PAL1/2/4, which is higher than 73% sequence identities of AtPAL3 when compared with AtPAL1 and 2 (Supplementary Fig. S2a), it is possible that SlKFBs may interact with AtPALs. Thus, we overexpressed SlKFB13 in Arabidopsis to test its impact on phenylpropanoid production. We also overexpressed SlKFB14, which interacts with SlPAL5 from Y2H and BiFC assays (Fig. 2, 3). As shown in Fig. 4, some Arabidopsis transgenic lines overexpressing SlKFB13 or SlKFB14 exhibited a red color under UV light, while others displayed a bluish color. Consistently, the lines showing a red color under UV light accumulated lower levels of phenylpropanoids, including three kaempferol glycosides, sinapolymalate, and sinapoylglucose, compared to the lines exhibiting a blue color (Fig. 4, Supplementary Fig. S3a). The reduced phenylpropanoid contents correlated with PAL activity and the expression levels of SlKFB13 and SlKFB14 (Fig. 4), suggesting the repressive roles of SlKFB13 and SlKFB14 on PAL activity. Notably, several Arabidopsis transgenic lines with strong expression of SlKFB13 and SlKFB14 displayed alteration in growth and development, such as stunted inflorescence growth (Supplementary Fig. S3b), similar to those observed in the Arabidopsis pal mutants (Huang et al. 2010).
PAL activity and phenylpropanoid contents in the transgenic lines overexpressing SlKFB13 or SlKFB14. a, b 3 weeks old plants expressing SlKFB13 (A) or SlKFB14 (B) photographed under visible light (top) and UV light (bottom), Plants exhibiting red fluorescence under UV light indicate a low level of sinapoylmalate compared to those with blue fluorescence. c, d The levels of sinapoylmalate, three kaempferol glycosides, and PAL enzyme activity in plants. The relative expression levels of SlKFB13 and SlKFB14 in representative high- or low-sinapoylmalate accumulation plants were shown with RT-PCR. AtTUB3 (AT5G62700) was used for an internal control. Kaempferol-3-O-glu-7-O-rha (K2), Kaempferol-3-O-rha-7-O-rha (K3), Kaempferol-3-O-[rha (1- > 2 glu)]-7-O-rha (K1)
To determine whether any interactions of SlKFBs with AtPALs contributed to the reduced PAL activity and phenylpropanoid contents, we used Y2H assays to examine the interactions between SlKFBs (SlKFB13 and SlKFB14) and Arabidopsis PALs (AtPAL1, AtPAL2, AtPAL3 and AtPAL4) (Rohde et al. 2004; Huang et al. 2010). SlKFB14 indeed interacted with AtPAL1, AtPAL2, and AtPAL4, while SlKFB13 interacted with AtPAL1 and AtPAL4, suggesting a possible role of SlKFBs in PAL stability (Supplementary Fig. S4). Interestingly, both SlKFBs did not interact with AtPAL3.
SlKFB18 does not function in flavonoid metabolism in tomato
In Arabidopsis, AtCHS (At5g13930) is the only CHS functioning in flavonoid biosynthesis as its loss-of-function mutant failed to make any flavonoids (Schmelzer et al. 1988; Shirley et al. 1995). We identified four CHS homologs in the tomato genome (SL4.0 Assembly), which showed over 60% sequence identities with the Arabidopsis CHS (AtCHS) (Supplementary Fig. S5). Among the four identified tomato CHS homologs, SlCHS1 (Solyc09g091510) and SlCHS2 (Solyc05g053550) have been previously characterized (Schijlen et al. 2007; España et al. 2014; Kong et al. 2020). Silencing SlCHS1 resulted in a notable reduction in flavonoid content (Schijlen et al. 2007; España et al. 2014; Kong et al. 2020). The other two SlCHS homologs were designated as SlCHS-like1 (Solyc12g098090) and SlCHS-like2 (Solyc05g053170) (Supplementary Fig. S5a). SlCHS1 and SlCHS2 showed approximately 85% sequence identities with AtCHS, while SlCHS-like1 and SlCHS-like2 exhibited sequence identities of 65% and 66% with AtCHS, respectively (Supplementary Fig. S5b). According to the public database (Ruprecht et al. 2017), SlCHS1 and SlCHS2 are expressed in most organs, while the expression of SlCHS-like1 and SlCHS-like2 are barely detected (Supplementary Fig. S5c). Thus, we used SlCHS1 and SlCHS2 to identify SlCHS-interacting SlKFBs.
To assess protein–protein interactions between SlKFB candidates and SlCHS1/2, we employed the Y2H system. Given that AtKFBCHS interacts with AtCHS (Zhang et al. 2017), we included them as a positive control for our Y2H assay. Previous studies have indicated that overexpression of CmKFB from muskmelon reduces flavonoid content in tomato, yet its target protein(s) remains unknown (Feder et al. 2015). It is possible that CmKFB functions in CHS degradation. Therefore, we also included CmKFB in our Y2H assay. As expected, AtKFBCHS interacted with AtCHS (Fig. 5). Interestingly, AtKFBCHS and CmKFB interacted with both SlCHS1 and SlCHS2 (Fig. 5). The interaction of AtKFBCHS and CmKFB with tomato CHS suggests that SlCHS1 and SlCHS2 may have the binding site for KFBs (Fig. 5). Our phylogenetic study identified only one SlKFB, SlKFB18, in clade 2, where CHS-targeting KFBs or flavonoid-related KFBs clustered (Fig. 1b). Despite having the highest sequence identity with known CHS-targeting KFBs (Supplementary Fig. S1), SlKFB18 did not interact with either SlCHS1 or SlCHS2. Similarly, SlKFB22, the KFB closest to SlKFB18, also did not interact with SlCHS1 and SlCHS2 (Figs. 1b and 5). We further investigated whether SlKFB18 interacts with two other flavonoid biosynthesis enzymes downstream of CHS in the flavonoid biosynthesis pathway, namely SlCHI (Solyc05g010320) and SlF3H (Solyc02g083860). However, SlKFB18 did not interact with either in our Y2H assay (Supplementary Fig. S6).
Y2H for the interaction between CHS and KFB. KFBs were fused with the activation domain (AD), and CHSs were fused with the binding domain (BD). SD media excluding leu, trp, his, and ade were used to assess protein interactions. SD media excluding leu, trp, his, and ade along with Aureobasidin A (AbA) were used to increase stringency of interactions. The pairs of AD-T and DB-P53, and Arabidopsis KFBCHS (AtKFBCHS; AT1G23390) and AtCHS (AT5G13930) were included as positive controls. The pair of AD and BD vectors was used as a negative control. Over-expression of CmKFB (XP 008446188) increased flavonoid production in tomato, but its binding partner has not yet been discovered. Thus, CmKFB was included in the assay. SlKFB18 and SlKFB22 did not interacted with either SlCHS1 or SlCHS2. But, AtKFBCHS and CmKFB physically interacted with both SlCHS1 and SlCHS2
We then tested these interactions in Nicotiana benthamiana using the bimolecular fluorescence complementation (BiFC) method. In the BiFC assay, we used truncated KFBs, AtKFBCHS (AtKFBCHS (∆)) and SlKFB18 (SlKFB18 (∆)), where the F-box domain was removed to prevent the degradation of target proteins after interaction. Consistent with Y2H results, no interaction between SlKFB18 and SlCHS1 was observed, while AtKFBCHS interacted with both AtCHS and SlCHS1 (Fig. 6).
The BiFC assay confirmed the interaction between AtKFBCHS and SlCHS1, but there was no interaction between SlKFB18 and SlCHS1. The BiFC was conducted with intact AtKFBCHS and SlKFB18 and truncated AtKFBCHS (AtKFBCHS (∆F)) and SlKFB18 (SlKFB18 (∆F)) that lack their F-box domains. YFPn- AtKFBCHS (lacking the F-box domain, consisting of amino acids 53–395) and YFPn-SlKFB (lacking the F-box domain, consisting of amino acids 52–370) were coexpressed with YFPc-AtCHS and YFPc-SlCHS1 in Nicotiana benthamiana leaves. The images were captured in bright field, YFP (captured using blue excitation light with a center wavelength of 470 nm and a FITC filter), and merged two-channel views under identical exposure conditions. More than five infiltrated leaves were examined, and similar images were obtained
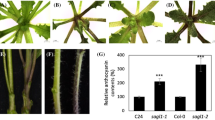
It is possible that SlKFB18 may interact with SlCHS in vivo or it may interact with SlCHS-likes or other flavonoid biosynthesis enzymes. To further investigate the potential involvement of SlKFB18 in flavonoid metabolism, we generated SlKFB18 overexpression lines (SlKFB18-OX1 and SlKFB18-OX2) and CRISPR-mediated SlKFB18 knock-out lines (SlKFB18CR-1 and SlKFB18CR-2) in tomato Micro-Tom. SlKFB18CR-1 and SlKFB18CR-2 have single base pair deletion mutations at the junction of the F-box domain and the Kelch domain of SlKFB18, which result in a frameshift and premature stop codon, leading to the production of a potential nonfunctional truncated protein that lacks all Kelch domains (Fig. 7a). However, the levels of quercetin-3-O-glucoside-6''-O-rhamnoside (rutin), a major flavonol in tomato, did not alter in the CRISPR knock-out lines (Fig. 7b). Additionally, we did not observe any visible alteration of plant growth and development in the mutants compared to wild type (Fig. 7c). The coloration of the hypocotyls, indicative of anthocyanin accumulation, was observed to be the same as in wild type (Fig. 7c). Although SlKFB18 is the only SlKFB in clade 2 (Fig. 1), we cannot exclude a possibility of functional redundancy. In Arabidopsis, overexpression of AtKFBCHS reduced flavonoid production (Zhang et al. 2017). Thus, we generated tomato transgenic lines overexpressing SlKFB18 driven by the 35S promoter. We isolated ten T1 transgenic lines showing resistance to hygromycin from tissue culture calli and analyzed the expression of SlKFB18, as well as the levels of rutin and chlorogenic acid (Supplementary Fig. S7). However, no statistically significant correlation was observed between the level of SlKFB18 expression and the accumulation of the two phenylpropanoids (Supplementary Fig. S7). We further analyzed T2 progeny from four overexpression lines, which exhibited over a 20 fold higher expression of SlKFB18 compared to the wild-type tomato (Fig. 7d). However, the levels of rutin and chlorogenic acid in the overexpression lines were comparable to those in the wild type and vector controls (Fig. 6e, f). Moreover, their morphology was indistinguishable from the wild type (Fig. 7g). We used Micro-Tom for the generation of transgenic lines, and the SlKFB18 sequence was retrieved from tomato reference genome (version ITAG4.0), which was generated with Heinz 1706 cultivar (Tomato Genome Consortium 2012). Notably, there was no sequence variation in SlKFB18 between Micro-Tom (MiBASE database) and the tomato reference genome (ITAG4.0) (Supplementary Fig. S8). Our biochemical and genetic data suggest that SlKFB18 is unlikely to be involved in flavonoid metabolism.
Flavonoid contents and morphology of SlKFB18 knock-out mutants and SlKFB18 overexpression lines remained unaltered compared to the wild type. a Two SlKFB18 knockout lines (SlKFB18CR-1 and SlKFB18CR-2) were generated with the CRISPR system and SlKFB18CR-1 and SlKFB18CR-2 have G and A deletion mutations in its exon, respectively. The gRNA targets a region encoding the Kelch domain, as shown in the SlKFB18 gene structure (top). Single base pair deletion mutations in SlKFB18CR-1 and SlKFB18CR-2 lead to premature translation (bottom). b The level of quercetin-3-O-glucoside-6''-O-rhamnoside (rutin) in the leaves of three-week-old SlKFB18CR-1 and SlKFB18CR-2 compared with wild type (WT). Error bars represent standard deviation. c Representative images of 3-week-old wild type, SlKFB18CR-1, and SlKFB18CR-2. d Expression of SlKFB18 was quantified in wild-type (WT) plants, vector controls (VC), and four T2 transgenic tomato lines overexpressing SlKFB18. Expression levels were measured based on three biological replicates for WT and VC, and two siblings from each transgenic line. T-test analysis results, denoted by asterisks (*), indicate a statistically significant difference between wild type and transgenic lines with a P-value of less than 0.05. e, f Rutin and chlorogenic acid contents were determined for the plants described in (d). Analysis was conducted on metabolite extracts from three-week-old plants. Both WT and VC groups comprised three biological replicates, and for the transgenic lines, three leaves from each of two siblings were sampled, resulting in a total of six replicates for each line. Error bars represent standard deviation. g Representative images of 5 weeks old wild type and SlKFB18 overexpression lines
We tested four SlKFBs that we have shown to regulate PAL stability by using Y2H assays with SlCHS1. None of the four SlKFBs interacted with SlCHS1, indicating the specificity of these SlKFBs in targeting PAL (Supplementary Fig. S9).
Discussion
Protein–protein interactions significantly impact various cellular functions, including protein stability (Struk et al. 2019). Despite the detrimental consequence of destabilized proteins, pinpointing interacting partners is challenging, given the influence of factors such as post-translational modifications and the presence of other molecules (Liddington 2004; Keskin et al. 2008). Phylogenetic and homology analyses are commonly used to infer evolutionary relationships among proteins and identify those with similar functions. This approach proves useful, as demonstrated in our identification of PAL-interacting SlKFBs from the tomato genome (Figs. 1b, 2). However, our homology study did not yield similar results for SlCHS-interacting SlKFBs. Despite its high sequence similarity with Arabidopsis KFBCHS and other flavonoid-regulating KFBs from three different plant species, SlKFB18 did not interact with SlCHSs (Figs. 1b, 5, 6) or the two flavonoid biosynthesis enzymes, CHI and F3H (Supplementary Fig. S6). Given that overexpression of SlKFB18 did not reduce flavonoid production in tomato, SlKFB18 unlikely functions as negative regulator in flavonoid production (Fig. 7, Supplementary Fig. S7). We also did not find any visible growth and developmental changes or alteration in fertility in either SlKFB18 overexpression lines or knock-out lines. According to expression data from a public database (Supplementary Fig. S10), SlKFB18 expresses in most organs, suggesting that SlKFB18 is not a pseudo gene and likely has functions beyond the regulation of flavonoid metabolism, which remains unknown. Given that both AtKFBCHS and CmKFB physically interacted with SlCHS1 and SlCHS2 (Fig. 5), SlCHS1 and SlCHS2 are capable of being recognized by Kelch domain-containing proteins. SlKFB22, which is closely related to clade 2, and SlKFB13, 14, 21, and 29, which are shown to interact with PAL, did not interact with SlCHS1 in our Y2H assay (Fig. 5, Supplementary Fig. S9). In a study with Paeonia, the ring-domain containing protein (PhRING-H2) is responsible for CHS ubiquitination and degradation (Gu et al. 2019). Although, we found no homolog of PhRING-H2 in the tomato genome, it is possible that, in addition to KFBs, other ubiquitination machinery could be involved in the CHS turnover mechanism in tomato.
SlKFB13, despite having the highest sequence homology with PAL-targeting AtKFBs, did not interact with SlPAL5, while SlKFB14, SlKFB21, and SlKFB29 did in both Y2H and BiFC assays (Figs. 1b, 2, 3). However, the overexpression of SlKFB13 in Arabidopsis resulted in a significant reduction of PAL activity and phenylpropanoid contents (Fig. 4). Under our growth condition, several Arabidopsis transgenic lines having strong expression of SlKFB13 or SlKFB14 exhibited dwarfism and immature siliques (Supplementary Fig. S3b), reminiscent of those observed in the Arabidopsis pal mutants (Huang et al. 2010). The presented biochemical and genetic data suggest that these four SlKFBs (13, 14, 21, 29) are affecting PAL stability. When targeted by these SlKFBs, SlPALs likely undergo degradation through a process that requires the F-box domain of the SlKFBs, which is similar to the Arabidopsis PAL ubiquitination and degradation process (Zhang et al. 2013, 2015a).
In Arabidopsis, the four Arabidopsis KFBPALs interact with all four AtPALs redundantly (Zhang et al. 2013, 2015a). Our data suggest that SlKFB13 may interact with tomato PALs, excluding SlPAL5. The four tomato SlKFB proteins could potentially interact with specific PAL enzymes, enabling a more finely tuned regulation of phenylpropanoid flux in specific organs or under particular conditions. Additionally, interactions between SlKFB13 and AtPALs were shown to be relatively weak compared to the interactions between SlKFB14 and AtPALs, although overexpression of both SlKFB13 and SlKFB14 led to the repression of phenylpropanoid biosynthesis (Supplementary Fig. S4, Fig. 4). It is possible that SlKFB13 may interact with additional targets besides PALs that require phenylpropanoid production in the Arabidopsis.
The F-box domain interacts with Skp1, a component protein of the E3 complex, while the kelch domain of KFB serves as the mediator for substrate protein interaction (Schumann et al. 2011). We found that four SlKFBs in clade 1 possessed an F-box domain and three Kelch domains, and both domains of these four SlKFBs are highly conserved when compared with four AtKFBPALs (Supplementary Fig. S11). Further study is required to understand what feature enables these SlKFBs discern different target proteins, at least for SlPAL5.
The regulation of PAL activity is precise and occurs in response to environmental stimuli and developmental cues, such as the need for the requirement of specific phenylpropanoids with unique in vivo roles or lignin in certain tissues (Edwards et al. 1985; Liang et al. 1989; Dixon and Paiva 1995; Pawlak‑Sprada et al. 2011; Kim et al. 2020). Also, the process of phenylpropanoid metabolism channels a significant portion of photosynthetically derived organic carbon to downstream products, with lignin as the primary sink (Novaes et al. 2010). Thus, the stability of PAL, the gateway enzyme for the entire phenylpropanoid production, is imperative for the allocation of available energy and carbon, which is essential for plant survival under unfavorable conditions (Kim et al. 2020). The identification of the SlKFBs targeting PAL suggests the conserved regulation mechanism for phenylpropanoid metabolism in tomato.
Given that phenylpropanoids confer health benefits to humans, as well as enhance stress tolerance and rigidity in plants, the identified PAL-interacting SlKFBs might be targets to augment phenylpropanoid content in tomato. The expression profiles of these PAL-targeting SlKFBs vary in organs, implying their possible unique roles or regulations (Supplementary Fig. S10). However, in Arabiodpsis, AtKFBPALs function redundantly and single knockout mutants of AtKFBPALs did not affect PAL activity and phenylpropanoid production (Zhang et al. 2013). It has been reported that AtKFBPALs regulate cytokinin signaling by interacting with the type-B ARR family members, transcriptional regulators of the cytokinin response (Kim et al. 2013) and TCP14 (Steiner et al. 2021). Whether tomato PAL-interacting KFBs also participate in cytokinin signaling remains unknown.
In this study, we identified 31 KFB-encoding genes in the tomato genome, which is a notably modest number compared to Arabidopsis, harboring over 100 KFBs (Zhang et al 2015a, b). Majority of Arabidopsis KFBs have yet been characterized. The data presented here revealed four SlKFBs functioning in the degradation of PAL. A comprehensive study of the remaining SlKFBs would further broaden our understanding of KFB functions in plants.
Data availability
All data supporting the findings of this study are available within the paper and its supplementary information.
References
Agati G, Brunetti C, Fini A et al (2020) Are flavonoids effective antioxidants in plants? twenty years of our investigation. Antioxidants. https://doi.org/10.3390/antiox9111098
Anwar R, Fatima T, Mattoo A (2019) Tomatoes: a model crop of solanaceous plants. Environ Sci. https://doi.org/10.1093/acrefore/9780199389414.013.223
Berger A, Latimer S, Stutts LR et al (2022) Kaempferol as a precursor for ubiquinone (coenzyme Q) biosynthesis: an atypical node between specialized metabolism and primary metabolism. Curr Opin Plant Biol 66:102165. https://doi.org/10.1016/j.pbi.2021.102165
Bondonno NP, Dalgaard F, Kyrø C et al (2019) Flavonoid intake is associated with lower mortality in the danish diet cancer and health cohort. Nat Commun 10:3651. https://doi.org/10.1038/s41467-019-11622-x
Brown DE, Rashotte AM, Murphy AS et al (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol 126:524–535. https://doi.org/10.1104/pp.126.2.524
Chandra HM, Shanmugaraj BM, Srinivasan B, Ramalingam S (2012) Influence of genotypic variations on antioxidant properties in different fractions of tomato. J Food Sci 77:C1174–C1178. https://doi.org/10.1111/j.1750-3841.2012.02962.x
Chapman JM, Muday GK (2021) Flavonols modulate lateral root emergence by scavenging reactive oxygen species in Arabidopsis thaliana. J Biol Chem 296:100222. https://doi.org/10.1074/jbc.RA120.014543
Chen W, Xiao Z, Wang Y et al (2021) Competition between anthocyanin and kaempferol glycosides biosynthesis affects pollen tube growth and seed set of Malus. Hortic Res 8:173. https://doi.org/10.1038/s41438-021-00609-9
Deng Y, Lu S (2017) Biosynthesis and regulation of phenylpropanoids in plants. CRC Crit Rev Plant Sci 36:257–290. https://doi.org/10.1080/07352689.2017.1402852
Dixon RA, Paiva NL (1995) Stress-Induced Phenylpropanoid Metabolism. Plant Cell 7:1085–1097. https://doi.org/10.1105/tpc.7.7.1085
Dong N-Q, Lin H-X (2021) Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J Integr Plant Biol 63:180–209. https://doi.org/10.1111/jipb.13054
Edwards K, Cramer CL, Bolwell GP et al (1985) Rapid transient induction of phenylalanine ammonia-lyase mRNA in elicitor-treated bean cells. Proc Natl Acad Sci 82:6731–6735. https://doi.org/10.1073/pnas.82.20.6731
España L, Heredia-Guerrero JA, Reina-Pinto JJ et al (2014) Transient silencing of CHALCONE SYNTHASE during fruit ripening modifies tomato epidermal cells and cuticle properties. Plant Physiol 166:1371–1386. https://doi.org/10.1104/pp.114.246405
Feder A, Burger J, Gao S et al (2015) A kelch domain-containing F-Box coding gene negatively regulates Flavonoid accumulation in muskmelon. Plant Physiol 169:1714–1726. https://doi.org/10.1104/pp.15.01008
Fernández-Del-Río L, Soubeyrand E, Basset GJ, Clarke CF (2020) Metabolism of the flavonol kaempferol in kidney cells liberates the B-ring to enter coenzyme Q biosynthesis. Molecules. https://doi.org/10.3390/molecules25132955
Fernandez-Pozo N, Menda N, Edwards JD et al (2015) The sol genomics network (SGN)–from genotype to phenotype to breeding. Nucleic Acids Res 43:D1036–D1041. https://doi.org/10.1093/nar/gku1195
Garibay-Hernández A, Kessler N, Józefowicz AM et al (2021) Untargeted metabotyping to study phenylpropanoid diversity in crop plants. Physiol Plant 173:680–697. https://doi.org/10.1111/ppl.13458
Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Meth Enzymol 350:87–96. https://doi.org/10.1016/S0076-6879(02)50957-5
Gray WM, Estelle I (2000) Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem Sci 25:133–138. https://doi.org/10.1016/s0968-0004(00)01544-9
Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57:761–780. https://doi.org/10.1146/annurev.arplant.57.032905.105248
Gu Z, Men S, Zhu J et al (2019) Chalcone synthase is ubiquitinated and degraded via interactions with a RING-H2 protein in petals of Paeonia “He Xie.” J Exp Bot 70:4749–4762. https://doi.org/10.1093/jxb/erz245
Guo J, Wang M-H (2009) Characterization of the phenylalanine ammonia-lyase gene (SlPAL5) from tomato (Solanum lycopersicum L.). Mol Biol Rep 36:1579–1585. https://doi.org/10.1007/s11033-008-9354-9
Gupta S, Van Eck J (2016) Modification of plant regeneration medium decreases the time for recovery of Solanum lycopersicum cultivar M82 stable transgenic lines. Plant Cell Tissue Organ Cult 127:417–423. https://doi.org/10.1007/s11240-016-1063-9
Han J, Ma K, Li H et al (2022) All-in-one: a robust fluorescent fusion protein vector toolbox for protein localization and BiFC analyses in plants. Plant Biotechnol J 20:1098–1109. https://doi.org/10.1111/pbi.13790
Hristova V, Sun S, Zhang H, Chan DW (2020) Proteomic analysis of degradation ubiquitin signaling by ubiquitin occupancy changes responding to 26S proteasome inhibition. Clin Proteomics 17:2. https://doi.org/10.1186/s12014-020-9265-x
Huang J, Gu M, Lai Z et al (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153:1526–1538. https://doi.org/10.1104/pp.110.157370
Keskin O, Gursoy A, Ma B, Nussinov R (2008) Principles of protein-protein interactions: what are the preferred ways for proteins to interact? Chem Rev 108:1225–1244. https://doi.org/10.1021/cr040409x
Kim HJ, Chiang Y-H, Kieber JJ, Schaller GE (2013) SCF(KMD) controls cytokinin signaling by regulating the degradation of type-B response regulators. Proc Natl Acad Sci USA 110:10028–10033. https://doi.org/10.1073/pnas.1300403110
Kim JI, Dolan WL, Anderson NA, Chapple C (2015) Indole glucosinolate Biosynthesis Limits phenylpropanoid Accumulation in Arabidopsis thaliana. Plant Cell 27:1529–1546. https://doi.org/10.1105/tpc.15.00127
Kim JI, Zhang X, Pascuzzi PE et al (2020) Glucosinolate and phenylpropanoid biosynthesis are linked by proteasome-dependent degradation of PAL. New Phytol 225:154–168. https://doi.org/10.1111/nph.16108
Kong D, Li S, Smolke CD (2020) Discovery of a previously unknown biosynthetic capacity of naringenin chalcone synthase by heterologous expression of a tomato gene cluster in yeast. Sci Adv. https://doi.org/10.1126/sciadv.abd1143
Li Y, Kim JI, Pysh L, Chapple C (2015) Four isoforms of arabidopsis 4-coumarate: CoA ligase Have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol 169:2409–2421. https://doi.org/10.1104/pp.15.00838
Liang XW, Dron M, Cramer CL et al (1989) Differential regulation of phenylalanine ammonia-lyase genes during plant development and by environmental cues. J Biol Chem 264:14486–14492
Liddington RC (2004) Structural basis of protein–protein interactions. Methods Mol Biol 261:3–14. https://doi.org/10.1385/1-59259-762-9:003
Liu J, Osbourn A, Ma P (2015) MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant 8:689–708. https://doi.org/10.1016/j.molp.2015.03.012
Løvdal T, Olsen KM, Slimestad R et al (2010) Synergetic effects of nitrogen depletion, temperature, and light on the content of phenolic compounds and gene expression in leaves of tomato. Phytochemistry 71:605–613. https://doi.org/10.1016/j.phytochem.2009.12.014
Mao W, Han Y, Chen Y et al (2022) Low temperature inhibits anthocyanin accumulation in strawberry fruit by activating FvMAPK3-induced phosphorylation of FvMYB10 and degradation of chalcone synthase 1. Plant Cell 34:1226–1249. https://doi.org/10.1093/plcell/koac006
Micek A, Godos J, Del Rio D et al (2021) Dietary flavonoids and cardiovascular disease: a comprehensive dose-response meta-analysis. Mol Nutr Food Res 65:e2001019. https://doi.org/10.1002/mnfr.202001019
Muhlemann JK, Younts TLB, Muday GK (2018) Flavonols control pollen tube growth and integrity by regulating ROS homeostasis during high-temperature stress. Proc Natl Acad Sci USA 115:E11188–E11197. https://doi.org/10.1073/pnas.1811492115
Muro-Villanueva F, Mao X, Chapple C (2019) Linking phenylpropanoid metabolism, lignin deposition, and plant growth inhibition. Curr Opin Biotechnol 56:202–208. https://doi.org/10.1016/j.copbio.2018.12.008
Nakabayashi K, Bartsch M, Ding J, Soppe WJJ (2015) Seed dormancy in arabidopsis requires self-binding ability of DOG1 protein and the presence of multiple isoforms generated by alternative splicing. PLoS Genet 11:e1005737. https://doi.org/10.1371/journal.pgen.1005737
Novaes E, Kirst M, Chiang V et al (2010) Lignin and biomass: a negative correlation for wood formation and lignin content in trees. Plant Physiol 154:555–561. https://doi.org/10.1104/pp.110.161281
Ohno S, Hori W, Hosokawa M et al (2018) Post-transcriptional silencing of chalcone synthase is involved in phenotypic lability in petals and leaves of bicolor dahlia (Dahlia variabilis) “Yuino.” Planta 247:413–428. https://doi.org/10.1007/s00425-017-2796-3
Pawlak-Sprada S, Arasimowicz-Jelonek M, Podgórska M, Deckert J (2011) Activation of phenylpropanoid pathway in legume plants exposed to heavy metals. Part I. Effects of cadmium and lead on phenylalanine ammonia-lyase gene expression, enzyme activity and lignin content. Acta Biochim Pol 58:211–216
Perez VC, Dai R, Bai B et al (2021) Aldoximes are precursors of auxins in arabidopsis and maize. New Phytol 231:1449–1461. https://doi.org/10.1111/nph.17447
Potter SC, Luciani A, Eddy SR et al (2018) HMMER web server: 2018 update. Nucleic Acids Res 46:W200–W204. https://doi.org/10.1093/nar/gky448
Prasanna P, Upadhyay A (2021) Flavonoid-based nanomedicines in alzheimer’s disease therapeutics: promises made, a long way to go. ACS Pharmacol Transl Sci 4:74–95. https://doi.org/10.1021/acsptsci.0c00224
Rohde A, Morreel K, Ralph J et al (2004) Molecular phenotyping of the pal1 and pal2 mutants of arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16:2749–2771. https://doi.org/10.1105/tpc.104.023705
Rosa-Martínez E, Bovy A, Plazas M et al (2023) Genetics and breeding of phenolic content in tomato, eggplant and pepper fruits. Front Plant Sci 14:1135237. https://doi.org/10.3389/fpls.2023.1135237
Ruegger M, Chapple C (2001) Mutations that reduce sinapoylmalate accumulation in arabidopsis thaliana define loci with diverse roles in phenylpropanoid metabolism. Genetics 159:1741–1749. https://doi.org/10.1093/genetics/159.4.1741
Ruprecht C, Proost S, Hernandez-Coronado M et al (2017) Phylogenomic analysis of gene co-expression networks reveals the evolution of functional modules. Plant J 90:447–465. https://doi.org/10.1111/tpj.13502
Saito K, Yonekura-Sakakibara K, Nakabayashi R et al (2013) The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol Biochem 72:21–34. https://doi.org/10.1016/j.plaphy.2013.02.001
Schijlen EGWM, de Vos CHR, Martens S et al (2007) RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol 144:1520–1530. https://doi.org/10.1104/pp.107.100305
Schmelzer E, Jahnen W, Hahlbrock K (1988) In situ localization of light-induced chalcone synthase mRNA, chalcone synthase, and flavonoid end products in epidermal cells of parsley leaves. Proc Natl Acad Sci USA 85:2989–2993. https://doi.org/10.1073/pnas.85.9.2989
Schumann N, Navarro-Quezada A, Ullrich K et al (2011) Molecular evolution and selection patterns of plant F-box proteins with C-terminal kelch repeats. Plant Physiol 155:835–850. https://doi.org/10.1104/pp.110.166579
Shao T, Qian Q, Tang D et al (2012) A novel gene IBF1 is required for the inhibition of brown pigment deposition in rice hull furrows. Theor Appl Genet 125:381–390. https://doi.org/10.1007/s00122-012-1840-8
Shin DH, Cho M, Choi MG et al (2015) Identification of genes that may regulate the expression of the transcription factor production of anthocyanin pigment 1 (PAP1)/MYB75 involved in Arabidopsis anthocyanin biosynthesis. Plant Cell Rep 34:805–815. https://doi.org/10.1007/s00299-015-1743-7
Shin D, Perez VC, Dickinson GK et al (2023) Altered methionine metabolism impacts phenylpropanoid production and plant development in Arabidopsis thaliana. BioRxiv. https://doi.org/10.1101/2023.05.29.542770
Shirley BW, Kubasek WL, Storz G et al (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8:659–671. https://doi.org/10.1046/j.1365-313x.1995.08050659.x
Shomali A, Das S, Arif N et al (2022) Diverse physiological roles of flavonoids in plant environmental stress responses and tolerance. Plants. https://doi.org/10.3390/plants11223158
Slika H, Mansour H, Wehbe N et al (2022) Therapeutic potential of flavonoids in cancer: ROS-mediated mechanisms. Biomed Pharmacother 146:112442. https://doi.org/10.1016/j.biopha.2021.112442
Soubeyrand E, Johnson TS, Latimer S et al (2018) The peroxidative cleavage of kaempferol contributes to the biosynthesis of the benzenoid moiety of ubiquinone in plants. Plant Cell 30:2910–2921. https://doi.org/10.1105/tpc.18.00688
Soubeyrand E, Latimer S, Bernert AC et al (2021) 3-O-glycosylation of kaempferol restricts the supply of the benzenoid precursor of ubiquinone (coenzyme Q) in Arabidopsis thaliana. Phytochemistry 186:112738. https://doi.org/10.1016/j.phytochem.2021.112738
Steiner E, Triana MR, Kubasi S et al (2021) KISS ME DEADLY F-box proteins modulate cytokinin responses by targeting the transcription factor TCP14 for degradation. Plant Physiol 185:1495–1499. https://doi.org/10.1093/plphys/kiab033
Struk S, Jacobs A, Sánchez Martín-Fontecha E et al (2019) Exploring the protein-protein interaction landscape in plants. Plant Cell Environ 42:387–409. https://doi.org/10.1111/pce.13433
Sun C, Deng L, Du M et al (2020) A transcriptional network promotes anthocyanin biosynthesis in tomato flesh. Mol Plant 13:42–58. https://doi.org/10.1016/j.molp.2019.10.010
Tan H, Man C, Xie Y et al (2019) A crucial role of GA-regulated flavonol biosynthesis in root growth of Arabidopsis. Mol Plant 12:521–537. https://doi.org/10.1016/j.molp.2018.12.021
Teale WD, Pasternak T, Dal Bosco C et al (2021) Flavonol-mediated stabilization of PIN efflux complexes regulates polar auxin transport. EMBO J 40:e104416
Tohge T, de Souza LP, Fernie AR (2017) Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J Exp Bot 68:4013–4028. https://doi.org/10.1093/jxb/erx177
Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635–641. https://doi.org/10.1038/nature11119
Tu S-H, Chen L-C, Ho Y-S (2017) An apple a day to prevent cancer formation: reducing cancer risk with flavonoids. J Food Drug Anal 25:119–124. https://doi.org/10.1016/j.jfda.2016.10.016
Verweij W, Spelt CE, Bliek M et al (2016) Functionally similar WRKY proteins regulate vacuolar acidification in petunia and hair development in arabidopsis. Plant Cell 28:786–803. https://doi.org/10.1105/tpc.15.00608
Vogt T (2010) Phenylpropanoid biosynthesis. Mol Plant 3:2–20. https://doi.org/10.1093/mp/ssp106
Wang Y, Liu W, Wang X et al (2020) MiR156 regulates anthocyanin biosynthesis through SPL targets and other microRNAs in poplar. Hortic Res 7:118. https://doi.org/10.1038/s41438-020-00341-w
Wedick NM, Pan A, Cassidy A et al (2012) Dietary flavonoid intakes and risk of type 2 diabetes in US men and women. Am J Clin Nutr 95:925–933. https://doi.org/10.3945/ajcn.111.028894
Wen W, Alseekh S, Fernie AR (2020) Conservation and diversification of flavonoid metabolism in the plant kingdom. Curr Opin Plant Biol 55:100–108. https://doi.org/10.1016/j.pbi.2020.04.004
Wen K, Fang X, Yang J et al (2021) Recent research on flavonoids and their biomedical applications. Curr Med Chem 28:1042–1066. https://doi.org/10.2174/0929867327666200713184138
Xian D, Guo M, Xu J et al (2021) Current evidence to support the therapeutic potential of flavonoids in oxidative stress-related dermatoses. Redox Rep 26:134–146. https://doi.org/10.1080/13510002.2021.1962094
Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20:176–185. https://doi.org/10.1016/j.tplants.2014.12.001
Yin R, Messner B, Faus-Kessler T et al (2012) Feedback inhibition of the general phenylpropanoid and flavonol biosynthetic pathways upon a compromised flavonol-3-O-glycosylation. J Exp Bot 63:2465–2478. https://doi.org/10.1093/jxb/err416
Yin R, Han K, Heller W et al (2014) Kaempferol 3-O-rhamnoside-7-O-rhamnoside is an endogenous flavonol inhibitor of polar auxin transport in Arabidopsis shoots. New Phytol 201:466–475. https://doi.org/10.1111/nph.12558
Zhang X, Liu C-J (2015) Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol Plant 8:17–27. https://doi.org/10.1016/j.molp.2014.11.001
Zhang X, Henriques R, Lin S-S et al (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1:641–646. https://doi.org/10.1038/nprot.2006.97
Zhang X, Gou M, Liu C-J (2013) Arabidopsis kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 25:4994–5010. https://doi.org/10.1105/tpc.113.119644
Zhang X, Gou M, Guo C et al (2015a) Down-regulation of kelch domain-containing F-box protein in Arabidopsis enhances the production of (poly)phenols and tolerance to ultraviolet radiation. Plant Physiol 167:337–350. https://doi.org/10.1104/pp.114.249136
Zhang Y, Butelli E, Alseekh S et al (2015b) Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat Commun 6:8635. https://doi.org/10.1038/ncomms9635
Zhang X, Abrahan C, Colquhoun TA, Liu C-J (2017) A proteolytic regulator controlling chalcone synthase stability and flavonoid biosynthesis in arabidopsis. Plant Cell 29:1157–1174. https://doi.org/10.1105/tpc.16.00855
Zhang D, Song YH, Dai R et al (2020) Aldoxime metabolism is linked to phenylpropanoid production in Camelina sativa. Front Plant Sci 11:17. https://doi.org/10.3389/fpls.2020.00017
Zhao T, Huang C, Li S et al (2023) VviKFB07 F-box E3 ubiquitin ligase promotes stilbene accumulation by ubiquitinating and degrading VviCHSs protein in grape. Plant Sci 331:111687. https://doi.org/10.1016/j.plantsci.2023.111687
Funding
This work was supported by the United States Department of Agriculture (USDA)-National Institute of Food and Agriculture Hatch project 7004334 and National Science Foundation Division of Integrative Organismal Systems -CAREER-2142898.
Author information
Authors and Affiliations
Contributions
D.S. and J.K. designed the experiments and wrote the manuscript. D.S., K.H.C., and E.T. conducted the experiments. C.Y.Y. provided the Y2H systems and guided Y2H experiments. All authors read and agreed with the manuscript.
Corresponding author
Ethics declarations
Competing interests
There is no competing interest declared.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shin, D., Cho, K.H., Tucker, E. et al. Identification of tomato F-box proteins functioning in phenylpropanoid metabolism. Plant Mol Biol 114, 85 (2024). https://doi.org/10.1007/s11103-024-01483-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11103-024-01483-4