Abstract
Alternaria leaf blight (ALB), caused by a necrotrophic fungus Alternaria brassicae is a serious disease of oleiferous Brassicas resulting in significant yield losses worldwide. No robust resistance against A. brassicae has been identified in the Brassicas. Natural accessions of Arabidopsis show a spectrum of responses to A. brassicae ranging from high susceptibility to complete resistance. To understand the molecular mechanisms of resistance/ susceptibility, we analysed the comparative changes in the transcriptome profile of Arabidopsis accessions with contrasting responses- at different time points post-infection. Differential gene expression, GO enrichment, pathway enrichment, and weighted gene co-expression network analysis (WGCNA) revealed reprogramming of phenylpropanoid biosynthetic pathway involving lignin, hydroxycinnamic acids, scopoletin, anthocyanin genes to be highly associated with resistance against A. brassicae. T-DNA insertion mutants deficient in the biosynthesis of coumarin scopoletin exhibited enhanced susceptibility to A. brassicae. The supplementation of scopoletin to medium or exogenous application resulted in a significant reduction in the A. brassicae growth. Our study provides new insights into the transcriptome dynamics in A. brassicae-challenged Arabidopsis and demonstrates the involvement of coumarins in plant immunity against the Brassica pathogen A. brassicae.
Key message
Alternaria brassicae responsive comparative transcriptomics and analysis of mutants deficient in phenylpropanoid pathway provide evidence for its involvement in resistance against the necrotrophic fungus A. brassicae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alternaria brassicae, a necrotrophic pathogen causes Alternaria leaf blight, a highly destructive disease of Brassicaceae crops. In India, where Brassica juncea is a major oilseed crop, A. brassicae infection on aerial plant parts (leaf, stem, silique) results in substantial losses in seed yield and oil quality (Saharan et al. 2016). Losses are particularly pronounced in the Himalayan foothills due to the high humidity and low-temperature conditions during the crop growing season. The lack of resistance against A. brassicae in the cultivated vegetable and oilseed Brassicas has hindered efforts to develop Alternaria leaf blight-resistant varieties. While some insights into the involvement of phytohormone crosstalk, chitinases, and phytoalexins have been reported in naturally tolerant close relatives of oilseed mustard: Camelina sativa and Sinapis alba (Pedras et al. 2001; Mir et al. 2020; Yadav et al. 2020a), the detailed mechanisms are still largely unknown.
Arabidopsis thaliana accessions display extensive variation in their response to A. brassicae, ranging from immunity to high susceptibility, making it a good model species to understand the defence mechanisms that are activated during infection in different resistant accessions (Mandal et al. 2018; Rajarammohan et al. 2018). Conventional biparental mapping and Genome-Wide Association Studies (GWAS) have elucidated the complex genetic architecture governing resistance to A. brassicae in Arabidopsis. These studies have identified multiple loci associated with resistance, each loci contributes a small or moderate effect to the overall response (Rajarammohan et al. 2017, 2018). Accumulation of ROS and cell death have been implicated to play a crucial role in defense against A. brassicicola and A. brassicae in A. thaliana (Su’udi et al. 2011; Mandal et al. 2019). Transcriptional induction of several Pathogenesis Related (PR) genes mainly PR12, phytohormone signalling genes, chitinases, and MAPK signal transduction genes MAP2K9 and MAPK6 have been reported in Arabidopsis upon infection with A. brassicae (Kannan et al. 2012; Chandrashekar et al. 2015, 2018; Mandal et al. 2019). However, the mechanistic insights into the molecular basis of resistance and susceptibility, and the transcriptional regulation of defense in A. thaliana against A. brassicae, need to be explored.
Transcriptomics has been applied for global expression analysis and identification of target genes in economically important crops in response to other Alternaria species. Comparative transcriptomic studies have identified key transcription factors like WRKY1, WRKY33, and ERF-2-like for their role in resistance against Alternaria sp. (Yang et al. 2015; Shinde et al. 2018; Song et al. 2019; Tao et al. 2022). Brouwer et al. 2020; demonstrated the role of SA signalling in resistance against A. solani using a time course microarray analysis. In another study, transcriptome and transgenic analysis identified the CmNPR1, an SA signalling gene, to be involved in resistance against Alternaria sp in Chrysanthemum morifolium (Zhao et al. 2020). A microarray-based global transcriptome profiling of chickpea challenged with A. brassicae revealed processes like stomatal closure, cuticular wax accumulation, production of secondary metabolites, ROS accumulation and localized cell death to be involved in non-host mediated resistance (Fatima et al. 2019).
Comparative genome-wide expression profiles of resistant and susceptible genotypes can help to understand the plant defence mechanisms invoked at different stages of pathogen invasion and establishment. Herein, an RNA-Seq approach was used to decipher the resistance mechanisms activated against A. brassicae interaction using two resistant viz. CIBC-5 and Ei-2 and a highly susceptible Zdr-1 accessions of Arabidopsis. To the best of our knowledge, this is the first study that uses a comparative transcriptome analysis to gain a holistic understanding of plant defense mechanisms activated in Arabidopsis – A. brassicae interaction. Our results reveal the association of resistance with the reprogramming of the phenylpropanoid pathway. We further provide evidence for the involvement of a coumarin – scopoletin, in resistance against this pathogen.
Experimental procedures
Plant and fungal growth and inoculation assays
The seeds for A. thaliana accessions CIBC-5 (CS28142), Zdr-1 (CS22588), and Ei-2 (CS1124) and the T-DNA insertion mutant seeds (SALK_132418C, SALK_151507C, and SALK_151976C) were obtained from Arabidopsis Biological Resource Center (ABRC). The mutants were confirmed for their homozygosity using gene-specific primers (Suppl. Figure S9; Suppl. Table S8). The seeds were sterilized, stratified for 3–4 days at 4ºC and grown in Murashige and Skoog (MS) media for 10–15 days before transplanting in a mix of soil composed of soilrite: vermiculite: perlite (1:1:1). The plants were grown in controlled environmental chambers at 22 °C with 10-h/14-h light/dark cycle and 60 − 70% humidity.
The Alternaria brassicae J3 isolate (Mandal et al. 2018) was sub-cultured and maintained on Radish Root Sucrose Agar (RRSA) medium at 20 ± 1 °C under a 12 h light/12 h dark cycle. For infection assays, spores collected from a 15-day-old plate were re-suspended in distilled water at a concentration of 103-104 spores/ml.
For transcriptome profiling experiments, five-week-old Arabidopsis plants of each genotype (CIBC-5, Ei-2, and Zdr-1) were sprayed with spores (104 spores/ml). The infected leaf samples from at least 6–8 plants were collected at different time points and stored at -80 °C. In parallel, an equal number of plants were inoculated with distilled water and control samples (mock) were collected at corresponding time points. A total of three independent biological replicates at each time point were used for RNA extraction and subsequent transcriptome sequencing.
For assaying the disease response in mutants, eight leaves per plant were drop inoculated with 5 µl of spore suspension (104 spores/ml) at four sites per leaf and the phenotype was recorded at 7 dpi. A total of six plant per genotype were evaluated. The number of developed lesions and the size of the lesion were measured, and the Cumulative Disease Index (CDI) was calculated as described in (Rajarammohan et al. 2018). The susceptible accession Zdr-1 was used a check and its CDI was used to normalize the CDI for each plant and denoted as Normalized CDI (NCDI). The significance of differences between Col-0 and the mutants was determined from three independent biological replicates using ANOVA.
RNA extraction, library preparation, and sequencing
RNeasy mini kit (Qiagen, Germany) was used for total RNA extraction from the leaf samples (both infected and mock) of three replicates from each of the three genotypes following the manufacturer’s protocol. DNA was removed from the samples using an On-column RNase–free DNase (Qiagen, Germany). The quantity of isolated RNA was checked using a NanoDrop ND–1000 spectrophotometer. The integrity of RNA was checked by agarose gel electrophoresis followed by a quality check using the Agilent 2200 tape station. The paired-end sequencing libraries were prepared using the TruSeq Stranded Total RNA Library Prep Kit as per the manufacturer’s instructions The libraries were sequenced on a HiSeq 2000 platform (AgriGenomPvt Ltd., Kochi, India). For real-time quantitative PCR, the first-strand cDNA was synthesized from 1 µg of total RNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Quantitative PCR was performed on a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific, USA) using SYBR Select Master Mix (Thermo Fisher Scientific, USA), cDNA, and specific primers (Suppl. Table S8). The relativeexpression of genes was plotted as − ΔΔCt (Livak and Schmittgen 2001) with the UBC8 (At5g41700) and TIP41 (At4g34270) gene as the endogenous control.
Differential gene expression analysis
Differentially Expressed Genes (DEGs) in response to A. brassicae infection were determined by pairwise comparison of mock and infected expression values for each genotype and time point using DESeq2 (DESeq2 R package 1.16.1). A false discovery rate (FDR) ≤ 0.05 and an absolute value of the|log2(Fold change)|≥1.5 were used as a threshold to determine significant DEGs. The DEGs between mock and infected samples at each time point were analyzed separately for each accession. The DEGs were annotated using The Arabidopsis Information Resource (TAIR) database. Gene ontology (GO) enrichment analysis of the DEGs was performed using TAIR GO (https://www.arabidopsis.org/tools/bulk/go) and agriGO (http://bioinfo.cau.edu.cn/agriGO/analysis.php) with hypergeometric test and a significance threshold of 0.05 after Benjamini and Hochberg FDR correction. The GO terms were further filtered with REVIGO (revigo.irb.hr) to remove redundant terms. The DEGs were mapped onto pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.genome.jp/kegg/pathway.html) and KEGG enrichment analysis (p-value ≤ 0.05) was performed using DAVID Functional Annotation Bioinformatics Microarray Analysis (https://ncifcrf.gov/). Functional pathway analysis was performed by MAPMAN IMAGE ANNOTATOR 3.6.0 (https://mapman.gabipd.org/mapman) tool based on log2(fold change) values obtained from DESeq2.
Weighted gene co-expression network analysis
A weighted gene co-expression network analysis was carried out using the WGCNA package in R (Langfelder and Horvath 2008). The normalized expression counts (for 25,290 genes) obtained from DESeq2 for all 36 samples were used. An appropriate soft threshold of 12 was applied based on the scale-free topology criterion. The following parameters were used to obtain weighted co-expression clusters network = unsigned, power = 12, minimum module size = 50, minimum height for merging modules = 0.25. Pearson’s correlation coefficient was used to calculate the correlation between the module eigengene and the samples. To estimate the association of modules with the trait, modules with a correlation coefficient > 0.6 and significant p-value (< 0.05) were selected for further analysis. Cytoscape software version 3.9.1 (https://cytoscape.org) was used to visualize co-expression networks.
In vitro bioassays for antifungal activity of scopoletin against A. brassicae
The inhibitory effect of scopoletin on the growth of A. brassicae was assessed on RRSA media containing 50, 100, and 200 µg/mL of scopoletin (Sigma-Aldrich, USA) and plates with methanol (1%) served as solvent controls. A 5 mm agar plug with actively growing mycelia from a 7-day-old RRSA plate was placed in the center of the plate and the radial fungal growth was recorded every 24 h.
For spore germination assays, A. brassicae spores were suspended in water supplemented with 100 µg ml–1 scopoletin or methanol (1% solvent control). Four to five drops with 30 to 40 spores were added on a glass slide which was placed in a humid chamber and incubated at 20 °C o/n to facilitate spore germination. The germinating spores were stained with trypan blue (0.4%) and observed using a bright-field microscope (Olympus Corporation, Japan). Spore germination and hyphal length for a minimum of 90 spores were recorded using ImageJ software. The experiment was repeated thrice and the significance of differences between treatment and control was analyzed using Student’s T-test. The solvent control and scopoletin (100 µg ml–1) was amended to the spore suspension (104 spores/ml), incubated for 3–4 hours to initiate germination and susceptible Zdr-1 plants were inoculated as described above. Cumulative Disease index was calculated at 7dpi. Eight leaves per plant were drop inoculated with 5 µl of spore suspension (104 spores/ml) at four sites per leaf. A total of four plants per treatment were evaluated in four independent experiments. The significance of differences between treatment and control was analyzed using Student’s T-test.
Quantification of scopoletin
For scopoletin estimation, 8–10 leaves per plant, from a set of six plants of each genotype (CIBC-5, Zdr-1, and Col-0) were inoculated with either six drops of 5 µl of spore suspension (5 × 103 spores/ml) or distilled water (mock) and leaf samples were collected at 2 dpi and 4 dpi. The experiment was performed thrice. Plant tissues were freeze-dried (Labconco, USA), 100 mg of each sample was pulverized at a frequency of 22 Hz for 2.5 min and 1mL of 80% methanol containing 4-methylumbelliferone (10 µM) as an internal standard was added. The tubes were incubated overnight at 4 °C, centrifuged twice at 13,000× g for 12 min and the supernatant was collected. Hydrolysis of scopoletin was performed as described by (Chezem et al. 2017). The supernatants were filtered with a 0.2 μm syringe filter (MDI Pvt Ltd, India), and analysed using a Waters Acquity UPLC H Class System (Waters Corp., Milford, MA). Scopoletin separation was performed on a bridged ethylene hybrid (BEH) C18 analytical column (1.7 μm, 2.1 mm × 50 mm, Waters Corp., Milford, MA). The mobile phase consisted of 2% Ammonium formate in formic acid (solvent A) and acetonitrile (solvent B) at a constant flow rate of 0.4 ml/min. A coupled UV Photodiode array detector (PDA) collected data at 340 nm for scopoletin and 4-methylumbelliferone (MeU). The standard curve was prepared by diluting 1 mg/ml of scopoletin (Sigma Aldrich, USA) to 0.05, 0.1, 0.25, 0.5, 1, 2.5, 5 µg/ml. The standard curve and percent recovery (estimated by the peak area of the internal standard) were used for the absolute quantification of total scopoletin amounts.
Results
Transcriptome sequencing of resistant and susceptible Arabidopsis accessions infected with A. brassicae
Arabidopsis accessions CIBC-5, Ei-2, and Zdr-1 respond differentially upon A. brassicae infection with Zdr-1 being susceptible while CIBC-5 and Ei-2 showed resistant response. The normalized disease index (NDI) for these accessions was reported to be 0.03 (CIBC-5), 0.02 (Ei-2) and 1.07 (Zdr-1) (Rajarammohan et al. 2018). The A. brassicae spores germinate on the leaf surface within 6–12 h post inoculation, however, the first macroscopically observable symptom was seen two (2) days post inoculation (dpi) as water-soaked lesions in both resistant and susceptible accessions. At 4 dpi, necrotic lesions start to develop only in the susceptible accession (Zdr-1) (Figs. 1a and 6a) and by 7dpi the lesions expand and are associated with a chlorotic halo. In the resistant accessions, CIBC-5 and Ei-2, lesions were highly restricted at both 4 dpi and 7 dpi (Mandal et al. 2018; Rajarammohan et al. 2017). Based on the disease progression, 2 dpi and 4 dpi were considered the most suitable time points for exploring the mechanisms of defense regulation in Arabidopsis against A. brassicae. A comparative transcriptome profiling was conducted on mock and infected leaf samples collected from CIBC-5, Ei-2, and Zdr-1 at early (2dpi) and late (4dpi) stages in three independent biological replicates (Fig. 1b). RNA Sequencing of 36 samples (3 genotypes × 2 time points × 2 treatments × 3 biological replicates) using the Illumina platform resulted in a total of > 2.4 billion reads, out of which > 93% of the reads could be mapped unambiguously to the reference genome (TAIR 10.1). A summary of the read counts and mapping is given in Suppl. Table S1.
Overview of RNA sequencing data: (a) Symptom development and contrasting responses of CIBC-5 and Ei-2 (resistant) and Zdr-1 (susceptible) at 2 day and 4 day post inoculation with A. brassicae. The red circles represent the formation of water-soaked lesions at 2dpi and the development of prominent necrotic lesions at 4dpi (b) Schematic representation of experimental design and sample collection for RNA sequencing analyses of A. thaliana accessions in response to A. brassicae. The experimental design consisted of three important traits, Genotype (resistant or susceptible), condition (mock or infected) and time (2dpi and 4dpi). Three independent experiments were conducted for RNA-Seq analysis (c) Principal component analysis of transcriptome expression of 36 samples: The PCA plot for RNA-Seq data shows the clustering of transcriptome data by condition (mock vs. infected) along PC1 (X-axis) and genotype (CIBC-5, Ei-2 and Zdr-1) along PC2 (Y- axis)
Principal Component analysis (PCA) was carried out to understand the variation among the samples and the relative importance of each trait (Genotype, treatment, and time). The analysis showed that the mock and infected samples were grouped separately. The variation in gene expression was primarily influenced by pathogen infection (PC1 56%). In addition the variation in resistant accessions at different time points post infection appeared as a separate cluster as compared to Zdr-1 (PC2 16%). The data from three different biological replicates were tightly grouped for each sample indicating high reproducibility within the experiments (Fig. 1c).
Dynamics of transcriptional reprogramming post A. brassicae infection
A pairwise comparative analysis of differentially expressed genes was performed at 2 dpi and 4 dpi in the three accessions and between the resistant (CIBC-5; Ei-2) and susceptible (Zdr-1) accessions. A comparable number of DEGs were identified in CIBC-5 (6767), Ei-2 (4874), and Zdr-1 (7131). The number of DEGs increased markedly from 2 dpi to 4 dpi in all three accessions- from 2019 to 4748 in CIBC-5; 1411 to 3463 in Ei-2; 2344 to 4787 in Zdr-1 indicating a major transcriptional reprogramming at 4dpi. A comparative analysis between CIBC-5 and Zdr-1 at both time points detected 54.3% (3688) common DEGs while Ei-2 and Zdr-1 had 44.8% (2908) common DEGs. 882 and 705 unique DEGs were identified in CIBC-5 and Ei-2 respectively, while the expression of 1183 genes was uniquely modulated in susceptible accession post infection (Fig. 2).
UpSet plot depicting the intersections of differentially expressed genes within resistant and susceptible accession. The horizontal bars on the left represent the total number of DEGs identified in each pairwise comparison (Infected vs mock) within an accession (CIBC-5, Ei-2 and Zdr-1) for different time points (2-, 4 dpi). DEGs shared between groups are represented by connected black dots, while the unconnected black dots represent unique genes. Vertical bars indicate the number of genes within each intersection. Grey points depict no intersections
Gene Ontology (GO) enrichment, KEGG mapping, and MapMan analysis were performed on both up and downregulated gene sets to classify DEGs into different functional categories. The up-regulated DEGs were significantly enriched in biological processes and pathways involved in “response to stress”, “response to chitin”, “defense response”, “secondary metabolic process” “glutathione” and “phenylpropanoid pathway”. The down-regulated gene sets showed an over-representation of biological processes and pathways related to “photosynthesis”, “phytohormone signalling” “cell wall biogenesis and organization” and “biosynthesis of secondary metabolites” (Suppl. Figures S1 & S2; Suppl. Table S2). MapMan analysis confirmed the up-regulation of DEGs involved in secondary metabolism, amino acids, and lipid metabolism category while the down-regulated DEGs were involved in photosynthesis and cell wall-related functions in all the accessions (Suppl. Figures S3, S4, and S5).
Co-expression analysis reveals association of phenylpropanoid pathway with resistance
A Weighted Gene Co-expression Network Analysis (WGCNA) was performed to identify gene transcriptional networks correlated with stages of infection within each accession. To ensure a scale-free network, the soft threshold power of 12 (β = 12) was selected to construct co-expression networks (Suppl. Figure S6a). The modules with similar expression were merged by the dynamic tree cut method resulting in 25 modules comprising 127 genes (M22) to 4877 genes (M2) (Fig. 3a; Suppl. Figure S6b; Suppl. Table S3). Based on Module Eigengene- Trait relationships, we identified modules that had a significant (p < 0.05) positive (r2 > 0.6) or negative (r2<-0.6) correlation with the phenotype. At 4dpi, M2 (r = 0.62) showed the highest correlation with CIBC-5, M4 (r = 0.68) and M5 (r = 0.93) with Ei-2 while M8 (r = 0.78) and M9 (r = 0.9) were significantly correlated with Zdr-1 (Fig. 3a). GO terms like secondary metabolic processes, defense response to fungus, and response to oxidative stress were over-represented in all the modules correlated with both the resistant and susceptible accessions while GO terms related to signal transduction, glutathione metabolism, and toxin metabolic process were specific to modules associated with resistant accessions (Fig. 3b; Suppl. Table S4).
Weighted gene co-expression network analysis (a) Heatmap depicting Module-trait relationships calculated for each trait. The traits of interest (x-axis) were correlated to each module (y-axis). The numbers in each cell represent the corresponding correlation value and the p-value is represented in brackets. The color in the heatmap reflects the degree of correlation of the module with respective samples. (b) Dot plots depicting significant GO Biological Process enriched in selected modules M2, M5, M4, and M9 from both resistant and susceptible accession at 4 dpi (c) Co-expression network of phenylpropanoid related genes in the M2 module. The network depicts edge interactions of weight > 0.25. The genes involved in scopoletin biosynthesis and regulation are marked as red circles. The genotypes and the time points are represented as; C2W: CIBC-5 mock 2 day; C2: CIBC-5 infected 2 day; C4W: CIBC-5 mock 4 day; C4: CIBC-5 infected 4 day; Z2W: Zdr-1 mock 2 day; Z2: Zdr-1 infected 2 day; Z4W: Zdr-1 mock 4 day; Z4: Zdr-1 infected 4 day; E2W: Ei-2 mock 2 day; E2: Ei-2 infected 2 day; E4W: Ei-2 mock 4 day; E4: Ei-2 infected 4 day
The M2 module with 4778 genes, specifically associated with CIBC-5, was examined further (Fig. 3a). KEGG pathway enrichment analysis revealed DEGs related to thephenylpropanoid biosynthesis pathway to be highly enriched (Fig. 3c; Suppl. Table S4). The DEGs with high connectivity of Kwithin >500 and gene significance > 0.4 were selected as candidate hub genes which included genes like At3g50280, At3g13610, At1g33030, At4g34230, At4g34050, At1g26380, At1g72680, At2g30490, At2g37040, At4g08770, At4g08780 from the phenylpropanoid pathway (Fig. 3c; Suppl. Table S5). Interestingly, the genes involved in scopoletin biosynthesis namely, At3g13610 encoding FERULOYL-CoA 6-HYDROXYLASE1 (F6′H1) and At4g34050 encoding CAFFEOYL COENZYME A O-METHYLTRANSFERASE 1 (CCoAOMT1) were among the top candidate hub genes. Additionally, transcription factor MYB15 [GS = 0.6] known to regulate the expression of F6’H1 and other phenylpropanoid-related genes was also found in the same module (Suppl. Table S5). Based on these results, we posited the importance of this branch of the phenylpropanoid pathway in resistance against A. brassicae.
Scopoletin deficient mutants exhibit increased susceptibility to A. brassicae
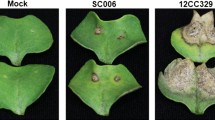
The role of phenylpropanoid compounds in defense against a large number of pathogenic infections is well documented (Yadav et al. 2020b). The expression of the phenylpropanoid pathway genes, specifically the scopoletin biosynthesis gene F6′H1 was highly enhanced at 4dpi in CIBC-5 as compared to Zdr-1. Similarly, MYB15 was induced in CIBC-5 as compared to Zdr-1 (Fig. 4a; Suppl. Figure S7a; Suppl. Figure S8). To evaluate if defense-induced expression of multiple genes of this pathway contributes towards resistance against A. brassicae, we performed infection assays on previously characterized T-DNA mutants f6’h1-1(SALK_132418C), ccoaomt1 (SALK_151507C), and myb15-1 (SALK_151976C) (Kai et al. 2008; Chezem et al. 2017). Since the mutants are available in the Col-0 background which is also resistant to A. brassicae (NDI-0.13 Rajammohan et al. 2018) (Fig. 6a), we estimated the levels of gene expression post- infection in Col-0. Consistent with our transcriptome analysis the level of transcripts of F6’H1, CCoAOMT1, and MYB15 in Col-0 increased post infection (4dpi) (Suppl. Figure S7b). All the mutant lines challenged with A. brassicae spores developed larger necrotic lesions as compared to Col-0 (Fig. 4b). A normalized cumulative disease index (NCDI) for mutants and Col-0 was calculated by considering the number and size of necrotic lesions developed at 7 dpi. The mutants showed a higher disease index of 0.69 ± 0.09 for f6’h1-1, 0.54 ± 0.08 for ccoaomt1, and 0.68 ± 0.08 for myb15-1 as compared to 0.24 ± 0.08 for Col-0 (Fig. 4b; Suppl. Table S6). Based on these results we speculated the role of the scopoletin biosynthesis pathway in imparting resistance against A. brassicae.
Scopoletin contributes to resistance against A. brassicae (a) Schematic representation of the differential expression of genes related to the scopoletin biosynthesis pathway. The genes are labeled as CCoAOMT1 (Caffeoyl-CoA-O methyltransferase), F6′H1 (Feruloyl CoA 6′-hydroxylase 1), MYB15 (Transcription Factor). Expression of genes in CIBC-5, Ei-2, and Zdr-1 at 2dpi and 4dpi is represented as heat map generated using log2FC. The colors specify the levels of expression (0–7 log scale) in each pairwise comparison between mock and infected samples while the grey-colored boxes indicate no differential expression. C2 represents CIBC-5 2dpi infected vs. mock; E2: Ei-2 2dpi infected vs. mock, Z2: Zdr-1 2dpi infected vs. mock, C4: CIBC-5 4dpi infected vs. mock; E4: Ei-2 4dpi infected vs. mock, Z4: Zdr-1 4dpi infected vs. mock. (b) Representative images of infected leaves of mutants- f6’h1-1, ccoaomt1, myb15-1 and wild-type (Col-0) at 7 dpi. The boxplot shows the mean normalized cumulative disease index (NCDI) of wild-type (Col-0) and mutants from three biological replicates. To calculate NCDI of each line, the disease index of mutants and Col-0 were normalized with respect to Zdr-1 (DI = 1)*
Scopoletin has antifungal activity against A. brassicae
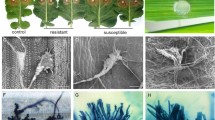
To test the effect of scopoletin on A. brassicae growth, different concentrations (50-, 100-, and 200 µg/ml) of scopoletin were added to RRSA plate, and radial fungal growth was recorded for 6 days at an interval of 24 h. A concentration-dependent reduction was observed in the fungal growth (Fig. 5a). The effect of scopoletin on spore germination and primary hyphal growth was also evaluated. The presence of scopoletin (100 µg/ml), did not affect the spore germination (Fig. 5b) however, primary hyphal length showed a marked reduction of 41% (0.056 ± 0.05 mm) as compared to the solvent control (0.09 ± 0.1 mm) (Fig. 5c & d). Co-application of scopoletin and fungal spores on a susceptible accession was effective in reducing the disease index significantly (Fig. 5e).
Inhibition of A. brassicae growth and spore germination in vitro: Graph depicting (a) the area of A. brassicae mycelium growth on RRSA supplied with 0, 50, 100, and 200 µg ml–1 scopoletin; data was collected each day till 7 days. Asterisks indicate the level of significant difference in area between solvent control and scopoletin supplemented plates (Student’s t-test: *P < 0.05) (b) percentage germination (c) primary hyphal length when spores were supplemented with 100 µg ml–1 along with solvent control. The primary hyphal length was measured by ImageJ software and a minimum of 90 spores were accessed per experiment. Asterisks indicate the level of significant difference in area between solvent control and scopoletin supplemented spores (Student’s t-test: *P < 0.01). The assays were performed in triplicates (d) Representative pictures depicting trypan blue stained spore germination and primary hyphal elongation in spores supplemented with 100µgml− 1 scopoletin and solvent control (e)A. brassicae spores were supplemented with solvent control or scopoletin (100 µg ml–1) and inoculated on leaves of Zdr1. The data represent four independent experiments. In each experiment eight leaves per plant with a minimum of four plants for each genotype were evaluated. Cumulative Disease Index (CDI), a measure of the number of lesions developed and the size of the lesion, was used to score the extent of disease severity (7dpi) which is represented as Box plots along with representative images
We estimated the total scopoletin levels in the resistant (CIBC-5 and Col-0) and susceptible accession (Zdr-1) in mock and infected samples. The total scopoletin level was observed to be ranging between 4.55 and 5.32 µM in all three accessions. Although a comparable absolute level of scopoletin was detected post infection in all the accessions, however a slight enhancement was observed in C2/C2W (2 dpi) (Fig. 6b).
In vivo levels of scopoletin in resistant and susceptible accessions: (a) Representative images showing contrasting disease responses of Arabidopsis accessions Zdr-1 (susceptible), CIBC-5 and Col-0 (resistant) after spray infection with A. brassicae spores (104spores/ml). Water soaked lesion at 2 dpi and necrotic lesions at 4dpi in Zdr-1 accession highlighted by circles (b) Graph depicting ratio of the level of scopoletin in infected samples as compared to the respective mock (infected/mock) in CIBC-5 and Col-0 and Zdr-1. The experiment was performed with three biological replicates. In each replicate, a total of six plants and 8–10 leaves from each plant were inoculated with A. brassicae spores or water (mock) and 100 mg of leaf tissues from each sample was analysed for scopoletin levels. The genotypes and the time points are represented as C2/C2W: CIBC-5 2dpi; Z2/Z2W: Zdr-1 2dpi; Col2/Col2W: Col-0 2dpi; C4/C4W: CIBC-5 4dpi; Z4/Z4W: Zdr-1 4dpi; Col4/Col4W: Col-0 4dpi
Discussion
The molecular basis of quantitative resistance to the necrotrophic pathogen A. brassicae is less understood due to the complexity of responses it triggers in the host. The availability of Arabidopsis accessions with contrasting responses to A. brassicae infection was leveraged to examine the resistance mechanisms activated in the model host. Here we examined the global transcriptional status of resistant (CIBC-5 and Ei-2) and susceptible (Zdr-1) Arabidopsis accessions during pathogen infection. In total, we identified 5774 and 12,998 DEGs at the early and late stages of infection. Following the appearance of differential symptoms at 4dpi, a larger number of DEGs (12,998) were detected at a later stage of infection.
As observed earlier fungus starts to penetrate through the stomata as early as 6–12 hpi infection (Mandal et al. 2018). We observed distinct upregulation of several Receptor-Like Kinases (RLKs) including Receptor Like Proteins (RLPs), Cysteine Rich Kinases (CRKs), and Wall Associated Kinases (WAKs) early during infection. RLPs (20, 35, 41) and CRKs (7, 32, 36) were highly expressed in resistant accessions and might be involved in PAMPS/ effectors/ toxin recognition. Several of these receptors like CRK36, RLP23, and RLP30 have been implicated in defense against necrotrophic fungal pathogens namely Alternaria brassicicola, Sclerotinia sclerotiorum, and Botrytis cinerea respectively (Lee et al. 2017; Ono et al. 2020; Zhang et al. 2013). GO enrichment analysis of the DEGs highlighted the downregulation of photosynthesis genes in the susceptible accession early during infection. Similar chloroplast degradation and associated downregulation in the photosynthetic processes were also reported in a compatible interaction between Brassica oleracea and A. brassicicola (Macioszek et al. 2020) and in pad3 susceptible mutant of Arabidopsis (Narusaka et al. 2003). Chloroplasts are a major source of ROS and phytohormone production that are targeted by several fungal effectors (Li and Kim 2022) ROS generation and associated cell death in the host effectively limits the spread of biotrophic pathogens; on the contrary, necrotrophic pathogens have evolved strategies to hijack this host response to their benefit (Torres et al. 2006). In Alternaria - A. thaliana interaction ROS and cell death were observed to be important determinants of susceptibility (Mandal et al. 2019; Su’udi et al. 2011). To maintain the delicate balance of ROS production and scavenging, Arabidopsis has efficient antioxidant machinery (Dvořák et al. 2021). An upregulation of genes like thioredoxin, glutaredoxin, peroxiredoxin, and glutathione involved in ROS scavenging was observed in the resistant accession CIBC-5 while some components of this machinery were downregulated in Zdr-1. Previous proteome studies have also reported enhanced expression of antioxidants, especially GSTUs post A. brassicicola infection in A. thaliana (Mukherjee et al. 2010). Several Transcription Factors (TFs) belonging to WRKYs, MYBs, AP2- ERFs known to be involved in defense against necrotrophs (Birkenbihl and Somssich 2011) were also found to be upregulated in the resistant accessions CIBC-5 and Ei-2.
The gene co-expression analysis followed by GO enrichment suggested a connection between induction of the phenylpropanoid pathway and resistance against A. brassicae. Activation of the phenylpropanoid pathway and accumulation of its derivatives has been strongly correlated with resistance in various Alternaria- host interactions like A. alternata- pear, A. cassia- Cassia obstuisifolia, A. alternata f. sp.mali- apple, A. solani- tomato (Hoagland 1990; Shinde et al. 2018; Geng et al. 2020; Guo et al. 2023). Additionally, the upregulation of the MYB class of transcription regulators: MYB − 15, -75, -90, and 113, involved in lignin, scopoletin, and anthocyanin biosynthesis and various downstream genes in the resistant accessions post-infection was also observed (Suppl. Figure S7a and Suppl. Table S7). Enrichment analysis of the genes in the M2 module, positively correlating with the resistant line at 4dpi, further underscored the possible role of the scopoletin biosynthesis pathway in resistance. Some earlier reports have also implicated scopoletin in resistance against fungal pathogens A. alternata (Sun et al. 2014) and Phakopsora pachyrhizi (Beyer et al. 2019).
Scopoletin is a major secondary metabolite generally synthesized in the Arabidopsis roots (Kai et al. 2008). In Arabidopsis, scopoletin biosynthesis pathway involves the conversion of caffeic acid to ferulic acid by CAFFEOYL COENZYME A O-METHYLTRANSFERASE 1 (CCoAOMT1). The further conversion of ferulic acid to scopoletin is regulated by FERULOYL-CoA 6-HYDROXYLASE1 (F6′H1), the key enzyme for scopoletin biosynthesis (Kai et al. 2008). The expression of F6′H1 and the metabolite scopoletin has been reported to be induced in leaves upon pathogen infection or treatment with 2,4-dichlorophenoxyacetic acid (Kai et al. 2006, 2008). Arabidopsis T-DNA mutants of major genes involved in scopoletin biosynthesis- CCoAOMT1 and F6’H1 result in 30% and 90% reduction respectively, in the metabolite content and exhibit enhanced susceptibility towards Pto DC3000 (Kai et al. 2008; Chezem et al. 2017). MYB15, a SG2 type R2R3- transcription factor, induced upon flg22 elicitation transcriptionally activates scopoletin production by regulating the expression of CCoAOMT1 and F6’H1. The basal level of total scopoletin in myb15 mutant is significantly reduced (Chezem et al. 2017). The enhanced susceptibility of myb 15 − 1, f6’h1-1, and ccoaomt1 mutants towards A. brassicae and improved tolerance upon topical application of scopoletin provides novel insights into the role of this coumarin in A. brassicae defense. Unlike F6′H1 and MYB15, differential expression of CCoAOMT1 between the resistant and susceptible accession was not observed however, it was identified as one of the hub genes in WGCNA.
Scopoletin has been reported to have antifungal activity against several fungal pathogens like S. sclerotiorum, Colletotrichum species, B. cinerea, A. alternata, P. pachyrhizi (Urdangarin et al. 1999; Carpinella et al. 2005; El Oirdi et al. 2010; Sun et al. 2014; Beyer et al. 2019). We also observed suppression of A. brassicae mycelial growth and primary hyphal elongation in the presence of scopoletin (100 µg/ml) in growth media however spore germination was not affected. Scopoletin might be suppressing A. brassicae infection at the penetration stage as well as during the establishment within the host. Beyer et al. 2019 also showed the inhibitory effect of scopoletin on germination and development of pre- penetration structures in P. pachyrhizi required for non-host penetration in Arabidopsis.
Despite the evidence for a correlation between scopoletin biosynthesis pathway and resistance against A. brassicae, the absolute levels of scopoletin post infection in the resistant and susceptible accessions were comparable. There might be a preferential accumulation of the phytoalexin at the site of fungal penetration which restricts the fungal spread. However, detection of such localised accumulation would require further detailed investigation. This was shown to be the case in sunflower where higher accumulation of scopoletin was observed in the medulla region of the stem in contrast to the cortex region upon challenge with a non-pathogenic fungus Helminthosporium carbonum (Tal and Robeson 1986). Chezem et al. 2017 also reported enhanced level of F6’H1 transcription post treatment with elicitor (flg22) and f6’h1 null mutants exhibited enhanced susceptibility to PstDC3000 infection. However, they did not find any enhancement in the coumarin levels post infection. The multiple branches of the phenylpropanoid pathway are interrelated and therefore the susceptibility observed in f6’h1-1 and myb15-1 could be a combinatorial effect of reduced scopoletin biosynthesis and additional perturbation of associated components.
The present work for the first time shows the role of the scopoletin branch of the phenylpropanoid pathway in defense against A. brassicae in Arabidopsis. These results overall serve as a foundation for more detailed research on the role of scopoletin and other phenylpropanoid derivatives in defense against A. brassicae. Taken together the results have unraveled a new aspect of resistance mechanism against A. brassicae in the model host Arabidopsis. Since F6’H1 is conserved across Brassicaceae, with three orthologs present in B. juncea var Varuna, functional characterization of B. juncea F6’H1 can be explored as a potential strategy for developing Alternaria leaf blight resistant varieties in the future. Given the broad-spectrum antifungal and antioxidant activity of scopoletin, it could be a promising candidate molecule to be developed as a natural fungicide.
Data availability
The sequencing data has been deposited in the NCBI sequence read archive (SRA) under Bio Project accession PRJNA902094.
References
Beyer SF, Beesley A, Rohmann PFW, Schultheiss H, Conrath U, Langenbach CJG (2019) The Arabidopsis non-host defence‐associated coumarin scopoletin protects soybean from Asian soybean rust. Plant J 99(3):397–413
Birkenbihl RP, Somssich IE (2011) Transcriptional plant responses critical for resistance towards necrotrophic pathogens. Front Plant Sci 2:76
Brouwer SM, Odilbekov F, Burra DD, Lenman M, Hedley PE, Grenville-Briggs L, Alexandersson E, Liljeroth E, Andreasson E (2020) Intact salicylic acid signalling is required for potato defence against the necrotrophic fungus Alternaria solani. Plant Mol Biol 104(1–2):1–19
Carpinella MC, Ferrayoli CG, Palacios SM (2005) Antifungal synergistic effect of scopoletin, a hydroxycoumarin isolated from Melia azedarach L. fruits. J Agric Food Chem 53(8):2922–2927
Chandrashekar N, Ali S, Rawat S, Grover A (2015) Gene expression profiling of Arabidopsis thaliana chitinase genes in response to Alternaria brassicae challenge. Indian Phytopathol 68(1):106–111
Chandrashekar N, Ali S, Grover A (2018) Exploring expression patterns of PR-1, PR-2, PR-3, and PR-12-like genes in Arabidopsis thaliana upon Alternaria brassicae inoculation. 3 Biotech 8:1–10
Chezem WR, Memon A, Li F-S, Weng J-K, Clay NK (2017) SG2-type R2R3-MYB transcription factor MYB15 controls defense-induced lignification and basal immunity in Arabidopsis. Plant Cell 29(8):1907–1926
Dvořák P, Krasylenko Y, Zeiner A, Šamaj J, Takáč T (2021) Signaling toward reactive oxygen species-scavenging enzymes in plants. Front Plant Sci 11:618835
El Oirdi M, Trapani A, Bouarab K (2010) The nature of tobacco resistance against Botrytis cinerea depends on the infection structures of the pathogen. Environ Microbiol 12(1):239–253
Fatima U, Bhorali P, Senthil-Kumar M (2019) Morpho-pathological and global transcriptomic analysis reveals the robust nonhost resistance responses in chickpea interaction with Alternaria brassicae. Mol Plant Microbe Interact. 32(12):1598–1613. https://doi.org/10.1094/MPMI-05-19-0117-R
Geng D, Shen X, Xie Y, Yang Y, Bian R, Gao Y, Li P, Sun L, Feng H, Ma F (2020) Regulation of phenylpropanoid biosynthesis by MdMYB88 and MdMYB124 contributes to pathogen and drought resistance in apple. Horticulture Research, 7
Guo Y, Li X, Li C, Jinyue R, Xu H, Ge Y (2023) Acibenzolar-S‐methyl activates phenylpropanoid pathway to enhance resistance against Alternaria alternata in pear fruit. J Sci Food Agric 103(2):829–836
Hoagland RE (1990) Alternaria cassiae alters phenylpropanoid metabolism in sicklepod (Cassia obtusifolia). J Phytopathol 130(3):177–187
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444(7117):323–329
Kai K, Shimizu B, Mizutani M, Watanabe K, Sakata K (2006) Accumulation of coumarins in Arabidopsis thaliana. Phytochemistry 67(4):379–386
Kai K, Mizutani M, Kawamura N, Yamamoto R, Tamai M, Yamaguchi H, Sakata K, Shimizu B (2008) Scopoletin is biosynthesized via Ortho-hydroxylation of feruloyl CoA by a 2‐oxoglutarate‐dependent dioxygenase in Arabidopsis thaliana. Plant J 55(6):989–999
Kannan P, Pandey D, Gupta AK, Punetha H, Taj G, Kumar A (2012) Expression analysis of MAP2K9 and MAPK6 during pathogenesis of Alternaria blight in Arabidopsis thaliana Ecotype Columbia. Mol Biol Rep 39:4439–4444
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9(1):1–13
Lee DS, Kim YC, Kwon SJ, Ryu C-M, Park OK (2017) The Arabidopsis cysteine-rich receptor-like kinase CRK36 regulates immunity through interaction with the cytoplasmic kinase BIK1. Front Plant Sci 8:1856
Li M, Kim C (2022) Chloroplast ROS and stress signaling. Plant Communications
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25(4):402–408 https://doi.org/10.1006/meth.2001.1262
Macioszek VK, Gapińska M, Zmienko A, Sobczak M, Skoczowski A, Oliwa J, Kononowicz AK (2020) Complexity of Brassica oleracea–Alternaria brassicicola susceptible interaction reveals downregulation of photosynthesis at ultrastructural, transcriptional, and physiological levels. Cells 9(10):2329
Mandal S, Rajarammohan S, Kaur J (2018) Alternaria brassicae interactions with the model Brassicaceae member Arabidopsis thaliana closely resembles those with mustard (Brassica juncea). Physiol Mol Biology Plants 24(1):51–59
Mandal S, Rajarammohan S, Kaur J (2019) ROS accumulation and associated cell death mediates susceptibility to Alternaria brassicae in Arabidopsis accessions. Physiol Mol Plant Pathol 107:51–59
Mir ZA, Ali S, Shivaraj SM, Bhat JA, Singh A, Yadav P, Rawat S, Paplao PK, Grover A (2020) Genome-wide identification and characterization of chitinase gene family in Brassica juncea and Camelina sativa in response to Alternaria brassicae. Genomics 112(1):749–763
Mukherjee AK, Carp M-J, Zuchman R, Ziv T, Horwitz BA, Gepstein S (2010) Proteomics of the response of Arabidopsis thaliana to infection with Alternaria brassicicola. J Proteom 73(4):709–720
Narusaka Y, Narusaka M, Seki M, Ishida J, Nakashima M, Kamiya A, Enju A, Sakurai T, Satoh M, Kobayashi M (2003) The cDNA microarray analysis using an Arabidopsis pad3 mutant reveals the expression profiles and classification of genes induced by Alternaria brassicicola attack. Plant Cell Physiol 44(4):377–387
Ono E, Mise K, Takano Y (2020) RLP23 is required for Arabidopsis immunity against the grey mould pathogen Botrytis cinerea. Sci Rep 10(1):13798
Pedras MSC, Zaharia IL, Gai Y, Zhou Y, Ward DE (2001) In planta sequential hydroxylation and glycosylation of a fungal phytotoxin: avoiding cell death and overcoming the fungal invader. Proceedings of the National Academy of Sciences, 98(2), 747–752
Rajarammohan S, Kumar A, Gupta V, Pental D, Pradhan AK, Kaur J (2017) Genetic architecture of resistance to Alternaria brassicae in Arabidopsis thaliana: QTL mapping reveals two major resistance-conferring loci. Front Plant Sci 8(February):1–9. https://doi.org/10.3389/fpls.2017.00260
Rajarammohan S, Pradhan AK, Pental D, Kaur J (2018) Genome-wide association mapping in Arabidopsis identifies novel genes underlying quantitative disease resistance to Alternaria brassicae. Mol Plant Pathol 19(7):1719–1732
Saharan GS, Mehta N, Meena PD, Dayal P (2016) Alternaria diseases of crucifers: biology, ecology and disease management. Springer
Shinde BA, Dholakia BB, Hussain K, Aharoni A, Giri AP, Kamble AC (2018) WRKY1 acts as a key component in improving resistance against Alternaria solani in wild tomato, Solanum arcanum Peralta. Plant Biotechnol J 16(8):1502–1513
Song N, Ma L, Wang W, Sun H, Wang L, Baldwin IT, Wu J (2019) An ERF2-like transcription factor regulates production of the defense sesquiterpene capsidiol upon Alternaria alternata infection. J Exp Bot 70(20):5895–5908
Su’udi M, Kim MG, Park S-R, Hwang D-J, Bae S-C, Ahn I-P (2011) Arabidopsis cell death in compatible and incompatible interactions with. Alternaria brassicicola Molecules Cells 31:593–601
Sun H, Wang L, Zhang B, Ma J, Hettenhausen C, Cao G, Sun G, Wu J, Wu J (2014) Scopoletin is a phytoalexin against Alternaria alternata in wild tobacco dependent on jasmonate signalling. J Exp Bot 65(15):4305–4315
Tal B, Robeson DJ (1986) The metabolism of sunflower phytoalexins ayapin and scopoletin: plant-fungus interactions. Plant Physiol 82(1):167–172
Tao H, Miao H, Chen L, Wang M, Xia C, Zeng W, Sun B, Zhang F, Zhang S, Li C, Wang Q (2022) WRKY33-mediated indolic glucosinolate metabolic pathway confers resistance against Alternaria brassicicola in Arabidopsis and Brassica crops. J Integr Plant Biol 64(5):1007–1019
Torres MA, Jones JDG, Dangl JL (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141(2):373–378
Urdangarin C, Regente MC, Jorrin J, De La Canal L (1999) Sunflower coumarin phytoalexins inhibit the growth of the virulent pathogen Sclerotinia sclerotiorum. J Phytopathol 147(7–8):441–443
Yadav P, Mir ZA, Ali S, Papolu PK, Grover A (2020a) A combined transcriptional, biochemical and histopathological study unravels the complexity of Alternaria resistance and susceptibility in Brassica coenospecies. Fungal Biology 124(1):44–53
Yadav V, Wang Z, Wei C, Amo A, Ahmed B, Yang X, Zhang X (2020b) Phenylpropanoid pathway engineering: an emerging approach towards plant defense. Pathogens 9(4):312
Yang X, Hu H, Yu D, Sun Z, He X, Zhang J, Chen Q, Tian R, Fan J (2015) Candidate resistant genes of sand pear (Pyrus pyrifolia Nakai) to Alternaria alternata revealed by transcriptome sequencing. PLoS ONE 10(8):e0135046
Zhang W, Fraiture M, Kolb D, Löffelhardt B, Desaki Y, Boutrot FFG, Tör M, Zipfel C, Gust AA, Brunner F (2013) Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25(10):4227–4241
Zhao X, Song L, Jiang L, Zhu Y, Gao Q, Wang D, Xie J, Lv M, Liu P, Li M (2020) The integration of transcriptomic and transgenic analyses reveals the involvement of the SA response pathway in the defense of chrysanthemum against the necrotrophic fungus Alternaria sp. Horticulture research, 7
Acknowledgements
The work was financially supported by SERB (CRG/2020/002928), DBT (BT/IN/Indo-UK/CGAT/12/DP/2014–15), and University of Delhi (IoE/2021/12/FRP). Fellowship support from ICMR to SH [F.No.45/17/2020-TRM/BMS] is highly appreciated. SR is supported by DST-INSPIRE Faculty fellowship grant (IFA18-LSPA118). MA acknowledges the support of CSIR-JRF scheme Grant Number: 09/045/(1637)/2019-EMR-1. We would like to thank Dr. Naveen C. Bisht and Dr. Pawan Kumar (NIPGR) for assistance in scopoletin quantification. We are grateful for the DST-FIST/UGC-SAP-supported instrumentation facilities at the Department of Genetics, and the CIF UDSC.
Author information
Authors and Affiliations
Contributions
JK and SR contributed in planning and designing the study. SH and SR analyzed the transcriptome data. MA and MK performed the WGCNA analysis. SH carried out the disease assays and functional characterizations. SH, SR, and JK wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Accession numbers
CIBC-5 (CS28142), Zdr-1 (CS22588), Ei-2 (CS1124), SALK_132418C, SALK_151507C, and SALK_151976C.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hamsa, S., Rajarammohan, S., Aswal, M. et al. Transcriptome responses of Arabidopsis to necrotrophic fungus Alternaria brassicae reveal pathways and candidate genes associated with resistance. Plant Mol Biol 114, 68 (2024). https://doi.org/10.1007/s11103-024-01453-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11103-024-01453-w