Abstract
Anther dehiscence, one of the essential steps in pollination and double fertilization, is regulated by a complex signaling pathway encompassing hormones and environmental factors. However, key components underlying the signaling pathway that regulate anther dehiscence remain largely elusive. Here, we isolated a rice mutant anther dehiscence defected 1 (Osadd1) that exhibited defects in anther dehiscence and glume open. Map-based cloning revealed that OsADD1 encoded a GARP (Golden2, ARR-B and Psr1) transcription factor. Sequence analysis showed that a single base deletion in Osadd1 mutant resulted in pre-termination of the GARP domain. OsADD1 was constitutively expressed in various tissues, with more abundance in the panicles. The major genes associated with anther dehiscence were affected in the Osadd1 mutant, and the expression level of the cellulose synthase-like D sub-family 4 (OsCSLD4) was significantly decreased. We demonstrate that OsADD1 regulated the expression of OsCSLD4 by binding to its promoter, and affects rice anther dehiscence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa) is one of the most important food crops in the world. A successful rice anther dehiscence requires the precise timing of dehiscence, formation of secondary wall thickening, degeneration of various anther tissues, and changes in carbohydrate metabolism and movement of water out of the anther (Goldberg et al. 1993; Ma 2005; Scott et al. 2004). In this process, plant hormones and external temperature and humidity, have close connections with anther dehiscence.
Auxin regulates anther dehiscence (Cecchetti et al. 2015, 2017; Estornell et al. 2018; Ghelli et al. 2018; Salinas-Grenet et al. 2018; Song et al. 2018). In Arabidopsis, the timing of anther dehiscence is controlled by the joint action of Auxin Response Factor8 (ARF8) two splicing variants, ARF8.4 and ARF8.2 (Ghelli et al. 2018). In rice, DAO encodes 2OG-Fe(II) dioxygenase catalyze the conversion of IAA into OxIAA. This process leads to the higher level of auxin in the dao mutant plants which exhibited anther indehiscence (Zhao et al. 2013). FT-Interacting Protein 7 (OsFTIP7) mediates the nucleocytoplasmic distribution of OSH1 that directly suppresses auxin biosynthetic gene OsYUCCA4, during the late development of anthers. Mutant of OsFTIP7 results in anther indehiscence due to the auxin regulating function of OsFTIP7 in rice anther (Song et al. 2018). In addition, jasmonic acid and gibberellin also affect plant anther dehiscence (Jibran et al. 2017; Kanno et al. 2016; Saito et al. 2015). Environmental stress can also affect anther dehiscence. Low temperature causes anther indehiscence, decreases spikelet and pollen fertility in four cold-sensitive cultivars (Zeng et al. 2017). In the flowering stage of rice, heat stress affects a series of flowering processes, including anther dehiscence, pollination, pollen germination and pollen tube growth (Prasad et al. 2006). An episode of high temperature (39 °C or above) treatment 1 day before flowering leads to rice anther indehiscence (Matsui and Omasa 2002). Despite of the reported connections between plant hormone/thermal stress and anther dehiscence, the molecular mechanisms underlying anther dehiscence remain unknown.
GARPs (Golden2, ARR-B and Psr1) are transcription factors involved in many biological processes, such as hormonal signaling, nutrient response, sensing processes, chloroplast biogenesis, plant development, clock oscillation, pathogens resistance, and so on (Safi et al. 2017). B-motif is the signature motif in GARP transcription factors, which somewhat resembles the MYB-like domain of MYB-related proteins (Sakai et al. 1998). Latest discoveries indicated that GARP transcription factors contain only one of the three regularly spaced tryptophan residues highly conserved in the MYB domain. In contrast to MYB-related proteins characterized by the Ser-His-Ala-Gln-Lys-Tyr/Phe-Phe (SHAQK(Y/F)F) amino acid sequence, GARP transcription factors contain a different consensus amino acid sequence Ser-His-Leu-Gln-Lys/Met-Tyr/Phe (SHLQ (K/M) (Y/F)) which is highly conserved among many species (Hosoda 2002; Riechmann et al. 2000; Safi et al. 2017).
MYB transcription factors are reported to be associated with anther dehiscence. MYB26 is a key MYB transcription factor that regulates anther development in Arabidopsis (Steiner-Lange et al. 2003; Yang et al. 2007, 2017). The myb26 is a mutant of male sterility due to non-dehiscent anthers. The secondary thickening in the anther was controlled by the precise localization of the MYB26 protein to the endothecium cell layer, and directly promoted the expression of NST1 and NST2 (Yang et al. 2017). In rice, Anther Indehiscence1 (AID1), encoding a single MYB DNA-binding domain protein, causes anther indehiscence (Zhu et al. 2004). Considering the similar domain in GARP and MYB, and the hormone regulation function of the GARP transcription factors, we speculate that GARP transcription factors participant in anther dehiscence process. The downstream genes regulated by GARP transcription factors remain unclear.
Strong connection was shown between cellulose synthase and cell wall structure of different organs (Hu et al. 2010; Li et al. 2009; Luan et al. 2011; Wu et al. 2010; Yoshikawa et al. 2013). OsCSLD4/NRL1/SLE1 is a cellulose synthase of the D-subfamily of Cellulose Synthase-like (CSL) proteins of glycosyltransferase family 2 (GT2). Oscsld4 exhibited rolled leaf, low plant height and seed fertility (Hu et al. 2010; Li et al. 2009; Wu et al. 2010; Yoshikawa et al. 2013). Particularly, Yoshikawa et al. (2013) found that only a few dehisced anthers and a small number of pollen grains were observed on the stigmas in Oscsld4. These studies revealed a possible link between CSL and rice anther dehiscence process.
In this study, a mutant, namely anther dehiscence defected 1 (Osadd1), was identified with anther dehiscence defected. Map-based cloning revealed that OsADD1 encodes a GARP transcription factor that is essential for regulating anther dehiscence. Our results demonstrated that OsADD1 interacts with OsCSLD4, and established the link between GARP transcription factors and anther dehiscence process.
Materials and methods
Plant materials
The Osadd1-1 was isolated from an ethyl methane sulfonate (EMS) mutant pool of indica cultivar 9311. The Osadd1-2 was isolated from an EMS mutant pool of japonica cultivar Ningjing4. The Oscsld4 was provided by Professor Wenzhen Liu (China National Rice Research Institute, Hangzhou, China). To map the OsADD1 locus, we constructed an F2 population derived from a cross of the Osadd1-1 mutant and N22. All plants were grown during the natural growing season at Nanjing Agricultural University, Nanjing, China.
Paraffin sections
The fresh spikelets were immersed directly in Carnoy’s fixative solution containing ethanol, chloroform, and acetic acid (6:3:1). After dehydrated through an ethanol series, xylene was used to induce transparency. Routine paraffin section procedures were used to embed the spikelets, which were cut into serial sections of 10 μm in thickness. The spikelets were dyed and mounted by toluidine blue and neutral balsam, respectively. Photographs and records were collected using Olympus BX43 microscope (Olympus, Japan).
Semi-thin cross sections
For anther semi-thin cross sections, the fresh spikelets were collected and fixed in FAA solution that contained a 3.7% formaldehyde/acetic acid (v/v). After dehydrated through an ethanol series, samples were placed in a Spurr’s resin and then ultrathin sectioned into 1 μm. Furthermore, sections were double-stained with 2% (w/v) uranyl acetate and 2.6% (w/v) lead citrate aqueous solution, and examined with Olympus BX43 microscope (Olympus, Japan).
Scanning electron microscopy (SEM)
The fresh spikelets at stage 13 were collected and fixed in 2.5% glutaraldehyde in a phosphate buffer at 4 °C for 4 h, then washed and incubated in 1% OsO4 at 4 °C for 12 h. After dehydration through an ethanol series, samples were embedded in Spurr’s resin prior to ultrathin sectioning. SEM was performed as described previously (Kang et al. 2005). Samples were examined with a Hitachi S-3400N scanning electron microscope.
Map-based cloning
Genetic analysis was performed using an F2 population (Osadd1-1/N22); 200 plants with the anther dehiscence defected phenotype were used for genetic mapping. Insert–delete (InDel) markers were developed based on the Nipponbare (japonica) and 9311 (indica) genome sequences (http://www.gramene.org/). Primers used for mapping are listed in Supplementary Table S1.
Sequence analysis
Gene prediction and structure analysis were performed using the GRAMENE database (http://www.gramene.org/). Homologous sequences of OsADD1 were identified using the protein BLAST search program of the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/). Target sequence alignments were conducted using ClustalX and the phylogenetic analysis was performed with MEGA 5.0 software by Neighbor-Joining method with 1000 bootstrap replicates.
Genetic complementation
For complementation of the Osadd1, a 1134 bp coding sequence fragment of OsADD1 was amplified from 9311 and cloned into the binary vector pCAMBIA1390 harbouring 35S promoter to generate the vector pCAMBIA1390-OsADD1. This vector was introduced into Agrobacterium tumefaciens strain EHA105, which was then used to infect Osadd1-1 calli as described previously (Hiei et al. 1994). Primers used for genetic complementation are listed in Supplementary Table S1.
Quantitative RT-PCR (qRT-PCR) analysis
Total RNA was isolated from different rice tissues using the RNA prep pure plant kit, and treated with DNase (Tiangen, Beijing). First-strand cDNA was synthesized using oligo(dT)18 primer and 1 µg of RNA, and then using PrimeScript Reverse Transcriptase (Takara, Dalian, China) for reverse transcription. qRT-PCR was performed in three biological repeats using an ABI 7500 real-time PCR system with SYBR Green Mix. Eight genes are selected for their mutants exhibited similar phenotype comparing with Osadd1 (OsCSLD4 Hu et al. 2010; Li et al. 2009; Wu et al. 2010; Yoshikawa et al. 2013, ROC5 Zou et al. 2011, OsZHD1 Xu et al. 2014, ACL2 Li et al. 2010, YABBY1 Dai et al. 2007, LC2 Zhao et al. 2010, NAL7 Fujino et al. 2008, SRL1 Li et al. 2017; Xiang et al. 2012). The rice ubiquitin gene was used as the normalizer control. Primers used for qRT-PCR are listed in Supplementary Table S1.
Subcellular localization
The OsADD1 was fused with green fluorescent protein (GFP) and inserted in the pAN580-GFP vector or pCAMBIA1305.1-GFP vector between the cauliflower mosaic virus (CaMV) 35S promoter and the nopaline synthase (NOS) terminator. The 35S-OsADD1-GFP plasmids were transiently expressed in rice protoplasts or tobacco (Nicotiana benthamiana) epidermal cells. Primers used for subcellular localization are listed in Supplementary Table S1.
Dual-luciferase assay
Full-length CDS of OsADD1 was cloned into pCAMBIA1305.1-GFP vector to act as the effector. The promoter fragment of OsCSLD4 was fused into pGreenII0800-LUC vector as the reporter. The vectors were individually transformed into the A. tumefaciens strain EHA105 using Hiei et al. (1994) methods. Transient co-expression of the effector and reporter constructs were infiltrated into the leaves of 4-week-old tobacco (N. benthamiana) plants for 2 days. Firefly LUC and REN activities were surveyed with a dual-luciferase reporter assay kit (Promega), and the LUC activity, normalized to REN activity, was determined. The Renilla luciferase (REN) gene driven by 35S promoter was used as a normalizer control. Transient transactivation with the reporter and the empty vector pCAMBIA1305.1-GFP was used as a negative control and its activity was taken as 1. Primers used for dual-luciferase are listed in Supplementary Table S1.
Electrophoretic mobility shift assay (EMSA)
The full length of OsADD1 coding sequence was fused into pGEX-4T-2 vector, and the fusion protein was expressed in Escherichia coli at 16 °C for 20–24 h in the presence of 0.1 mM isopropyl β-d-1-thiogalactopyranoside. GST-OsADD1 protein was purified using Amylose Resin (New England Biolabs) according to the manufacturer’s instructions. The promoter fragment of OsCSLD4 was synthesized using EMSA Probe Biotin Labeling Kit. The LightShift™ Chemiluminescent EMSA Kit was used to perform EMSA following the manufacturer’s instructions. 60 fM biotin-labeled DNA probes were incubated with 2 μg purified proteins (GST-OsADD1) in a total volume of 20 μL. The reaction mixtures were incubated at 25 °C for 30 min and loaded onto a 6% (w/v) native polyacrylamide gel. Electrophoresis was conducted at 100 V for 1.5 h in 0.5 × TBE buffer (44.5 mM Tris, 44.5 mM boric acid, and 1 mM EDTA, pH 8.3) at 4 °C. The gel was sandwiched and transferred to a positively charged Nylon Membranes (Roche) in 0.25 × TBE buffer at 200 mA for 45 min at 4 °C. Biotin labeled DNA was detected using Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Scientific) and X-ray film. Primers used for EMSA are listed in Supplementary Table S1.
Results
Osadd1 mutants exhibit defected anther dehiscence
We screened mutants with defected anther dehiscence from the progeny generated by EMS treatment in the 9311 cultivar background. One of the mutants was identified and nominated as anther dehiscence defected 1 (Osadd1) (Fig. 1a). The wild-type (WT) anthers became plain after anthesis and pollen shading (Fig. 1b–d). Unlike the WT, most anthers of Osadd1 remained yellow after anthesis and no splits could be observed in the surface of the anthers (Fig. 1c, d). In addition, Osadd1 also exhibited rolled leaves, opening glume after grain filling (Fig. 1e) and significantly decreased seed setting rate (35 ± 10%) compared to WT (92 ± 3%) due to failure of pollen dispersal. We also identified another mutant, designated as Osadd1-2, showing more severe phenotypes comparing to Osadd1 with complete anther indehiscence, opening glume, and no matured seeds (Supplementary Fig. S1).
Comparison of the wild type (WT) and Osadd1-1 mutant. a Comparison of the WT (left) and Osadd1-1 (right) plants after heading. b Comparison of the panicles of the WT (left) and Osadd1-1 (right) mutant at the heading stage. c and d Comparison of the WT (left) and Osadd1-1 (right) anther after flowering. e Comparison of the mature seeds of the WT (up) and Osadd1-1 (down). Scale bars = 10 cm in a, 2 cm in b, 2 mm in c, 1 mm in d, and 2 mm in e
Phenotypic characteristics of spikelets and anthers of Osadd1
As the Osadd1 mutants showed defective opening glume and anther dehiscence, a transverse section observation of spikelet was conducted. In Fig. 2a, b, the hook region of rice spikelets was marked in the red square. The palea and lemma could hook together in the WT, but not in the Osadd1 (Fig. 2a–d). The cell morphology of outermost layer from Osadd1 lemma was similar with the innermost layer, which was different from WT (Fig. 2a, b black arrow), and the Osadd1 palea was thicker than that of WT (Fig. 2a, b white asterisk).
Transverse sections showing spikelet of the WT and Osadd1-1 mutant. a and b Transverse sections of the spikelet middle region of the WT (a) and Osadd1-1 mutant (b) at the heading stage. Red square, hook region; black arrow, outermost layer of lemma; white asterisk, thicken palea. c and d A higher magnification image of spikelet from WT (c) and Osadd1-1 mutant (d). Scale bars = 1 mm in a, b and 0.5 mm in c, d
We then observed the mature anthers from WT and Osadd1 via SEM and transverse section observation. The morphology of Osadd1 anthers showed distinct difference from WT (Fig. 3a, b). Some splits were observed on the surface of the WT anther which revealed the mature pollen grains inside (Fig. 3c), while there were no splits observed on the surface of the anthers from Osadd1 (Fig. 3d). As for transverse section observation of WT and Osadd1, there was no obvious distinction between WT and Osadd1 until anther development stage 13 (Supplementary Fig. S2). The two adjacent pollen sacs of WT anther became connected and the anther dehiscence occurred to facilitate the release of pollen grains at this stage, while the majority of Osadd1 pollen sacs remained closed which resulted in anther dehiscence defected (Fig. 3e, f). These observations exhibited cell structure defects in Osadd1 anthers.
Electron micrographs and transverse sections showing anther dehiscence of the WT and Osadd1-1 mutant. a and b Scanning electron micrograph showing that wild-type (a) anther was dehiscent and Osadd1-1 (b) mutant was not. Red square, splits on the surface of anthers. c and d A higher scanning electron magnification image of WT (c) and Osadd1-1 (d) anther. e and f Transverse sections of WT (e) and Osadd1-1 (f) anther at the dehiscence stage. Scale bars = 0.5 mm in a, b, 50 μm in c, d and 0.1 mm in e, f
Isolation of OsADD1 by map-based cloning
In order to isolate OsADD1, we initially mapped the OsADD1 locus between two InDel molecular markers X1 and X6 on the long arm of rice chromosome 9 (Fig. 4a). To more precisely localize OsADD1, 200 additional mutants from a F2 mapping population were identified and analyzed using another 4 polymorphic InDel molecular markers. Finally, the OsADD1 locus was narrowed to a 1.2M region between markers X3 and X4. By sequencing the Osadd1-1 mutant genomic DNA, we found that a single base deletion on the first exons of Os09g0395300, which led to the complete loss of encoded GARP domain (Fig. 4a). While Osadd1-2 exhibited a single base deletion on the second exon and resulted in a truncation of GARP domain (Fig. 4a). Previous study showed that the gene Os09g0395300 encodes a 377-amino acids GARP transcription factor involved in leaf development (Zhang et al. 2009).
Map-based cloning of OsADD1 gene and complementation test. a Fine-mapping of the OsADD1 gene. The OsADD1 locus was mapped to a 1.2 Mb region between markers X3 and X4 on the long arm of chromosome 9. Lines indicate introns and boxes indicate exons. The GARP domain is marked in the genomic structure of OsADD1. Sequence analysis showed that Osadd1-1 and Osadd1-2 carry a single base mutation. b Phenotypes of WT (left) and Osadd1-1-COM (right) plants at maturity. c Comparison of WT (left), Osadd1-1 (middle) and Osadd1-1-COM (right) spikelets. d Comparison of WT (left), Osadd1-1 (middle) and Osadd1-1-COM (right) anther after flowering. e Comparison of the mature seeds of WT (left), Osadd1-1 (middle) and Osadd1-1-COM (right). Scale bars = 10 cm in b, 2 mm in c, 1 mm in d and 2 mm in e
To confirm the mutation of OsADD1 was responsible for Osadd1 phenotype, we performed a functional complementation experiment. A binary plasmid carrying the OsADD1 coding region and driving by the ubiquitin promoter was transformed into calli derived from Osadd1 mutant. The complemented lines displayed normal anther dehiscence and high seed setting rates, which were similar to those of the WT (Fig. 4b–e). Together, our results confirmed that Os09g0395300 represented OsADD1 and was responsible for the mutant phenotype.
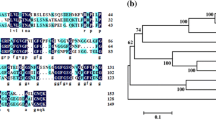
We searched public databases using BLAST with the OsADD1 protein sequence. A phylogenetic relationship analysis revealed that the OsADD1 proteins of 18 species are related in evolution closely, including Arabidopsis thaliana, Oryza brachyantha, O. sativa, Panicum hallii, Sorghum bicolor, Setaria italic, Zea mays, etc. Our result revealed that OsADD1 was widely existed in various species (Supplementary Fig. S3). Sequence alignment revealed that the GARP domain in OsADD1 homologous genes was highly conserved in many species, although there were large differences in the rest part of OsADD1 (Supplementary Fig. S4).
Expression pattern and subcellular localization of OsADD1
To discover the tissue expression patterns of OsADD1, we detected mRNA levels in various organs of WT by qRT-PCR. OsADD1 is expressed in both vegetative and reproductive tissues, higher expression levels in spikelets (Fig. 5a), which indicates the important role of OsADD1 during rice plant growth process. The expression pattern is consistent with the organs that exhibited different phenotypes comparing with WT.
Spatio-temporal expression analysis of OsADD1. a Relative expression of OsADD1 in different organs. Transcript levels were normalized relative to OsUbi1. Anthers were classified into different stages. BS7, before Stage 7. Data are presented as the mean of three biological replicate ± SD. Error bars indicate SD (n = 3). b and c Subcellular location of OsADD1. Green fluorescence indicates OsADD1-GFP, red fluorescence indicates nucleus, yellow fluorescence indicates images merged from the two fluorescence. b OsADD1-GFP fusion protein in rice protoplasts. c OsADD1-GFP fusion protein in tobacco (Nicotiana benthamiana) epidermal cells. Bars = 10 μm in b and c
To study subcellular localization of OsADD1, we fused the full-length OsADD1 coding region to GFP under the control of 35S promoter and transiently expressed the construct in mesophyll protoplasts of rice leaves. The GFP signal was observed in nucleus and membrane due to the putative transcription function of OsADD1 (Fig. 5b). In order to verify this result, we transfected tobacco (N. benthamiana) epidermal cells with the fusion protein, and the GFP fluorescence was also observed in the nucleus (Fig. 5c).
Genes associated with anther dehiscence and rolled leaves were affected in Osadd1
OsADD1 is a putative GARP transcription factor that may regulate gene expression by binding to the promoters of the target genes, and then control anther dehiscence at anthesis stage. We measured the expression level of eight genes (OsCSLD4, ROC5, OsZHD1, ACL2, YABBY1, LC2, NAL7, and SRL1) in anthers of WT and Osadd1, because the mutants of these genes shared similar phenotypes comparing with Osadd1 (Dai et al. 2007; Fujino et al. 2008; Li et al. 2010; Xiang et al. 2012; Xu et al. 2014; Yoshikawa et al. 2013; Zhao et al. 2010; Zou et al. 2011). All of the eight genes expression levels were down-regulated in Osadd1 (Fig. 6). Among these genes, the expression level of OsCSLD4 decreased the most.
Expression levels of anther dehiscence related genes in wild type and Osadd1-1 anther. Transcript levels were normalized relative to OsUbi1. Data are presented as the mean of three biological replicate ± SD. Error bars indicate SD (n = 3). Student’s t test was used for statistical analysis (*P ≤ 0.05, **P ≤ 0.01)
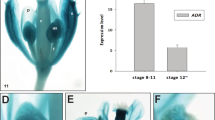
OsCSLD4 is predicted to catalyze the biosynthesis of non-cellulosic polysaccharides such as the β-d-glycan backbone of hemicelluloses. Previous reported shows that the cellulose level was affected in cd1/Oscsld4 mutants (Luan et al. 2011). We thus measured the cellulose level of panicles from WT and Osadd1 after heading stage, which was significantly reduced in Osadd1 (Fig. 7a). Then we analyzed the tissues expression pattern of OsCSLD4, which was highly similar to the tissues expression pattern of OsADD1 (Fig. 7b). nrl1/Oscsld4 exhibited phenotypes which were similar to Osadd1, such as reduced dehisced anthers, rolled leaf, opening glume and low seed setting rates (34.8 ± 14%) (Supplementary Fig. S5), these phenotypes were consistent with previous reports (Wu et al. 2010; Yoshikawa et al. 2013), and implied a possible interaction between OsCSLD4 and OsADD1.
Cellulose content measurement and spatio-temporal expression analysis of OsCSLD4. a Measurement of cellulose contents of panicles from WT and Osadd1 after heading stage. b Relative expression of OsCSLD4 in different organs. Transcript levels were normalized relative to OsUbi1. Anthers were classified into different stages. BS7, before Stage 7. Data are presented as the mean of three biological replicate ± SD. Error bars indicate SD (n = 3). Student’s t test was used for statistical analysis (**P ≤ 0.01)
OsADD1 directly regulates the expression of OsCSLD4
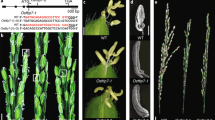
To further investigate the relationship between OsADD1 and OsCSLD4, we first measured the expression level of OsADD1 in WT and Oscsld4 with no obvious difference (Supplementary Fig. S6). Then we conducted dual-LUC assays to testify if OsADD1 could activate OsCSLD4 transcription. The ~ 3 kb upstream segment of OsCSLD4 promoter was cloned into pGreenII0800-LUC as the reporter and the coding region of OsADD1 was cloned into pCAMBIA1305.1-GFP as the effector (Fig. 8a). Two vectors co-transformed into tobacco (N. benthamiana) protoplasts and after 2 days the activities of LUC and REN were measured. The LUC:REN ratios were significantly elevated in the protoplasts transformed with the effector and reporter compared with the control (Fig. 8b).
OsADD1 binds to and activates the promoter of OsCSLD4. a Schematic diagram of the effector and reporter constructs used for transient expression assay. MCS multiple cloning site. P35S and T35S are promoter and terminator of CaMV 35S, respectively. LUC and REN are firefly luciferase and Renilla luciferase, respectively. b Dual luciferase assays of the promoter activity using tobacco protoplasts. LUC:REN ratio of the control was taken as 1 for normalization. Error bars indicate ± SD (n = 3). Student’s t-test was used for statistical analysis (**P ≤ 0.01). c The length of OsCSLD4 promoter region and the location of biotin labeled probe for electrophoretic mobility shift assay (EMSA). Red line, biotin-labeled probe site. d EMSA using affinity-purified fusion protein glutathione S-transferase OsADD1-GST incubated with biotin-labeled probe of OsCSLD4 fragment
After the transcription activation function of OsADD1 was proved through LUC assay, we aimed to seeking for the precise site of OsCSLD4 promoter which was binding by OsADD1. By using promoter analysis tool provided by PlantPAN 2.0 (http://plantpan2.itps.ncku.edu.tw/), we found that there is an ATAT motif in – 2384 to − 2381 of OsCSLD4 promoter region which could bind directly by OsADD1. EMSA was then carried out to testify if this motif could be binding by OsADD1. We constructed GST-OsADD1 fusion protein expressed in E. coli and synthesized probes with/without biotin labeled from − 2397 to − 2368 bp upstream from OsCSLD4 promoter region for EMSA (Fig. 8c). The recombinant protein GST-OsADD1 can bind with the 30-bp oligonucleotide derived from OsCSLD4 promoter. With the addition of the unlabeled competitor, the color of retarded band became lighter (Fig. 8d). These results indicated that OsADD1 could directly bind to the promoter region of OsCSLD4 in vivo and in vitro.
Discussion
Anther dehiscence is the final stage of anther development, and releases mature pollen grains for pollination, fertilization and seed production. It is one of the essential steps during reproductive growth in plants (Song et al. 2018). In our study, we presented convincing evidence that OsADD1, a GARP transcription factor, directly regulates the cellulose synthase-like D sub-family 4 (OsCSLD4) and controls anther dehiscence in rice reproductive development. This molecular mechanism of anther dehiscence is not only conducive to revealing the male reproductive development process, but also beneficial for artificially control anther dehiscence, thereby saving breeding costs and improving breeding quality in production.
The anther dehiscence process mainly involves three special tissues: the endothecium, the septum and the rupture. Thickening of the endothecium is necessary step for anther dehiscence. In A. thaliana, one of the mutants from Cinnamoyl CoA Reductase1, named ccr1g, showed defects in synthesize lignin monomers, which resulted in reduced thickening of the endothecium and anther indehiscence (Thevenin et al. 2011). Also, the Arabidopsis thaliana Receptor-like Protein Kinase 2 (rpk2) mutant showed male sterility due to the lack of thickening of the endothecium which led to anther indehiscence (Mizuno et al. 2007). OsCSLD4 encodes a cellulose synthase-like gene which is essential for cell-wall formation. Previous report showed that the cell-wall structure of root and culm was thickened in Oscsld4 (Li et al. 2009). Subsequently, an allelic mutant of OsCSLD4 (named sle1) showed rolled leaf blades, few anthers dehiscence and significant structure changes of sle1 anthers’ cell-wall (Yoshikawa et al. 2013). Considering the connection between OsADD1 and OsCSLD4, and the cell-wall structure changes observed in Osadd1, the reason for anther indehiscence occurred in Osadd1 and Oscsld4 could be the same, i.e. lacking the driving force from fibrous structures in the endothecium cells, along with the development of cavities and the rupture of septa (Matsui et al. 1999).
In the past, GARP was classified into MYB transcription factors because of the resemblance domain and the similar function of these two transcription factor family. MYB transcription factors affect many aspects of plant growth including anther anthesis. In Arabidopsis, MYB26 can direct regulate two NAC domain transcription factors, NST1 and NST2, and induce their expression for further regulating genes associated with cellulose and lignin biosynthesis which eventually involved in cell-wall secondary thickening and affected anther dehiscence (Yang et al. 2017). But the exact cellulose related genes participated in this pathway have not been found to show the interactions with NST1/2. In our study, we demonstrated that OsCSLD4, as the precise downstream genes of OsADD1, participated in cell-wall secondary thickening pathway, could affect rice anther structure and dehiscence process.
GARP transcription factors participated in many aspects of plant physiological and biochemical processes (Safi et al. 2017), including hormonal signaling, which indicating the importance of GARP transcription factors in plant growth. Some GARP transcription factors were reported to have close relation with plant hormone. For example, HRS1 encodes a putative Golden2-like transcription factor in Arabidopsis, and hrs1-1 exhibited germination defects. These deficiencies can be reverted when hrs1-1 crossed with abi3, abi4 and abi5 mutants, which demonstrated that HRS1 acts upstream of the ABA signaling pathway during germination (Wu et al. 2012). KAN1, one of GARP transcription factor had close connection with auxin (Huang et al. 2014; Ilegems et al. 2010; Izhaki and Bowman 2007). Plant hormone has huge impact on plant development, with GARP transcription factors’ participation in hormonal signaling and the tight relation between plant hormone and anther dehiscence process. There could be another anther dehiscence regulation pathway in Osadd1 mutants needs OsADD1 to regulate hormone signaling. Hormone regulation may also account for the other phenotypes of Osadd1, such as opening glume and low seed setting rate in addition to anther dehiscence defected (Fig. 1).
Intriguingly, Osadd1-2 from japonica background showed more severe phenotype comparing with Osadd1-1 from indica background (reduced plant height and no mature seeds). This indicated that the effect of OsADD1 is distinct from those in different varieties, the working mechanism of OsADD1 remain further investigated in japonica background.
In summary, our results demonstrated that OsADD1 can directly bind to the promoter of OsCSLD4, regulate anther dehiscence, and then control pollen dispersal (Supplementary Fig. S7).
Key message
We demonstrated that OsADD1 can directly bind to the promoter of OsCSLD4 and regulate anther dehiscence in rice.
References
Cecchetti V, Brunetti P, Napoli N, Fattorini L, Altamura MM, Costantino P, Cardarelli M (2015) ABCB1 and ABCB19 auxin transporters have synergistic effects on early and late Arabidopsis anther development. J Integr Plant Biol 57:1089–1098
Cecchetti V, Celebrin D, Napoli N, Ghelli R, Brunetti P, Costantino P, Cardarelli M (2017) An auxin maximum in the middle layer controls stamen development and pollen maturation in Arabidopsis. N Phytol 213:1194–1207
Dai M, Zhao Y, Ma Q, Hu Y, Hedden P, Zhang Q, Zhou DX (2007) The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol 144:121–133
Estornell LH, Landberg K, Cierlik I, Sundberg E (2018) SHI/STY genes affect pre- and post-meiotic anther processes in auxin sensing domains in Arabidopsis. Front Plant Sci 9:150
Fujino K, Matsuda Y, Ozawa K, Nishimura T, Koshiba T, Fraaije MW, Sekiguchi H (2008) NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol Genet Genomics 279:499–507
Ghelli R et al (2018) A newly identified flower-specific splice variant of AUXIN RESPONSE FACTOR8 regulates stamen elongation and endothecium lignification in Arabidopsis. Plant Cell 30:620–637
Goldberg RB, Beals TP, Sanders PM (1993) Anther development: basic principles and practical applications. Plant Cell 5:1217
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Hosoda K (2002) Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14:2015–2029
Hu J et al (2010) Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol Biol 73:283–292
Huang T et al (2014) Arabidopsis KANADI1 acts as a transcriptional repressor by interacting with a specific cis-element and regulates auxin biosynthesis, transport, and signaling in opposition to HD-ZIPIII factors. Plant Cell 26:246–262
Ilegems M, Douet V, Meylan-Bettex M, Uyttewaal M, Brand L, Bowman JL, Stieger PA (2010) Interplay of auxin, KANADI and Class III HD-ZIP transcription factors in vascular tissue formation. Development 137:975
Izhaki A, Bowman JL (2007) KANADI and Class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19:495
Jibran R, Tahir J, Cooney J, Hunter DA, Dijkwel PP (2017) Arabidopsis AGAMOUS regulates sepal senescence by driving jasmonate production. Front Plant Sci 8:2101
Kang HG, Park S, Matsuoka M, An G (2005) White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C4-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J 42:901–911
Kanno Y et al (2016) AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat Commun 7:13245
Li M et al (2009) Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J 60:1055–1069
Li L, Shi ZY, Li L, Shen GZ, Wang XQ, An LS, Zhang JL (2010) Overexpression of ACL1 (abaxially curled leaf 1) increased bulliform cells and induced abaxial curling of leaf blades in rice. Mol Plant 3:807–817
Li WQ et al (2017) CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. Plant J 92:904–923
Luan W, Liu Y, Zhang F, Song Y, Wang Z, Peng Y, Sun Z (2011) OsCD1 encodes a putative member of the cellulose synthase-like D sub-family and is essential for rice plant architecture and growth. Plant Biotechnol J 9:513–524
Ma H (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56:393–434
Matsui T, Omasa K (2002) Rice (Oryza sativa L.) cultivars tolerant to high temperature at flowering: anther characteristics. Ann Bot 89:683–687
Matsui T, Omasa K, Horie T (1999) Mechanism of anther dehiscence in rice (Oryza sativa L.). Ann Bot 84:501–506
Mizuno S et al (2007) Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J 50:751–766
Prasad PVV, Boote KJ, Allen LH, Sheehy JE, Thomas JMG (2006) Species, ecotype and cultivar differences in spikelet fertility and harvest index of rice in response to high temperature stress. Field Crops Res 95:398–411
Riechmann JL et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105
Safi A, Medici A, Szponarski W, Ruffel S, Lacombe B, Krouk G (2017) The world according to GARP transcription factors. Curr Opin Plant Biol 39:159–167
Saito H et al (2015) The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis. Nat Commun 6:6095
Sakai H, Aoyama T, Bono H, Oka A (1998) Two-component response regulators from Arabidopsis thaliana contain a putative DNA-binding motif. Plant Cell Physiol 39:1232–1239
Salinas-Grenet H et al (2018) Modulation of auxin levels in pollen grains affects stamen development and anther dehiscence in Arabidopsis. Int J Mol Sci. https://doi.org/10.3390/ijms19092480
Scott RJ, Spielman M, Dickinson HG (2004) Stamen structure and function. Plant Cell 16:S46
Song S, Chen Y, Liu L, See YHB, Mao C, Gan Y, Yu H (2018) OsFTIP7 determines auxin-mediated anther dehiscence in rice. Nat Plants 4:495–504
Steiner-Lange S et al (2003) Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J 34:519–528
Thevenin J et al (2011) The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol Plant 4:70–82
Wu C, Fu Y, Hu G, Si H, Cheng S, Liu W (2010) Isolation and characterization of a rice mutant with narrow and rolled leaves. Planta 232:313–324
Wu C et al (2012) HRS1 acts as a negative regulator of abscisic acid signaling to promote timely germination of Arabidopsis seeds. PLoS ONE 7:e35764
Xiang JJ, Zhang GH, Qian Q, Xue HW (2012) SEMI-ROLLED LEAF1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol 159:1488–1500
Xu Y et al (2014) Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice. Planta 239:803–816
Yang C, Xu Z, Song J, Conner K, Vizcay Barrena G, Wilson ZA (2007) Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19:534–548
Yang C et al (2017) Transcription factor MYB26 is key to spatial specificity in anther secondary thickening formation. Plant Physiol 175:333–350
Yoshikawa T, Eiguchi M, Hibara K, Ito J, Nagato Y (2013) Rice SLENDER LEAF 1 gene encodes cellulose synthase-like D4 and is specifically expressed in M-phase cells to regulate cell proliferation. J Exp Bot 64:2049–2061
Zeng Y, Zhang Y, Xiang J, Uphoff NT, Pan X, Zhu D (2017) Effects of low temperature stress on spikelet-related parameters during anthesis in Indica–Japonica hybrid rice. Front Plant Sci 8:1350
Zhang G-H, Xu Q, Zhu X-D, Qian Q, Xue H-W (2009) SHALLOT-LIKE1 Is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21:719
Zhao SQ, Hu J, Guo LB, Qian Q, Xue HW (2010) Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res 20:935–947
Zhao Z et al (2013) A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev Cell 27:113–122
Zhu Q-H, Ramm K, Shivakkumar R, Dennis ES, Upadhyaya NM (2004) The ANTHER INDEHISCENCE1 gene encoding a single MYB domain protein is involved in anther development in rice. Plant Physiol 135:1514
Zou LP et al (2011) Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice. Plant Physiol 156:1589–1602
Acknowledgements
This research was supported by the National Transform Science and Technology Program (2016ZX08001004-002) and National Key Research and Development Program of China (2016YFD0101107, 2016YFD0101801), the Key Laboratory of Biology, Genetics and Breeding of Japonica Rice in the Mid-lower Yangtze River, the Ministry of Agriculture, China, Jiangsu Plant Gene Engineering Research Center, the Jiangsu Collaborative Innovation Center for Modern Crop Production. This study is funded by China Postdoctoral Science Foundation. We are grateful to Professor Wenzhen Liu (China National Rice Research Institute, China) for providing the seeds of Oscsld4/nrl1 mutant.
Author information
Authors and Affiliations
Contributions
Y.J.X., S.M.Y. and Z.G.Z. were involved in conceiving the project, designing the experiments and analyzing the data; W.Y.K., Y.J.X., S.M.Y. and Y.C. were involving in the map-based cloning of the OsADD1 gene; Q.Y.T., W.T.B. and H.Z. were involved in the generation of the transgenic plants; C.L.W. was involved in OsADD1 transient expression in rice protoplasts; Y.J.X. was involved in LUC assay and EMSA; Y.J.X. was involved in writing this article; J.L. and C.M.W. were involved in revising this article; J.L., Z.G.Z. and J.M.W. were involved in data discussions.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xiao, Y., You, S., Kong, W. et al. A GARP transcription factor anther dehiscence defected 1 (OsADD1) regulates rice anther dehiscence. Plant Mol Biol 101, 403–414 (2019). https://doi.org/10.1007/s11103-019-00911-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-019-00911-0