Abstract
We report the characterization of three UBA2 genes (UBA2a, -b, and -c; corresponding to At3g56860, At2g41060, and At3g15010) encoding Arabidopsis thaliana proteins with high homology to Vicia faba AKIP1 and other heterogeneous nuclear ribonucleoprotein (hnRNP)-type RNA-binding proteins. In vitro RNA binding assays revealed that the three UBA2 proteins interact efficiently with homoribopolymers. Biolistic transient expression of UBA2-GFPs demonstrated that the three UBA2 proteins localize to the nucleus. Expression analysis by RNA gel blot, RT-PCR, and promoter::GUS assays showed that UBA2 transcripts are present in all organs. UBA2 genes are subject to alternative splicing affecting only the 3′-untranslated regions (UTRs): six different splice variants were detected for UBA2a, and two each were found for UBA2b and UBA2c. RT-PCR and quantitative real-time RT-PCR analysis showed that the levels of UBA2 transcripts are regulated by wounding in a splice variant-specific manner: splice variants UBA2a.1 and UBA2c.1 increased following mechanical wounding. Wounding effects on gene expression are transduced by methyl jasmonate (MeJA)-dependent and oligogalacturonide (OGA)-dependent pathways. However, neither MeJA nor OGA treatment altered levels of any of the UBA2 transcripts, and other plant hormones implicated in wound responses, ethylene and abscisic acid (ABA), also had no effect on accumulation of UBA2 transcripts. Taken together, these results imply that the three UBA2 genes encode hnRNP-type nuclear RNA-binding proteins that function in a novel wound signal transduction pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Posttranscriptional processes, including pre-mRNA splicing, alternative splicing, capping, polyadenylation, mRNA stability and mRNA transport, are crucial in gene expression control. Numerous plant RNA-binding proteins have been identified at the sequence level and are good candidates to exert these functions. Many of these proteins are characterized by the presence of RNA-recognition motifs (RRM), also called consensus sequence RNA-binding domains (CS-RBD). An RRM contains two short consensus sequences consisting of RNP1 (octamer) and RNP2 (hexamer) embedded in a ∼80 amino acid region that is better conserved at the structural than at the sequence level (Lorković and Barta 2002).
The Arabidopsis genome encodes 196 proteins containing one to four RRMs (Lorković and Barta 2002). The biological functions of only a few of these RRM-containing proteins have been investigated. Three RRM-containing proteins are involved in flowering control in Arabidopsis: FCA (FLOWERING TIME CONTROL PROTEIN ALPHA) and FPA each have one RRM and FLK (FLOWERING LATE KH DOMAIN) has three RRMs (Macknight et al. 1997; Schomburg et al. 2001; Lim et al. 2004). These three proteins are involved in processing of the transcript of the key regulator of flowering, Flowering Locus C (FLC; Simpson 2004). It is noteworthy that FCA has also been characterized as an abscisic acid receptor and that ABA can exert a direct negative control on FCA function in flowering control (Razem et al. 2006). Vicia faba AKIP1 (VfAKIP1), which contains two RRMs, is part of the ABA signaling pathway that regulates stomatal closure. AKIP1 binds a dehydrin transcript when phosphorylated in response to ABA (Li et al. 2002).
Several glycine-rich RNA-binding proteins (GR-RBPs), which contain one RRM, are hypothesized to play roles in post-transcriptional gene regulation under various stress conditions (Albà and Pagès 1998; Sachetto-Martins et al. 2000). For example, the transcript level of GR-RBP4 in Arabidopsis increases with cold stress and decreases with salt stress and dehydration (Kwak et al. 2005). Despite this interesting correlation, overexpression of GR-RBP4 in Arabidopsis did not affect cold or freezing tolerance and the role of GR-RBP4 is still unknown. The strongest functional evidence for GR-RBP participation in stress response has been obtained for the Arabidopsis atRZ-1a gene. The transcript level of this gene is upregulated by cold and atRZ-1a overexpression leads to faster seed germination, better seedling growth under cold stress, and enhanced freezing tolerance in Arabidopsis (Kim et al. 2005).
Here we report characterization of the UBA2 class of Arabidopsis RNA-binding proteins, which are the proteins in the Arabidopsis genome with greatest similarity to Vicia faba AKIP1. This Arabidopsis protein family has three members, UBA2a, UBA2b and UBA2c, with overall similarity to metazoan hnRNPs of A/B and D types. These classes of hnRNPs are RRM-containing proteins that in mammals are known to be involved in many aspects of RNA metabolism, including pre-mRNA splicing, mRNA localization, stability, export to the cytoplasm, and translational control (Dreyfuss et al. 1996, 2002; Krecic and Swanson 1999; Mili et al. 2001; Reed and Magni 2001). UBA2a has been previously identified by Lambermon and coworkers (2002) for its ability to interact with UBP1, a hnRNP-like protein involved in both mRNA splicing and stability (Lambermon et al. 2000). UBA2b and UBA2c were subsequently identified in silico by sequence homology to UBA2a (Lambermon et al. 2002) but have not yet been studied.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia (Col-0) was used in this study. Seeds were surface-sterilized in 70% ethanol for 3 min once and then in 95% ethanol for 1 min three times. Seeds were dried on filter paper and transferred to half-strength Murashige and Skoog medium (Murashige and Skoog 1962) supplemented with 1% sucrose and 1.0% agar (Sigma). Plates were placed at 4°C for 2 days for stratification and then placed vertically under short-day growth conditions (8 h light/16 h dark cycle) at 22°C for 10–12 days. Healthy seedlings were transplanted to soil (Miracle-Gro) on trays and covered with a plastic lid. Plants were grown to maturity under short-day conditions at 22°C at a light intensity of 120 μmol m−2 s−1. Plants were watered with distilled water once a week.
Cloning and constructs of UBA2 cDNAs
Total RNA was isolated from rosette leaf tissues using Trizol reagent (Invitrogen, Carlsbad, CA) and first-strand cDNA was synthesized from 5 μg of total RNA using Superscript II RNase H− Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Full-length cDNAs of UBA2a, -b and -c were amplified from first-strand cDNA using Platinum Pfx DNA polymerase according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA) with the following primers: for UBA2a, A1-NcoI primer (5′-CCATGGCAAAGAAGAGAAAGCTCGAAGGAG-3′) and A1-C-terminal primer (5′-CATGGGTCACTAAATGTATCATGTATCC-3′); for UBA2b, A2-NcoI primer (5′-CCATGGCAAAGAAGAGAAAGCTCGAATCTG-3′) and A2-C-terminal primer (5′-CATGGGTCGTTAGATTAGCCACTCAGG-3′); for UBA2c, A3-NcoI primer (5′-CCATGGATATGATGAAGAAGCGTAAGC-3′) and A3-C-terminal primer (5′-CCCAACGGTCCACCAAACTACTGAAAA-3′). The resulting cDNA products were cloned into the pCR-Blunt II-TOPO vector using the Zero Blunt PCR cloning kit according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). The constructs were confirmed by sequencing.
Homoribopolymer binding assays
Each Nco I–Not I fragment of full-length cDNA in the pCR-Blunt II-UBA2 clones was cloned into the Nco I and Not I restriction sites of pENTR11 gateway vector (Invitrogen, Carlsbad, CA). Three UBA2 expression clones with the N-terminal glutathione S-transferase (GST)-tagged recombinant proteins were generated by performing a recombination reaction between the pENTR-UBA2 entry clones and gateway destination vector pDEST15 (Invitrogen, Carlsbad, CA). GST-UBA2 proteins, purified on glutathione resin according to the manufacturer’s instructions (Clontech, MountainView CA), were incubated with sepharose-bound homoribopolymers (Sigma) in binding buffer (10 mM Tris–HCl, pH 7.4, containing 250 mM, 500 mM, or 1,000 mM NaCl, 250 mM MgCl2, 0.8% Triton X-100, 0.5 mM phenylmethylsulphonylfluoride (PMSF) and Complete EDTA free protease inhibitor (Roche). After washing four times with binding buffer to remove nonspecific binding, the homoribopolymer beads were boiled in SDS sample buffer and proteins in the buffer were resolved by SDS-polyacrylamide gel electrophoresis and silver nitrate staining.
UBA2-GFP constructs
For transient expression analyses, UBA2 cDNA inserts were fused in-frame with GFP at the C-terminus following the protocol of Lambermon et al. (2002), with some modifications. The UBA2a-GFP clone was a kind gift from Dr Zdravko Lorković. The pUBA2a-GFP clone was generated as described by Lambermon et al. (2002). The coding regions of UBA2b and UBA2c were amplified by PCR from the pCR-Blunt-UBA2 plasmids using the following primers: for pUBA2b-GFP, A2-SalI 5′primer (5′-GTCGAC AATAAACCATGACAAAGAAGAGAAAGCTCGAA-3′) and A2-BamHI 3′primer (5′-GGATCCACGACCCATGTAAGGACCACC-3′); for pUBA2c-GFP, A3-SalI 5′primer (5′-GTCGAC AATAAACCATGGATATGATGAAGAAGCGTAAGC-3′) and A3-BamHI 3′primer (GGATCCGTAGTTTGGTGGACCGTTGGG-3′). The GFP vector contains a double CaMV 35S promoter (Lambermon et al. 2000). The 5′ primers introduced a Sal I restriction site (in bold type), followed by the plant consensus translation initiation sequence (in italic type), whereas the 3′ primers introduced a BamH I restriction site (in bold type) in place of the stop codon. The PCR products were first cloned into the pCR-Blunt II-TOPO vector and then the fragments were cut with Sal I and BamH I and ligated into the GFP vector, from which the UBA2a insert had been removed with the same restriction enzymes, resulting in pUBA2b-GFP or pUBA2c-GFP.
Biolistic transient assay of UBA2-GFP localization
The first fully expanded leaves of 4- to 5-week-old Vicia faba plants were used for particle bombardment with pUBA2-GFP plasmids as previously described (Li et al. 2000, 2002; Ng et al. 2004). Leaves were bombarded with 1.5 μg plasmid DNA and immediately floated abaxial side down on 25 ml of 10 mM MES, 50 mM KCl, pH 6.2 buffer with or without the addition of 50 μM ABA. Leaves were incubated at room temperature in closed Petri dishes in the dark for 18–20 h before examination.
Epidermal peels were taken and examined using an Olympus BX-60 epi-fluorescence digital microscope equipped with a Hamamatsu Orca-100 camera and Simple PCI software. Images (tif format) were collected using a 60×/1.4 oil immersion plan-apochromat lens and a FITC/RSGFP/Bodipy/Fluo 3/DiO Filter Cube. Nuclei having areas of intense punctuate signal regardless of the size or number of “speckles” were designated as “speckled”. “Smooth” nuclei had a uniform intensity of signal throughout the entire nucleus. In the “intermediate” nuclei, there were no clearly delineated punctuate areas, but the signal was not uniform in intensity throughout the entire nucleus. At least three independent experiments were performed for each construct and in each of these experiments at least 50 cells were assessed for each of the two conditions (control or +ABA).
To assess the relative level of expression, epidermal peels were examined using a confocal LSM 510 Meta microscope (Zeiss) equipped with a 20×/0.75 plan-apochromat lens and a 40×/0.75 plan-apochromat water immersion lens. Samples were excited using a 488 nm 25 mW laser at 15% power. Signal was collected using the lambda function of the imaging software over an emission range of 510–565 nm. Peak emission (Relative Fluorescence Units) was compared while keeping excitation energy, pinhole, and gain settings constant.
RNA extraction and RNA gel blot analysis
Total RNA was extracted from the frozen samples using the Plant RNeasy extraction kit (Qiagen, Valencia, CA). Total RNA was treated with DNase I on column to remove genomic DNA contamination according to the manufacturer’s protocol (Qiagen, Valencia, CA). The concentration of RNA was quantified by spectrophotometric measurement. For RNA gel blot analysis, 8 μg of total RNA was denatured and separated on a 1.2% agarose-formaldehyde gel, and transferred onto a nylon membrane (Hybond-N plus, Amersham Biosciences, NJ). The filters were prehybridized for 30 min and incubated for 16 h at 60–65°C in Church’s hybridization solution. The RNA blots were hybridized with gene-specific probes in Church’s hybridization solution (Church and Gilbert 1984) as described previously (Kim et al. 2003). The membranes were washed three times for 10 min in 2× SSC with 0.1% SDS and three times for 10 min in 0.2× SSC with 0.1% SDS at 60–65°C. Equal RNA loading was visualized by staining the rRNAs with ethidium bromide.
The probes were synthesized with α-[32P]dCTP via a random primer labeling system (Qiagen, Valencia, CA). The labeled probes were cleaned using Bio-spin P-30 columns (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions. An EcoR I–Hind III fragment (about 460 bp) corresponding to amino acids 1–159 of the predicted UBA2a protein was used as a UBA2a gene-specific probe. An EcoR I–Pst I fragment (about 300 bp) for UBA2b and an EcoR I–Pst I fragment (about 830 bp) for UBA2c were used as gene-specific probes (Fig. 4). To detect the previously known splice variants of UBA2a transcripts, 3′-UTR regions specific to UBA2a.1 and to UBA2a.2 and UBA2a.3 were amplified by PCR using specific primer pairs and were added as probes for RNA gel blot hybridization (Fig. S1). (We noted later that the 3′-UTR probe originally designed against UBA2a.2 sequence may also hybridize with other splice variants that were subsequently discovered, including UBA2a.3 and UBA2a.4).
Reporter gene analysis of UBA2 expression patterns
Promoter regions of UBA2a (1,120 bp upstream of the start codon plus 47 bp of the coding region for a fusion in frame with uidA CDS), UBA2b (1,182 + 29 bp) and UBA2c (1,126 + 65 bp) were amplified using Pfx DNA polymerase according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA) with the following primers: for UBA2a promoter, pA1F 5′-GGATCCCTACTCTTGGAATTGACTCAAC-3′and pA1R 5′-TCAGCTTCGTTAGATTCTTCTC-3′; for UBA2b promoter, pA2F 5′-GGATCCCGACATTCACATATCAACACC-3′ and pA2R 5′-TCAGATTCGAGCTTTCTCTTC-3′; for UBA2c promoter, pA3F 5′-GGATCCTCCTCCGCCTCATATGGGTG-3′ and pA3R 5′-CCGCCACCGTTGGTATTGAG-3′. The PCR products were cloned into the pCR4-Blunt-TOPO vector using the Zero Blunt TOPO PCR cloning kit (Invitrogen, Carlsbad, CA) and subcloned into the pENTR11 vector between the BamH I and Not I restriction sites present in the polylinkers of both vectors. The promoters were finally transferred by LR recombination reaction using the Gateway™ technology kit (Invitrogen, Carlsbad, CA) into the pGWB3 vector that contains the uidA gene without start codon. The constructs were introduced into Agrobacterium C58C1 strain, and used to transform Arabidopsis plants by the floral-dip method (Clough and Bent 1998). GUS staining with 5-bromo-4-chloro-3-indolyl-ß-d-glucuronic acid was performed as described by Carrington (1995).
Plant treatments
In initial experiments (Fig. S1) for mechanical wounding treatment we used detachment of rosette leaves from 5-week-old Arabidopsis plants. Detached leaves were placed in incubation buffer (10 mM MES, 50 mM KCl, pH 6.2). Subsequently, leaf discs were collected from rosette leaves of Arabidopsis plants using a cork borer (10 or 6 mm in diameter for RT-PCR and Q-PCR analyses, respectively). Tissue samples were kept in the incubation buffer for various periods, harvested and immediately immersed in liquid nitrogen. For hormone treatments, 5-week-old plants were sprayed with solutions containing 0.01% (∼424 μM) methyl jasmonate (95% methyl jasmonate solution, Sigma-Aldrich) or 100 μM ABA (A.G. Scientific, San Diego) and water was used as control. For ethylene treatment, 5-week-old plants were exposed to 5 ppm ethylene gas in a tightly sealed plastic container and harvested at various time points. Ethylene gas concentration in the box was verified using a Hewlett-Packard gas chromatograph model 5840A equipped with a flame ionization detector and a column of activated alumina (Clark et al. 1997).
For treatment with OGAs, the protocol of Rojo et al. (1999) was followed. Seeds were surface-sterilized and sown in 12-well (30 seeds per well) tissue culture clusters (Costar Corp., Cambridge, MA), containing 2 ml per well of sterile MS medium (Murashige and Skoog salts; JRH Bioscience, Inc., Lenexas, Kansas) supplemented with 0.5% sucrose, and grown with shaking (150 rpm) in a culture room under 16 h day/8 h night diurnal cycles for 10 days. Prior to treatments, the liquid medium was removed from the wells and replaced with 2 ml of fresh medium with or without 250 μg ml−1 OGAs. OGAs, prepared as described in Spiro et al. (1993), were kindly provided by Dr. Mark Spiro.
Touch stress was applied by bending leaves (from independent plants) 10 times back and forth (Lee et al. 2005). Leaf discs and the middle third (9 mm wide bands along the apical-distal axis) of wounded leaves (area surrounding the holes), bent leaves, unwounded leaves from wounded plants to assay for systemic induction, and leaves from unstressed plants (control) were collected 2 and 6 h after wounding for Q-PCR analysis.
For bacterial inoculation, the avirulent pathogen Pseudomonas syringae pv tomato (P. s. t.) DC3000 (avrRpt2) bacterial strain was grown at 28°C on Pseudomonas agar F (Sigma, St. Louis) media supplemented with 100 μg ml−1 rifampicin and 25 μg ml−1 kanamycin. Leaves of 5-week-old plants were syringe-inoculated with bacteria suspended in 10 mM MgCl2 at a concentration of 1 × 106 cfu ml−1 as described by Jambunathan and McNellis (2003).
RT-PCR analysis
For RT-PCR experiments RNA was treated with DNase I on column according to the manufacturer’s specifications (Qiagen, Valencia, CA), or treated with RQI DNase (Promega) followed by a phenol/chloroform extraction. One μg of total RNA was used in the RT-PCR system according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA) together with gene-specific primers. PCR was performed with denaturation at 96°C for 10 min, followed by 25 cycles of denaturation at 94°C for 15 s, annealing at 58°C for 30 s, and extension at 72°C for 1 min. A final extension at 72°C for 10 min was performed to complete the reaction. Control RT-PCR was performed with the same amount of total RNA using a primer pair specific to the Actin2 (At3g18780) gene. Twelve μl of each RT-PCR product was analyzed on a 1.0% (w/v) agarose gel to visualize the amplified cDNAs.
To obtain additional insight into the splice variants of the three UBA2 transcripts, we performed RT-PCR analyses using UBA2-splice variant-specific primers on RNA samples from wounded (leaf disc) tissue. Positions of primers relative to the sequences of the UBA2 transcripts are indicated by arrows in Fig. 6a. The primers used in this study are as follows: for UBA2a, A1-specific (5′-CTATCGCTGCTGCAGCTGTTTCAG-3′) and A1-C-terminal primers (5′-GGATACATGATACATTTAGTGACCCATG-3′); for UBA2a.1, A1-specific and A1-a type primers (5′-GACTTCATTTAGCATCAGCTCCTT-3′); for UBA2a.2, A1-C-For (5′-CAAGGCGGTACAAGTAGAGGGCAAC-3′) and A1-b type primers (5′-CACGCCTTTATAAACTCCTCTGAA-3′); for UBA2a.3, A1-C-For and A1-c type primers (5′-AATAATATTCAAAGCTCGTACTCTTGTG-3′); for UBA2b, A2-specific (5′-GGGAACCCTGTTGTGGCTCCTG-3′) and A2-C-terminal primers (5′-CCTGAGTGGCTAATCTAACGACCCATG-3′); for UBA2b.1, A2-specific and A2-a type primers (5′-GGTACCCCATAGATTTTTGTTGG-3′); for UBA2c, A3-For (5′-CTGGTAAATCTAGAGGCTTTGCAT-3′) and A3-C-terminal primer (5′-TTTTCAGTAGTTTGGTGGACCGTTGGG-3′); for UBA2c.1, A3-For and A3-a type primers (5′-CAATCAGGTAATCACCACTAAGCA-3′).
Quantitative real-time RT-PCR analysis
Four-week-old plants were transferred to continuous light 5 days before stress application to avoid any inadvertent diurnal stimulus (see “Plant treatments”). Total RNA was extracted using Trizol (Invitrogen) and treated with RQI DNase (Promega), according to the manufacturers’ instructions. After purification with chloroform–phenol–isoamyl alcohol, 1 μg of RNA was reverse-transcribed using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). Real-time PCR analysis was performed in 25 μl reactions containing 5 μl of 1:12.5 diluted cDNA samples, 12.5 μl IQ SYBR Green Supermix (Bio-Rad), and 0.2 μM of each primer, using an iQ5 Real-Time PCR detection system (Bio-Rad). These quantitative assays were performed twice on independent biological duplicates and the results of the four experiments were averaged to determine the fold change in RNA expression using the threshold cycles method. The amplicon of Actin2/8 (forward primer 5′-GGTAACATTGTGCTCAGTGGTGG-3′ and reverse primer 5′-AACGACCTTAATCTTCATGCTGC-3′) was used to normalize the data (Charrier et al. 2002). The different transcripts were amplified using 20 s of elongation time and specific primer pairs and annealing temperature: for UBA2a all splice variants: all a forward primer 5′-CAGGGAATGGAGTTGGAATG-3′, all a reverse primer 5′-TTAGTGACCCATGTAAGGAGT-3′, 60.3°C; UBA2a.1: a.1for 5′-CCACTCAGATATCAGAAATCAG-3′, a.1rev 5′-TTATGAGTTTGGCCTTAGGA-3′, 58.0°C; UBA2a.2: a.2for 5′-CCATCCACTCAGTCAAACTT-3′, a.2rev 5′-CTCCTCTGAAACATTAAGGGAA-3′, 59.8°C; UBA2a.3: a.3for 5′-TTGTTTATTGGTCCTAGTCCCT-3′, a.3rev 5′-ATTCAAAGCTCGTACTCTTGTG-3′, 60.9°C; UBA2a.4: a.4for 5′-CATCCACTCAGGTAATTGACT-3′, a.4rev 5′-CAGAACCAATTGCATTCAGA-3′, 58.7°C; UBA2a.5: a.5for 5′-CCATCCACTCAGTCAAACTT-3′, a.5rev 5′-GGTACATGCTCTCCTTCAAT-3′, 59.5°C; UBA2b all splice variants: all b forward primer 5′-TTTAAGAGCAAGCTGAACGG-3′, all b reverse primer 5′-GCTACCAATTCATAGATTCATCCC-3′, 60.9°C; UBA2b.1: b.1for 5′-GCCACTCAGGTAATTTAAGAGC-3′, b.1rev 5′-GCTACCAATTCATAGATTCATCCC-3′, 61.3°C; UBA2b.2: b.2for 5′-CGCTTTCACTATGTTTGAATCC-3′, b.2rev 5′-GTGGTAATCAAGATAAGCCGT-3′, 60.2°C; UBA2c all splice variants: all c forward primer 5′-ATAGCTGCTTTGACCTATCTG-3′, all c reverse primer 5′-AATCAGGAAGCCCAATAGAG-3′, 59.1°C; UBA2c.1: c.1for 5′-TACGGTCTTTCATCATCTGCT-3′, c.1rev 5′-CATCTTCACCCAATATGCTGTC-3′, 61.3°C; UBA2c.2: c.2for 5′-TAACGCATGAAAGATAGCCAG-3′, c.2rev 5′-ATCAACACCCATTCACAACTC-3′, 60.5°C; WR3: WR3-Qfor 5′-GATCCTCTTTGCTTCACTTCTC-3′, WR3-Qrev 5′-TAATGTTCAACGTATCCTTGCC-3′, 61.0°C. Absence of genomic DNA contamination was verified by running the PCRs on aliquots of the original DNase-treated RNAs, without reverse transcription.
AGI locus numbers
Arabidopsis Genome Initiative locus numbers for the genes described in this work are as follows: UBA2a (At3g56860), UBA2b (At2g41060), UBA2c (At3g15010), WR3 (At5g50200), VSP1 (At5g24780), HEL1 (At3g04720), Rab18 (At5g66400), PR-1 (At2g19990), Actin2 (At3g18780).
Results
Identification of hnRNP-type RNA-binding proteins from Arabidopsis
Arabidopsis cDNAs corresponding to the genes UBA2a (At3g56860), UBA2b (At2g41060), and UBA2c (At3g15010) were isolated by RT-PCR. The identities to VfAKIP1 of UBA2a, -b, and -c at the amino acid levels are 49.4%, 49.7%, and 30.4%, respectively (Fig. 1). Each of these hnRNP-type RNA binding proteins contains two RRM domains, each containing two short highly conserved RNA binding regions (RNP1 and RNP2), and the three proteins each have a C-terminal auxiliary glycine (G)-rich domain (Fig. 1). UBA2a and -b proteins have an N-terminal aspartate (d)/glutamate (E)-rich domain that is absent from UBA2c (Fig. 1).
Sequence alignment of three UBA2 hnRNP proteins and VfAKIP1. Sequences alignment of the four proteins was performed using Clustal W and sequence similarities were highlighted using Genedoc software. The dashed lines show the RNP1 (octamer) and RNP2 (hexamer) domains and the solid lines indicates the RRMs. The arrows following and preceding the letters D and G delimit the D/E-rich domain of UBA2a, UBA2b and AKIP1 and the G-rich domains of the four proteins
Homoribopolymer binding assays
To assess whether these three putative RNA binding proteins can indeed bind RNA, as suggested by their structure and their similarity to VfAKIP1, purified recombinant GST-UBA2 proteins were used for in vitro RNA binding experiments with homoribopolymers (Hirose et al. 1993; Kwak et al. 2005). Figure 2 shows the interaction of recombinant GST-UBA2 proteins with poly(A), poly(C), poly(G) and poly(U) in the presence of 0.25, 0.5 and 1 M NaCl. GST-UBA2a bound all RNA homoribopolymers, with a stronger binding to poly(G) and poly(U) at low salt concentration. GST-UBA2b barely bound poly(A), weakly bound poly(C), and bound poly(G) and poly(U) more strongly. GST-UBA2c has an overall higher affinity for the four RNA homoribopolymers as revealed by stronger bands at 0.5 and 1 M NaCl relative to the other two UBA2 proteins. Purified GST protein did not show affinity for the ribonucleotide homopolymers under identical conditions.
In vitro RNA binding anaysis. GST-UBA2 proteins were incubated with the indicated Sepharose-homoribopolymers and washed with buffers of increasing stringency (250, 500, and 1,000 mM NaCl). The proteins interacting with the synthetic RNA were subsequently revealed by silver nitrate-stained SDS-PAGE analysis
Subcellular localization of UBA2 proteins and ABA-induction of UBA2 nuclear speckles
To examine the subcellular localization of UBA2 proteins, the three UBA2-GFP fusion constructs were transiently expressed in leaves of Vicia faba under control of the 35S promoter (Li et al. 2000). Fluorescence microscopy of leaf tissues revealed that all three UBA2-GFP fusion proteins localized to the nuclei of leaf cells (Fig. 3). In less than 5% of transformed cells, localization to the cytosol was also observed (data not shown). Cytosolic localization did not correlate with expression level (fluorescence intensities quantified using the lambda function of a LSM 510 Meta system and the accompanying software; data not shown). However, most, although not all, of the cells exhibiting cytosolic localization also exhibited aberrant cellular morphology such as a deformed nucleus.
Subcellular localization and effect of ABA on nuclear speckles of UBA2-GFP proteins. Subcellular localization of (a) UBA2a-GFP, (b) UBA2b-GFP, and (c) UBA2c-GFP transiently expressed in Vicia faba. After an 18 h period to allow expression of the construct and treatment (no ABA indicated by solid bars, 50 μM ABA indicated by open bars), epidermal peels from Vicia faba leaves were examined. Each panel represents the mean of percentages from three independent experiments for each construct. For all three constructs the percent of speckled nuclei in the ABA treated leaves was significantly greater (P < 0.05) than for the control (Student’s t-test)
As ABA induces subnuclear relocalization of the related hnRNP, Vicia faba AKIP1, from a diffuse localization in the nucleus to punctuate “nuclear speckles” (Li et al. 2002; Ng et al. 2004), we assessed this phenomenon for the UBA2 proteins. We observed that UBA2a-GFP and UBA2b-GFP both showed localization in nuclear speckles upon ABA treatment (Fig. 3a, b). Interestingly, we found that UBA2c-GFP fusion protein formed speckles in the majority of nuclei even without ABA treatment (Fig. 3c; 74% for control vs. 84% for ABA). By contrast, in the absence of ABA treatment, only a minority of nuclei with UBA2a-GFP or UBA2b-GFP expression exhibited nuclear speckles. However, for all three constructs, the percent of speckled nuclei in the ABA treated leaves was significantly greater (P < 0.05) than for the control (Student’s t-test).
Expression patterns of UBA2 genes in Arabidopsis tissues
To study the expression patterns of the three UBA2 hnRNP-type RNA-binding genes in Arabidopsis, northern blot analysis was employed using total RNA isolated from leaves, roots, stems, flower buds, and cauline leaves from 8 to 10 weeks old plants. As shown in Fig. 4, UBA2a and UBA2c are expressed in all tissues and UBA2b is present at very low abundance in all tissues assayed. In all tissues, at least two bands were found to hybridize with a UBA2a probe (EcoR I–Hind III fragment, about 460 bp), and an additional hybridizing band was also detected with a UBA2c probe (EcoR I–Pst I fragment, 830 bp) in root and flower tissues. These bands were later found to correspond to splice variants (see next section).
Northern blot analysis of UBA2 mRNA expression. Accumulation patterns of UBA2 transcripts in mature rosette leaves (leaf), roots, stems, flower buds (flower), and cauline leaves of 8 to 10-week-old plants. Arrows indicate the presence of bands subsequently confirmed to be splice variants. An EcoR I–Hind III fragment corresponding to amino acids 1–159 of the predicted UBA2a protein was used as a UBA2a gene-specific probe. An EcoR I–Pst I fragment (∼300 bp) for UBA2b and an EcoR I–Pst I fragment (∼830 bp) for UBA2c were used as gene-specific probes
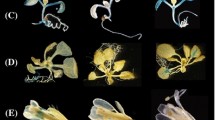
UBA2 expression patterns were investigated at the tissue level using GUS staining after fusion of each of the three corresponding promoters with the uidA gene (Fig. 5). By this technique, UBA2a showed a higher expression in the youngest leaves, in flowers and in 2-day-old imbibed embryos (Fig. 5a, d, e). UBA2b expression was detected in flowers, and in the shoot meristem area and in adjacent developing organs (Fig. 5b, f, g). UBA2c expression was observed in young leaves, the root apical and lateral meristems and the radicle of 2-day-old imbibed embryos (Fig. 5c, h–j).
Expression patterns of UBA2-promoter::GUS constructs. UBA2a is expressed in the young rosette leaves (a), flowers (d), and the embryo (e); UBA2b is expressed in the shoot apical meristem and adjacent young organs (b, g) and in the flowers (f); UBA2c is expressed in the young rosette leaves (c), strongly in the root apical (h) and lateral (i) meristems of seedlings and in the embryo radicle (j). Panels a, b, c, g, h, and i are images taken from 3-week-old plants, panels e and j are images of embryos after imbition in water for 48 h, and panels d and f are images of open flowers
Expression analysis of UBA2 splice variants
During further expression analysis of UBA2 transcripts by northern blotting, we observed that accumulation of UBA2a and UBA2c transcripts was enhanced by leaf detachment from soil-grown 5-week-old Arabidopsis plants (Supplemental Fig. 1), suggesting induction of these UBA2 genes by wounding. Through TAIR database comparisons of cDNA and EST sequences to genomic sequences, we found that three types of UBA2a splice variants (At3g56860.1, At3g56860.2, and At3g56860.3), one splice variant of UBA2b (At2g41060.1), and two splice variants of UBA2c (At3g15010.1 and At3g15010.2) have been previously reported. Remarkably, all of the previously reported splice variants from each gene, as well as new variants that we discovered in the course of this analysis (see below) arise solely as a result of unique processing of the 3′-UTR. Thus, all splice variants for each UBA2 gene have identical coding sequences and are expected to give rise to an identical protein product.
Schematic structures of splice variants detected for each UBA2 gene are shown in Fig. 6a–c based on both database analysis and our experimental results. We initially discovered the new splice variants by RT-PCR, during attempts to monitor variations in accumulation of each splice variant then present in TAIR database. First, an additional transcript, which we named UBA2a.4 (At3g56860.4, 2,688 bp), was detected in addition to UBA2a.2 when UBA2a.2-specific primers were used. Two new splice variants, named UBA2a.5 (At3g56860.5) and UBA2a.6 (At3g56860.6), were also amplified with UBA2a.3-specific primers. We confirmed the identities of the UBA2a.4, UBA2a.5, and UBA2a.6 splice variants by cloning and sequencing the PCR products (data not shown). We also found a new splice variant of UBA2b (Fig. 6b, e), in addition to the known UBA2b.1 variant (1,782 bp). This additional transcript was named UBA2b.2 (2,302 bp); it was amplified when UBA2b.1-specific primers were used.
RT-PCR analysis of UBA2 splice variants following mechanical wounding. Diagrams of splice variants for the three UBA2 genes (a–c). Time course of gene expression in response to mechanical wounding (d–f). Leaf discs from fully expanded rosette leaves were taken and incubated in buffer. At various times thereafter, leaf discs were harvested for subsequent RNA extraction. Total transcripts of UBA2a, -b and -c (topmost panels in d–f) were amplified using primers in the coding regions. Amplification of each splice variant was accomplished by using gene-specific primer sets (indicated by arrows in panel a). Primer sets for Actin2 gene were used as a control to confirm equal RNA amounts used for cDNA synthesis. Arrowheads indicate the position where possible genomic DNA contamination would occur
To investigate the regulation of alternative splicing by wounding, we performed RT-PCR experiments using total RNA isolated from wounded tissues (i.e. leaf discs of rosette leaves) and different pairs of primers specific to the different splice variants (black arrows in Fig. 6a–c). To confirm the effectiveness of leaf disc excision as a wounding treatment, the expression of WOUND-RESPONSIVE GENE3 (WR3) (At5g50200) was tested under identical conditions (Rojo et al. 1998). The WR3 gene was greatly induced by wounding damage via leaf disc excision (Fig. 6d). Actin2 gene primers were used to confirm that equal amounts of RNA were used for cDNA synthesis throughout the time course (Fig. 6d).
RT-PCR experiments (Fig. 6d–f) and our northern blot analysis performed at the same stage of rosette development (Supplemental Fig. 1) consistently showed that total transcripts of UBA2a and UBA2c, which were amplified from all splice variants by a pair of primers within the coding sequence, are induced in response to mechanical wounding (top panels in each of Fig. 6d–f). The expression level of the UBA2a.1 splice variant (1,899 bp, At3g56860.2) began to increase at 2 h with a maximum level at 8 h. UBA2a total transcripts and UBA2a.2, UBA2a.4, and UBA2a.5 splice variants showed a transient decrease at 30 min.
In multiple experiments, none of the UBA2b-related transcripts showed alterations in amount following wounding, as assessed by this RT-PCR method. As shown in Fig. 6f, of the two UBA2c splice variants, the UBA2c.1 variant (At3g15010.1) but not the UBA2c.2 variant (At3g15010.2) increased following wounding.
To quantify and extend these results, splice variant-specific quantitative real-time PCR (Q-PCR) experiments were performed. We were able to design splice variant-specific Q-PCR primer pairs for all the splice variants of all the UBA2 genes, except for UBA2a.6. In these experiments, transcript levels were evaluated not only in wounded tissues (Fig. 7), but also in distal leaves (Supplemental Fig. 2), to assess whether there was a systemic induction of UBA2 transcript levels after wounding. In addition, because transcript increases following wounding could reflect a touch response, we also applied an established methodology for mechanical stimulation, consisting of gently bending a leaf several times (Lee et al. 2005), was applied, to assess whether a touch stimulus was sufficient for UBA2 gene induction (Supplemental Fig. 2).
RT-QPCR analysis of UBA2 splice variants following mechanical wounding. Accumulation of overall transcripts and individual splice variants of UBA2a, UBA2b, UBA2c, and accumulation of WR3 transcript in leaf discs 2 h (black bars) and 6 h (gray bars) after disc collection. RNA levels are given relative to control leaves collected at the same times and after normalization with an Actin2/8 amplicon
In our Q-PCR experiments, increases greater than 2-fold were considered significant. As shown in Fig. 7, overall UBA2a transcript levels were increased about 4.5-fold in wounded leaf discs at 6 h, due primarily to increases in UBA2a.1 transcripts, with a lesser contribution from UBA2a.4. Overall UBA2c transcript levels increased approximately 3-fold at this time point, with the major contribution from UBA2c.1. Total transcript levels of UBA2b and UBA2b splice variants increased slightly more than 2-fold following wounding; such an increase may not have been detectable by the less sensitive RT-PCR method. Overall, there was very good agreement between our RT-PCR and Q-PCR results, although the time course of the wounding effects seemed somewhat more rapid in the RT-PCR experiments. As expected, WR3 responded strongly to wounding in leaf discs, with a 13.7-fold increase observed at the 6 h time point.
With regard to the touch stimulus, this stimulus does not appear to be effective for UBA2b or UBA2c (Supplemental Fig. 2). UBA2a transcripts exhibit an approximately 2-fold increase 2–6 h after touch, but experimental variability precludes a definitive conclusion as to whether UBA2a is touch-induced. WR3 exhibits moderate touch-induction (3.9- and 3.6-fold, 2 and 6 h after treatment, respectively). None of the three UBA2 transcripts show systemic induction following wounding; WR3 shows moderate systemic induction (Supplemental Fig. 2).
UBA2 transcript response to wounding-related stimuli
Upon wounding, JA biosynthesis is induced, resulting in higher endogenous JA levels (Creelman et al. 1992; Pena-Cortes et al. 1995). Two separate pathways in wound signal transduction have been reported in Arabidopsis; one is JA-dependent and the other is JA-independent (Titarenko et al. 1997; León et al. 1998; Rojo et al. 1998). Thus, some JA-inducible genes are activated by wounding through JA, while other genes, including choline kinase (CK) and WR3, are activated by wounding independently of JA. To test whether the induction of UBA2 transcripts by wounding functions through the JA signal transduction pathway, we monitored UBA2 transcript levels following MeJA treatment by RT-PCR. The expression patterns of several splice variants, including all variants that were shown to respond to wounding in our RT-PCR experiments, are shown in Fig. 8. While the JA-responsive gene, VSP1 (At5g24780; Ellis and Turner 2001), was dramatically induced in response to our MeJA treatment, no significant induction of any UBA2 total transcript or splice variant by MeJA treatment was seen that was greater than that seen with the water control. Use of splice variant-specific primers further confirmed that none of the six UBA2a splice variants were regulated by MeJA (data not shown). These results imply that the induction of UBA2a.1 and UBA2c.1 transcripts upon wounding occurs via a JA-independent signaling pathway.
UBA2 mRNA expression is not hormonally regulated. Arabidopsis plants were treated with MeJA (0.01%) (a), ethylene (5 ppm) (b) or ABA (100 μM) (c) and rosette leaves were collected at the indicated time points after treatment. As a control, water was applied to the plants. Primer sets for Actin2 gene were used as a control to confirm equal RNA amounts used for cDNA synthesis. Arrowheads indicate the position where possible genomic DNA contamination would occur
Ethylene and ABA are necessary for activation of the proteinase inhibitor II gene (Pin2) upon wounding in tomato (O’Donnell et al. 1996) and potato (Pena-Cortes et al. 1995), and we also observed that ABA induces UBA2a and UBA2b relocalization into nuclear speckles (Fig. 3a, b). Therefore, we also examined whether UBA2 transcript levels are responsive to these hormones (Fig. 8b, c). However, neither ABA nor ethylene induced the expression of any UBA2 genes, although the ethylene-responsive transcript, HEL1 and the ABA-responsive transcript, Rab18 were clearly induced as expected. Ozone treatment also did not change levels of any of the UBA2 transcripts (data not shown).
Gene induction in response to oligosaccharides via oligosaccharide-dependent (OSD) pathways is one JA-independent pathway of wounding and pathogen response (Rojo et al. 1999; León et al. 2001). Thus, we assessed UBA2 transcript levels after applying a standard oligogalacturonide (OGA) treatment (Rojo et al. 1999). However, no elevation of transcript levels for any of the UBA2 genes was observed, while the OGA-regulated transcripts, CK1 and WR3, accumulated as expected (Fig. 9).
UBA2 mRNA expression is not regulated by OGAs. Ten-day-old Arabidopsis seedlings grown in liquid culture were treated with 250 μg ml−1 OGAs or water and collected at the indicated time points (in h) for RT-PCR analyses of UBA2a (all splice variants), UBA2a.1, UBA2b (all splice variants), UBA2c (all splice variants) and UBA2c.1 transcript accumulation. RT-PCR was performed on Actin2 transcript to confirm equal RNA amounts used for RT-PCR, and on CK1 and WR3 transcripts to assess the effectiveness of the treatment. In control experiments (not shown) in which the reverse transcriptase step was omitted, no genomic DNA contamination was detected
We also examined whether UBA2 transcripts are responsive to bacterial pathogen infection. For bacterial infection, an avirulent bacterial strain of Pseudomonas syringae pv tomato DC3000 (avrRpt2) was inoculated in leaves of soil-grown 5-week-old Arabidopsis plants. However, no induction of any UBA2 transcript was detected in response to bacterial inoculation (Fig. 10). A positive control, the PR-1 gene (At2g19990), showed a dramatic induction pattern at 24 h post-inoculation of bacteria, as expected. We note that accumulation of UBA2a.1 and UBA2c.1 transcripts at 6 h was detected, however, such accumulation was also observed in the mock inoculation and thus presumably results from wounding incurred during the inoculation process.
UBA2 transcripts are not induced by avirulent bacterial pathogen. Rosette leaves of 5-week-old Arabidopsis plants were challenged with P.syringae pv tomato DC3000 (avrRpt2) and harvested at the indicated time points for RNA extraction. Primer sets for Actin2 gene were used as a control to confirm equal RNA amounts used for cDNA synthesis. Arrows indicate the second splice variant (UBA2b.2) derived from the UBA2b gene and revealed during PCR when UBA2b.1-based primers were used. Arrowheads indicate the position where possible genomic DNA contamination would occur
Discussion
UBA2 proteins are RNA-binding proteins
Forty-four hnRNP-like proteins are found in the Arabidopsis genome according to sequence analysis, including the three UBA2 proteins (Wang and Brendel 2004). Arabidopsis UBA2 proteins share highest similarity with hnRNP D and hnRNP A/B mammalian proteins. hnRNP proteins play roles in diverse aspects of mRNA metabolism, including pre-mRNA splicing, mRNA localization, mRNA stability, nuclear export of mRNA, and translational control (Dreyfuss et al. 1996, 2002; Krecic and Swanson 1999; Mili et al. 2001; Reed and Magni 2001). As noted by Lorković and Barta (2002), UBA2 proteins are plant-specific hnRNP proteins with no real orthologues in metazoan genomes (Lambermon et al. 2002).
Our RNA homoribopolymer binding assays showed that UBA2 proteins can indeed interact in vitro with RNAs. UBA2a and UBA2b show a possible preference for U-rich and G-rich sequences, while UBA2c has strong affinity for all four homoribopolymers. The binding specificities of UBA2a are different than those reported by Lambermon and colleagues (2002) who, using UV cross-linking experiments, showed a strong specificity of UBA2a for poly(U) alone. The difference between this result and ours may be due to the fact that, in the UV cross-linking experiment, homoribopolymers were used as competitors of an arbitrary CaMV 3′-UTR RNA while, in the homoribopolymer binding assays, binding preferences were measured directly at more stringent (higher salt) concentrations and without any competitor (this paper; Domon et al. 1998; Lambermon et al. 2002).
Affinity without specificity for the four homoribopolymers has been previously shown for GR-RBP4, an RNA-binding protein whose transcript level increases in response to abiotic stresses (Kwak et al. 2005), while poly(U) and poly(G) selectivity have been observed for other RRM-containing RNA-binding proteins such as Physcomitrella patens PpGRP1, PpGRP2 and PpGRP3 whose expression is induced by cold, Nicotiana sylvestris RGP-1, maize MA16 which is expressed during late embryogenesis and responds to ABA, and Arabidopsis AtRZ-1a which is involved in cold stress response and freezing tolerance (Hirose et al. 1993; Ludevid et al. 1992; Nomata et al. 2004; Kim et al. 2005). The role of the RRMs in UBA2a RNA-binding capacity has been demonstrated by the fact that the RNA binding ability of UBA2a was decreased by approximately 90% upon site-directed mutation of the aromatic residues in the RNP1 motifs to alanines (Lambermon et al. 2002).
UBA2 proteins localize to the nucleus
Our results show that all three UBA2-GFP proteins localize to the nucleus. Nuclear localization has also been observed for UBA2a both by GFP-fusion and immunodetection (Lambermon et al. 2002; Riera et al. 2006) and for UBA2c by proteomic analysis (Brown et al. 2004). Many mammalian hnRNPs shuttle between the nucleus and cytoplasm (Dreyfus et al. 2002). We occasionally observed localization of each of the UBA2-GFPs in the cytosol; however, because in only a few of these cases did the cell appear undamaged, it remains premature to conclude that such localization occurs in the normal situation in planta.
Within the nucleus, we observed that UBA2c-GFP was detected both throughout the nucleoplasm and also as strong fluorescent dots constituting a speckled pattern. Nuclear speckles, also known as interchromatin granule clusters or ‘splicing factor compartments’, are dynamic nuclear structures of variable size (from 0.4 to 1.5 μm in diameter in Arabidopsis (Fang et al. 2004)) and irregular shape (Lamond and Spector 2003). Situated in close proximity to transcriptionally active genes (Shopland et al. 2003), speckles are believed to be storage or modification/assembly sites of spliceosomal components recruited to transcription sites (Lamond and Spector 2003; Fang et al. 2004) and could also be the site of transcription-, splicing- and transport-related processes (Shopland et al. 2002 and references therein). Some of the plant RRM-containing splicing factors (Lopato et al. 2002; Ali et al. 2003; Docquier et al. 2004; Fang et al. 2004; Tillemans et al. 2005) and the Vicia faba UBA2 orthologue AKIP1 (Li et al. 2002; Ng et al. 2004) have been observed in speckles. We previously reported a rapid increase in the number of VfAKIP1 nuclear speckles in response to ABA, reflecting the activation of this hnRNP in response to a hormonal signal (Li et al. 2002; Ng et al. 2004). In the present studies, ABA-induction of UBA2 localization in apparent nuclear speckles was also observed for UBA2a and UBA2b, as also reported for UBA2a by Riera et al. (2006). By contrast, UBA2c typically exhibited localization in speckle-like structures even in the absence of ABA treatment (Fig. 3c), an observation also reported in the Arabidopsis nucleolar protein database (http://bioinf.scri.sari.ac.uk/cgi-bin/atnopdb/home; Brown et al. 2004). ABA-induced subnuclear speckling of VfAKIP1 is inhibited both by inhibitors of transcription and by Ca2+ chelators (Ng et al. 2004), suggesting that interaction between VfAKIP1 and its mRNA targets is needed for its ABA-dependent relocalization. It would be interesting to test the dependence of UBA2a and UBA2b subnuclear organization on calcium-regulated gene transcription. In addition, while the subnuclear structures visualized by the GFP-tagged UBA2a, UBA2b, and UBA2c proteins have the characteristic appearance of nuclear speckles, it will be important to confirm that the UBA2 proteins indeed co-localize with splicing factors or other proteins known to typify these subnuclear domains.
Expression of UBA2 splice variants
The transcript accumulation of UBA2a and UBA2c genes was detected by northern blot analyses in all organs tested. UBA2b transcripts were barely detectable by northern blot but could be detected by RT-PCR, indicating a lower expression level relative to the other two UBA2 genes. GUS staining experiments showed that UBA2a and UBA2c are expressed in stamens. The expression detected in the leaves appears stronger in the young leaves for UBA2a and UBA2c. A weak but clear GUS staining was observed in the shoot meristem area for UBA2b and a strong staining was detected in the root apical and lateral meristems for UBA2c.
An interesting feature of some RNA processing genes is that their mRNAs are themselves subjected to posttranscriptional controls. Three tobacco RNA-binding Glycine-rich Protein-1 genes (RGP-1a, 1b and 1c) are alternatively spliced, two of them in a tissue-specific manner (Hirose et al. 1993). The FCA transcript is subject to alternative splicing (Macknight et al. 2002) and in this process the FCA protein negatively regulates its own mRNA by promoting premature cleavage and polyadenylation (Quesada et al. 2003). This FCA pre-mRNA processing is negatively regulated by ABA (Razem et al. 2006). AtGRP7 protein and transcript are under the control of the circadian clock (Heintzen et al. 1997). AtGRP7 can bind AtGRP7 mRNA in vitro and the overexpression of AtGRP7 depresses the oscillation of AtGRP7 and AtGRP8 transcripts and promotes a change in splice site within these mRNAs, generating unstable transcripts with a premature stop codon (Heintzen et al. 1997; Staiger et al. 2003; Schöning et al. 2007).
Our analysis reveals that alternative splicing is also a mechanism of UBA2 gene expression control. Splice variants for two of the genes were present in Genbank and we have confirmed these and also identified new splice variants for UBA2a and UBA2b by RT-PCR. Alternative splicing is apparent only in the 3′-UTRs and in fact none of the UBA2 genes contain introns in their coding regions. We observed the preferential accumulation of one of the splice variants of UBA2a (UBA2a.1) and one of the splice variants of UBA2c (UBA2c.1) in response to wounding (see below). This result shows that UBA2 gene expression is at least partly controlled at the posttranscriptional level through alternative splicing of the 3′-UTR. In addition, given environmental (wounding) regulation of splice variant abundance, it is possible that still more splice variants of the UBA2 genes await discovery in plants grown under different environmental regimes.
The different UTRs of the splice variants, by their sequences, structures, and polyadenylation sites, contain a unique set of information that may influence the stability and fate of each transcript. Cis-acting sequences important for RNA stability are indeed known to frequently reside in 3′-UTRs (Hollams et al. 2002) and evidence that 3′-UTRs can have a major role in posttranscriptional regulation through control of mRNA stability has been obtained in plants (Chan and Yu 1998; Ortega et al. 2006). One previous example in which an environmental stress has been shown to regulate alternative splicing in the 3′-UTR is provided by the CLT cold-responsive gene of trifoliate orange (Jia et al. 2004). Recently it has been shown that a riboswitch present in the 3′-UTR of the Arabidopsis thiamin biosynthetic gene THIC senses thiamin pyrophosphate (TPP) and thereby controls the formation of transcripts with alternatively spliced 3′ UTRs; this alternative splicing in turn affects transcript stability and protein production (Bocobza et al. 2007; Wachter et al. 2007).
Bioinformatic analyses reveal that about 9% of transcripts contain introns in their UTR regions (Zhu et al. 2003). Alternative splicing in the 5′ and 3′ UTRs represents 22–29% of the total alternative splicing events (Ner-Gaon et al. 2004; Reddy 2007) and this high rate of alternative splicing of introns present in UTR regions suggests a regulatory role for such events in RNA metabolism (Ner-Gaon et al. 2004). However, of this 22–29% only 6% are estimated to occur in 3′-UTRs (Reddy 2007), suggesting that the UBA2 transcript variants exhibit regulation by a relatively unusual mechanism.
Regulation of UBA2 splice variant accumulation by wounding
UBA2a and UBA2c transcript accumulation is enhanced after mechanical wounding. Based on RT-PCR analysis using primers that enabled identification of specific splice variants, we were able to attribute the overall accumulation of UBA2a transcript primarily to just one of the six splice variants, UBA2a.1, which is generated by an intron retention and a premature polyadenylation site. Wounding increase the level of the spliced form of UBA2c (UBA2c.1). Because the RT-PCR product corresponding to the unspliced transcript (UBA2c.2) would merge with genomic DNA contamination, if it were present in our RT-PCR analyses, it was difficult to definitely monitor the level of this transcript (Fig. 6f). However, the absence of genomic contamination in our Q-PCR experiments (assessed by the absence of any amplification product in reactions performed on non-reverse transcribed RNAs; data not shown) demonstrated that UBA2c.2 transcript levels in fact did not increase significantly following wounding (Fig. 7).
UBA2a (UBA2a.1 splice variant) and UBA2c (UBA2c.1 splice variant) are wounding responsive genes. They thus join a small group of plant genes encoding RRM-containing proteins whose transcript accumulation is affected upon wounding: maize MA16 and CHEM2 (Gómez et al. 1988; Didierjean et al. 1996), carrot DcGRP-1 (Sturm 1992; gene subsequently named by Sachetto-Martins et al. 2000), tomato clone B1-20 (chloroplast mRNA-binding protein; Vian et al. 1999), and tobacco clone MTL-8 (daSilva et al. 2002).
While alternative splicing in response to biotic stressors and temperature is a well-documented phenomenon, only a few transcripts to date have been reported to be alternatively spliced in response to wounding (reviewed in Reddy 2007). Titarenko et al. (1997) observed a switch in the accumulation of splicing variants for VSP2 and JR1 genes, between 2 and 8 h after wounding, and it is interesting to speculate that UBA2 gene expression might be under a similar control. The peach homologue of the ethylene receptor, PpETR1, has a splice variant by intron retention that responds more rapidly (1 h) than the fully spliced transcript (4 h) to wounding in the fruit (Bassett et al. 2002). The significance of this regulation has not been addressed.
Because the alternative splicing of UBA2 genes affects only the 3′-UTRs, the specific accumulation patterns of UBA2a.1 and UBA2c.1 splice variants when UBA2a and UBA2c expression increases following wounding raises the possibility that UBA2a and UBA2c transcripts are themselves targets of UBA2 proteins. UBA2 proteins could either be involved in alternative splicing or preferentially stabilize transcripts as a result of the differential 3′-UTR composition.
An estimated ∼6% of alternative splicing events in Arabidopsis occur in the 3′-UTR (Reddy 2007), and such events have been documented to affect transcript stability and translation in mammalian systems (e.g. Chowdhury et al. 2005; Banihashemi et al. 2006; Thiele et al. 2006). Lambermon et al. (2002) originally identified UBA2a, one of the three proteins studied in this report, by two-hybrid screens with UBP1, a hnRNP-like protein involved in mRNA splicing and stability (Lambermon et al. 2000). Their results suggested that a major function of UBA2a may be to stabilize RNAs: UBA2a overexpression in N. plumbaginifolia protoplasts increased the steady-state level of a reporter RNA but did not affect splicing of the particular reporter constructs tested (Lambermon et al. 2002). It is of interest that Arabidopsis RNases are, like UBA2s, wound-induced (LeBrasseur et al. 2002). One speculative hypothesis is that increased stability conferred to particular transcripts by UBA2 induction following wounding counterbalances or protects these transcripts against the increase in general RNase activity that occurs following wounding.
Regulation of UBA2 transcripts by wounding-associated signals
Transcriptome data from microarray analyses show that many wound-responsive genes are also pathogen responsive genes (Cheong et al. 2002). However, this does not appear to be the case for the UBA2 genes as we did not observe any significant variation in the UBA2 transcript levels in response to an avirulent strain of Pseudomonas syringae (avrRpt2) when compared to the control plant.
We could not definitely ascertain whether mechanical stimulation without wounding was sufficient for transcript induction: in some experiments (e.g. Fig. 7) spraying with water appeared to increase transcript levels, and this was the method by which the original TOUCH genes were identified (Braam and Davis 1990). However, use of a standard touch stimulus consisting of leaf bending (Lee et al. 2005) did not reliably elevate levels of any of the UBA2 transcripts (Supplemental Fig. 2).
Neither MJ, ABA nor ethylene seem to participate in transducing the wound stimulus, as UBA2 transcript levels were unresponsive to application of these hormones, despite the high concentrations used. These three hormones were indeed tested because of their demonstrated roles in wounding response. Two distinct pathways have been identified in wounding response in plants, the JA-dependent and the OSD pathways (Rojo et al. 1999; León et al. 2001). Ethylene acts as a cross regulator between these two pathways and at the local response level, represses the JA-dependent pathway (Rojo et al. 1999). ABA does not appear to be a primary signal in wound signaling (Birkenmeier and Ryan 1998) and its role may be related to the desiccation resulting from wounding (Reymond et al. 2000; Delessert et al. 2004). The oligosaccharides involved in the OSD pathway are oligogalacturonides (OGAs) probably released from cell walls by mechanical or enzymatic disruption of pectic components (Benhamou et al. 1990). On the one hand, OGAs induce the expression of JA-independent wounding response genes such as acyl CoA oxidase (Aco), CK and WR3. On the other hand, OGAs repress JA-dependent gene expression in locally damaged tissues through an ethylene production- and perception-dependent pathway (Titarenko et al. 1997; Rojo et al. 1999; Cheong et al. 2002). However, overall UBA2 transcript levels, as well as levels of the wounding-responsive splice variants, were unresponsive to OGAs. Therefore, the position of the UBA2 transcripts in the current models of gene regulation in response to wounding in Arabidopsis (Rojo et al. 1999; León et al. 2001) is still unknown. It is of interest that wound-induced accumulation of RNS1 and other RNase activities was similarly shown to occur independently of JA and oligosaccharides (LeBrasseur et al. 2002). These results suggest that transcripts of RNA-binding proteins could be regulated by wounding via a novel pathway.
Conclusions
Our homoribopolymer-binding assays demonstrated the RNA-binding capabilities of each of the UBA2s, as predicted from sequence analysis. Our observation of the nuclear-localization of UBA2a, UBA2b, and UBA2c suggests that these hnRNPs do not affect translational processes, but rather participate in nuclear-localized RNA processing events. The functions of UBA2a and UBA2b but not UBA2c proteins may be regulated by ABA, given that ABA reorganizes distribution of the first two proteins within the nucleus. This observation, along with the greater sequence similarity of UBA2a and UBA2b as compared to UBA2c suggests that UBA2a and UBA2b may function redundantly; indeed Riera et al. (2006) detected no alteration in ABA-responsiveness of uba2a T-DNA mutants.
We characterized previously unknown UBA2 splice variants and demonstrated that specific splice variants of UBA2a and UBA2c are increased in a hormone-independent manner by wounding. Our results imply targeted post-transcriptional regulation of these specific splice variants by the wounding stimulus, possibly by a mechanism involving interaction between UBA2 3′-UTRs and UBA2 proteins themselves, given precedence for such self-regulation by the Arabidopsis RNA-binding proteins FCA and AtGRP7. The signaling mechanism that initiates such regulation following wounding appears novel, as none of the wounding-associated plant hormones suffice to alter UBA2 expression levels. In the future, functional analyses by reverse genetics and the identification of the RNA targets of UBA2 proteins will further our understanding of the biological and biochemical functions of the UBA2 proteins in stress responses.
References
Albà MM, Pagès M (1998) Plant proteins containing the RNA-recognition motif. Tr Plant Sci 3:15–21
Ali GS, Golovkin M, Reddy ASN (2003) Nuclear localization and in vivo dynamics of a plant-specific serine/arginine-rich protein. Plant J 36:883–893
Banihashemi L, Wilson GM, Das N, Brewer G (2006) Upf1/Upf2 regulation of 3′ untranslated region splice variants of AUF1 links nonsense-mediated and A + U-rich element-mediated mRNA decay. Mol Cell Biol 26:8743–8754
Bassett CL, Artlip TS, Callahan AM (2002) Characterization of the peach homologue of the ethylene receptor, PpETR1, reveals some unusual features regarding transcript processing. Planta 215:679–688
Benhamou N, Chambreland H, Pauzé FJ (1990) Implication of pectic components in cell surface interactions between tomato root cells and Fusarium oxysporum f. sp. radicis-lycopersici. Plant Physiol 92:995–1003
Birkenmeier FG, Ryan CA (1998) Wound signaling in tomato plants. Evidence that ABA is not a primary signal for defense gene activation. Plant Physiol 117:687–693
Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A (2007) Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev 21:2874–2879
Braam J, Davis RW (1990) Rain-, wind- and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell 60:357–364
Brown JWS, Shaw PJ, Shaw P, Marshall DF (2004) Arabidopsis nucleolar protein database (AtNoPDB). Nucl Acids Res 33:633–636
Carrington JC (1995) Targeting of proteins to the nucleus. In: Galbraith DW, Bourque DP, Bohnert HJ (eds) Methods in plant cell biology. Academic Press, San Diego, pp 283–293
Chan M-T, Yu S-M (1998) The 3′ untranslated region of a rice α-amylase gene functions as a sugar-dependent mRNA stability determinant. Proc Natl Acad Sci USA 95:6543–6547
Charrier B, Champion A, Henry Y, Kreis M (2002) Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase polymerase chain reaction. Plant Physiol 130:577–590
Cheong YH, Chang H-S, Gupta R, Wang X, Zhu T, Jan S (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol 129:661–677
Chowdhury B, Tsokos CG, Krishnan S, Robertson J, Fisher CU, Warke RG, Warke VG, Nambiar MP, Tsokos GC (2005) Decreased stability and translation of T cell receptor zeta mRNA with an alternatively spliced 3′-untranslated region contribute to zeta chain down-regulation in patients with systemic lupus erythematosus. J Biol Chem 280:18959–18966
Clark DG, Richards C, Hilioti Z, Lind-Iversen S, Brown K (1997) Effect of pollination on accumulation of ACC synthase and ACC oxidase transcripts, ethylene production and flower petal abscission in geranium (Pelargonium x hortorum L.H. Bailey). Plant Mol Biol 34:855–865
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81:1991–1995
Creelman RA, Tierney ML, Mullet JE (1992) Jasmonic acid/methyl jasmonate accumulate in wounded soybean hypocotyls and modulate wound gene expression. Proc Natl Acad Sci USA 89:4938–4941
daSilva I, Angelo PCS, Molfetta JB, Ferraz MT, daSilva LLP, Goldman GH, Goldman MHS (2002) A tobacco cDNA reveals two different transcription patterns in vegetative and reproductive organs. Braz J Med Biol Res 35:861–868
Delessert D, Wilson I, Van Der Straeten D, Dennis E, Dolferus R (2004) Spatial and temporal analysis of the local response to wounding in Arabidopsis leaves. Plant Mol Biol 55:165–181
Didierjean L, Frendo P, Nasser W, Genot G, Marivet J, Burkard G (1996) Heavy-metal-responsive genes in maize: identification and comparison of their expression upon various forms of abiotic stress. Planta 199:1–8
Docquier S, Tillemans V, Deltour R, Motte P (2004) Nuclear bodies and compartmentalization of pre-mRNA splicing factors in higher plants. Chromosoma 112:255–266
Domon C, Lorković ZJ, Juan Valcárcel J, Filipowicz W (1998) Multiple forms of the U2 small nuclear ribonucleoprotein auxiliary factor U2AF subunits expressed in higher plants. J Biol Chem 273:34601–34610
Dreyfuss G, Hentze M, Lamond AI (1996) From transcript to protein. Cell 85:963–972
Dreyfuss G, Kim VN, Kataoka N (2002) mRNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3:195–205
Ellis C, Turner JG (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13:1025–1033
Fang Y, Hearn S, Spector DL (2004) Tissue-specific expression and dynamic organization of SR splicing factors in Arabidopsis. Mol Biol Cell 15:2664–2673
Gómez J, Sánchez-Martínez D, Stiefel V, Rigau J, Puigdomènech P, Pagès M (1988) A gene induced by the plant hormone abscisic acid in response to water stress encodes a glycine-rich protein. Nature 334:262–264
Heintzen C, Nater M, Appel K, Staiger S (1997) AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc Natl Acad Sci USA 94:8515–8520
Hirose T, Sugita M, Sugiura M (1993) cDNA structure, expression and nucleic acid-binding properties of three RNA-binding proteins in tobacco: occurrence of tissue-specific alternative splicing. Nucl Acids Res 21:3981–3987
Hollams MH, Giles KM, Thomson AM, Leedman PJ (2002) mRNA stability and the control of gene expression: implications for human disease. Neurochem Res 27:957–980
Jambunathan N, McNellis TW (2003) Regulation of Arabidopsis COPINE 1 gene expression in response to pathogens and abiotic stimuli. Plant Physiol 132:1370–1381
Jia Y, del Rio HS, Robbins AL, Louzada ES (2004) Cloning and sequence analysis of a low temperature-induced gene from trifoliate orange with unusual pre-mRNA processing. Plant Cell Rep 23:159–166
Kim CY, Koo YD, Jin JB, Moon BC, Kang CH, Kim ST, Park BO, Lee SY, Kim ML, Hwang I, Kang KY, Bahk JD, Lee SY, Cho MJ (2003) Rice C2-domain proteins are induced and translocated to the plasma membrane in response to a fungal elicitor. Biochemistry 42:11625–11633
Kim YO, Kim JS, Kang H (2005) Cold-inducible zinc finger-containing glycine-rich RNA-binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. Plant J 42:890–900
Krecic AM, Swanson MS (1999) hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol 11:363–371
Kwak KJ, Kim YO, Kang H (2005) Characterization of transgenic Arabidopsis plants overexpressing GR-RBP4 under high salinity, dehydration, or cold stress. J Exp Bot 56:3007–3016
Lambermon MHL, Simpson GG, Kirk DA, Hemmings-Mieszcak M, Klahre U, Filipowicz W (2000) UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J 19:1638–1649
Lambermon MHL, Fu Y, Wieczorek Kirk DA, Dupasquier M, Filipowicz W, Lorković ZJ (2002) UBA1 and UBA2, two proteins that interact with UBP1, a multifunctional effector of pre-mRNA maturation in plants. Mol Cell Biol 22:4346–4357
Lamond AI, Spector DL (2003) Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol 4:605–612
LeBrasseur ND, MacIntosh GC, Pérez-Amador MA, Sitoh M, Green PJ (2002) Local and systemic wound-induction of RNase and nuclease activities in Arabidopsis: RNS1 as a marker for a JA-independent systemic signaling pathway. Plant J 29:393–403
Lee D, Polisensky DH, Braam J (2005) Genome-wide identification of touch- and darkness-regulated Arabidopsis genes: a focus on calmodulin-like and XTH genes. New Phytol 165:429–444
León J, Rojo E, Titarenko E, Sanchez-Serrano JJ (1998) Jasmonic acid-dependent and -independent wound signal transduction pathways are differentially regulated by Ca2+/calmodulin in Arabidopsis thaliana. Mol Gen Genet 258:412–419
León J, Rojo E, Sánchez-Serrano JJ (2001) Wound signalling in plants. J Exp Bot 52:1–9
Li J, Wang XQ, Watson MB, Assmann SM (2000) Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science 287:300–303
Li J, Kinoshita T, Pandey S, Ng CK-Y, Gygi SP, Shimazaki K, Assmann SM (2002) Modulation of an RNA-binding protein by abscisic-acid-activated protein kinase. Nature 418:793–797
Lim M-H, Kim J, Kim Y-S, Chung K-S, Seo Y-H, Lee I, Kim J, Hong CB, Kim H-J, Park C-M (2004) A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16:731–740
Lopato S, Forstner C, Kalyna M, Hilscher J, Langhammer U, Indrapichate K, Lorković ZJ, Barta A (2002) Network of interactions of a novel plant-specific Arg/Ser-rich protein, atRSZ33, with atSC35-like splicing factors. J Biol Chem 277:39989–39998
Lorković ZJ, Barta A (2002) Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucl Acids Res 30:623–635
Ludevid MD, Freire MA, Gómez J, Burd CG, Albericio F, Giralt E, Dreyfuss G, Pagès M (1992) RNA binding characteristics of a 16kDA glycine-rich protein from maize. Plant J 2:999–1003
Macknight R, Bancroft I, Page T, Lister C, Schmidt R, Love L, Westphal L, Murphy G, Sherson S, Cobbett C, Dean C (1997) FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89:737–745
Macknight R, Duroux M, Laurie R, Dijkwel P, Simpson G, Dean C (2002) Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 14:877–888
Mili S, Shu HJ, Zhao Y, Pinol-Roma S (2001) Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: candidate intermediates in formation and export of mRNA. Mol Cell Biol 21:7307–7319
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15:473–497
Ner-Gaon H, Halachmi R, Svaldi-Goldstein S, Rubin E, Ophir R, Fluhr R (2004) Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J 39:877–885
Ng CK-Y, Kinoshita T, Pandey S, Shimazaki K, Assmann SM (2004) Abscisic acid induces rapid subnuclear reorganization in guard cells. Plant Physiol 134:1327–1331
Nomata T, Kabeya Y, Sato N (2004) Cloning and characterization of glycine-rich RNA-binding protein cDNAs in the moss Physcomitrella patens. Plant Cell Physiol 45:48–56
O’Donnell PJ, Calvert C, Atzorn R, Wasternack C, Leyser HMO, Bowles DJ (1996) Ethylene as a signal mediating the wound response of tomato plants. Science 274:1914–1917
Ortega JL, Moguel-Esponda S, Potenza C, Conklin CF, Quintana A, Sengupta-Gopalan C (2006) The 3′ untranslated region of a soybean cytosolic glutamine synthetase (GS1) affects transcript stability and protein accumulation in transgenic alfalfa. Plant J 45:832–846
Pena-Cortes H, Fisahn J, Willmitzer L (1995) Signals involved in wound-induced proteinase inhibitor II gene expression in tomato and potato plants. Proc Natl Acad Sci USA 92:4106–4113
Quesada V, Macknight R, Dean C, Simpson GG (2003) Autoregulation of the site of 3 end formation in FCA pre-mRNA prevents precocious flowering. EMBO J 22:3142–3152
Razem FA, El-Kereamy A, Abrams SR, Hill RD (2006) The RNA-binding protein FCA is an abscisic acid receptor. Nature 439:290–294
Reddy ASN (2007) Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol 58:267–294
Reed R, Magni K (2001) A new view of mRNA export: separating the wheat from the chaff. Nat Cell Biol 3:E201–E204
Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12:707–719
Riera M, Redko Y, Leung J (2006) Arabidopsis RNA-binding protein UBA2a relocalizes into nuclear speckles in response to abscisic acid. FEBS Lett 580:4160–4165
Rojo E, Titarenko E, Leon J, Berger S, Vancanneyt G, Sanchez-Serrano JJ (1998) Reversible protein phosphorylation regulates jasmonic acid-dependent and -independent wound signal transduction pathways in Arabidopsis thaliana. Plant J 13:153–165
Rojo E, León J, Sánchez-Serrano JJ (1999) Cross-talk between wound signalling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J 20:135–142
Sachetto-Martins G, Franco LO, de Oliveira DE (2000) Plant glycine-rich proteins: a family or just proteins with a common motif? Biochim Biophys Acta 1492:1–14
Schomburg FM, Patton DA, Meinke DW, Amasino RM (2001) FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell 13:1427–1436
Schöning JC, Streitner C, Page DR, Hennig S, Uchida K, Wolf E, Furuya M, Staiger D (2007) Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J 52:1119–1130
Shopland LS, Johnson CV, Lawrence JB (2002) Evidence that all SC-35 domains contain mRNAs and that transcripts can be structurally constrained within these domains. J Struct Biol 140:131–139
Shopland LS, Johnson CV, Byron M, McNeil J, Lawrence JB (2003) Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J Cell Biol 162:981–990
Simpson GG (2004) The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr Opin Plant Biol 7:570–574
Spiro MD, Kates KA, Koller AL, O’Neill MA, Albersheim P, Darvill A (1993) Purification and characterization of biologically active 1,4-linked α-D-oligogalacturonides after partial digestion of polygalacturonic acid with endopolygalacturonase. Carbohydr Res 247:9–20
Staiger D, Zecca L, Wieczorek Kirk DA, Appel K, Eckstein L (2003) The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J 33:361–371
Sturm A (1992) A wound-inducible glycine-rich protein from Daucus carota with homology to single-stranded nucleic acid-binding proteins. Plant Physiol 99:1689–1692
Thiele A, Nagamine Y, Hauschildt S, Clevers H (2006) AU-rich elements and alternative splicing in the beta-catenin 3′UTR can influence the human beta-catenin mRNA stability. Exp Cell Res 312:2367–2378
Tillemans V, Dispa L, Remacle C, Collinge M, Motte P (2005) Functional distribution and dynamics of Arabidopsis SR splicing factors in living plant cells. Plant J 41:567–582
Titarenko E, Rojo E, León J, Sánchez-Serrano JJ (1997) Jasmonic acid-dependent and independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol 115:817–826
Vian A, Henry-Vian C, Davies E (1999) Rapid and systemic accumulation of chloroplast mRNA-binding protein transcripts after flame stimulus in tomato. Plant Physiol 121:517–524
Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR (2007) Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell 19:3437–3450
Wang B-B, Brendel V (2004) The ASRG database: identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol 5:R102
Zhu W, Schlueter SD, Brendel V (2003) Refined annotation of the Arabidopsis genome by complete expressed sequence tag mapping. Plant Physiol 132:469–484
Acknowledgments
This research was supported by NSF grant MCB-03-45251. We thank Tzuu-fen Lee for providing total RNA samples inoculated by P. s. t. DC3000 (avrRpt2). We thank Dr. Kathleen Brown for help with gas chromatography. We thank Dr. Zdravko Lorković for giving us the UBA2a-GFP clone, Dr. Mark Spiro for providing the OGAs, and Dr. Philip Bevilacqua for helpful comments on the research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jérôme Bove and Cha Young Kim contributed equally to this work.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Bove, J., Kim, C.Y., Gibson, C.A. et al. Characterization of wound-responsive RNA-binding proteins and their splice variants in Arabidopsis. Plant Mol Biol 67, 71–88 (2008). https://doi.org/10.1007/s11103-008-9302-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-008-9302-z