Abstract
The Glycine max sucrose binding protein (GmSBP2) promoter directs vascular tissue-specific expression of reporter genes in transgenic tobacco. Here we showed that an SBP2-GFP fusion protein under the control of the GmSBP2 promoter accumulates in the vascular tissues of vegetative organs, which is consistent with the proposed involvement of SBP in sucrose transport-dependent physiological processes. Through gain-of-function experiments we confirmed that the tissue-specific determinants of the SBP2 promoter reside in the distal cis-regulatory domain A, CRD-A (position −2000 to −700) that is organized into a modular configuration to suppress promoter activity in tissues other than vascular tissues. The four analyzed CRD-A sub-modules, designates Frag II (−1785/−1508), Frag III (−1507/−1237), Frag IV (−1236/−971) and Frag V (−970/−700), act independently to alter the constitutive pattern of −92pSBP2-mediated GUS expression in different organs. Frag V fused to −92pSBP2-GUS restored the tissue-specific pattern of the full-length promoter in the shoot apex, but not in other organs. Likewise, Frag IV confined GUS expression to the vascular bundle of leaves, whereas Frag II mediated vascular specific expression in roots. Strong stem expression-repressing elements were located at positions −1485 to −1212, as Frag III limited GUS expression to the inner phloem. We have also mapped a procambium silencer to the consensus sequence CAGTTnCaAccACATTcCT which is located in both distal and proximal upstream modules. Fusion of either repressing element-containing module to the constitutive −92pSBP2 promoter suppresses GUS expression in the elongation zone of roots. Together our results demonstrate the unusual aspect of distal sequences negatively controlling tissue-specificity of a plant promoter.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The sucrose binding protein (SBP) was initially identified in soybean cotyledons by its capacity to bind the sucrose analogue 6′-deoxy-6′-(4-azido-2-hydroxy)-benzamide-sucrose (Ripp et al. 1988). Members of the SBP family have also been identified in pea (Castillo et al. 2000), faba bean (Heim et al. 2001), spinach (Warmbrodt et al. 1989, 1991) and Medicago truncatula (Contim et al. 2003). At the amino acid level, SBP is most related to cupin domain-containing proteins and vicilin-like seed storage proteins (about 35% identity and 50% similarity) from other plant species. In addition to primary sequence conservation, homology-based molecular modeling has shown that SBPs are capable of folding into a cupin tertiary structure and they have therefore been classified as members of the cupin superfamily (Dunwell et al. 2004).
The cupin superfamily corresponds to a functionally diverse family of proteins that comprises seed storage proteins and enzymes as well as proteins that bind different sugars (Dunwell et al. 2000). The Glycine max SBP (GmSBP) binds sucrose (Rocha et al. 2007) and exhibits a low sucrose transport activity in the yeast heterologous system (Grimes and Overvoorde 1996; Overvoorde et al. 1996, 1997; Elmer et al. 2003). Furthermore, GmSBP interacts with GTP in a sucrose translocation independent manner and has been shown to be involved in sucrose-dependent physiological processes in transgenic tobacco plants (Delú-Filho et al. 2000; Pedra et al. 2000; Pirovani et al. 2002). In fact, SBP repression studies in tobacco resulted in some phenotypes typically consistent with inhibition of sucrose transport-translocation between source and sink tissues (Riesmeier et al. 1994; Kühn et al. 1996), such as sugar accumulation in source leaves, photosynthesis inhibition, stunted growth, delayed development, inhibition of sucrose exudation rate from detached source leaves and significant reductions in sucrose and hexose content in sink organs (Delú-Filho et al. 2000; Pedra et al. 2000; Waclawovsky et al. 2006b). Nevertheless, the observed variations in photosynthetic metabolism, sucrose exudation and growth in the tobacco transgenic plants are transient and are restricted to the vegetative phase of development (Waclawovsky et al. 2006b). This temporal restriction of the metabolic effects resulting from SBP repression indicates that SBP is probably functionally associated with temporal changes of sink strength and development rather than being directly involved in sucrose transport from source leaves.
In soybean, SBP is encoded by a small gene family, which is represented by at least two non-allelic genes, GmSBP1 (GeneBank accession number—L06038) and GmS64 cDNAs (AF191299), also designated GmSBP2 (Pirovani et al. 2002). These have extensive sequence similarity but are found at different loci in the soybean genome (Contim et al. 2003). A third isolated GmSBP cDNA is 99% identical to the previously isolated GmS64/GmSBP2 gene and may represent an allelic form (Pirovani et al. 2002; Elmer et al. 2003). Among other legumes, SBP genes have been characterized in pea (Castillo et al. 2000) and Vicia faba (VfSBPL, Heim et al. 2001). However, GmSBP from soybean and VfSBPL from fava bean display distinct expression patterns. While the expression of VfSBPL has been demonstrated to be confined to seeds, GmSBP1 transcripts have also been detected in young sink leaves (Grimes et al. 1992; Heim et al. 2001). Furthermore, the VfSBPL promoter-mediated expression of a reporter gene has been shown to be restricted to cotyledons (Heim et al. 2001). In contrast, the GmS64/GmSBP2 promoter drives expression of linked reporter genes to the vascular tissue of roots, stems and leaves from tobacco transgenic lines (Contim et al. 2003). We have recently shown that the GmSBP2 promoter is functionally organized into a proximal region, with the combinatorial modular configuration of plant promoters, and a distal domain, which restricts gene expression to the vascular tissues in vegetative organs (Waclawovsky et al. 2006a). Here we further characterize the distal region on the GmSBP2 promoter and demonstrate that it harbors discrete repressing cis-regulatory domains that act in a context-independent manner to control the tissue-specific expression of the GmSBP2 promoter. We also analyze the expression of a SBP-GFP translational fusion under the control of the GmSBP2 promoter to confirm the vascular accumulation of SBP protein in vegetative organs.
Materials and methods
Construction of SBP2 promoter–reporter gene constructs
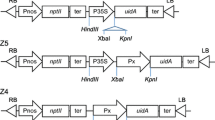
The −2000pSBP2-GUS, −136pSBP2-GUS and −92pSBP2-GUS constructs, which contain a GUS cDNA under the control of 5′-flanking sequences of pgsS641.1 (GmSBP2 genomic clone) up to positions −2000, −136 and −92 relative to the translational initiation codon, respectively, have been described previously (Contim et al. 2003; Waclawovsky et al. 2006a). The CRD-A region of pgsS641.1, spanning nucleotide position −2000 to −700 (Waclawovsky et al. 2006a), was amplified with the appropriate primers (Table 1), and inserted into −136pSBP2-GUS, −92pSBP2-GUS and pCAMBIA1381Z (CAMBIA, Black Mountain, Australia) to generate CRD-A/−136pSBP2-GUS (also designated pUFV652), CRD-A/−92pSBP2-GUS (pUFV651) and CRD-A-GUS (pUFV744), respectively (Fig. 1A). Deletions of the SBP2 CRD-A sequences were obtained by PCR-based mutagenesis using Pfu DNA polymerase (Stratagene, La Jolla, CA, USA) and pgS641.1 as the DNA template. The deleted fragments encompassing sequences −1785 to −1508 (Frag II), −1507 to −1237 (Frag III), −1236 to −971 (Frag IV) and −970 to −700 (Frag V) from the SBP2 promoter were amplified using the appropriate primers (Table 1), digested with EcoRI and HindIII and inserted in the same sites of −92pSBP2-GUS (Waclawovsky et al. 2006a), resulting in the clones pUFV797 (Frag II/−92pSBP2-GUS), pUFV798 (Frag III/−92pSBP2-GUS), pUFV799 (Frag IV/−92pSBP2-GUS) and pUFV800 (Frag V/−92pSBP2-GUS) (Fig. 1B).
DNA constructs used to transform Nicotiana tabacum. Numbers indicate the position relative to the translation start codon. (A) Schematic representation of 5′ flanking sequences of GmSBP2 (gsS641.1) fused to GUS. The CRD-A (−2000a to −700) fragment were generated by PCR-based mutagenesis and cloned into the appropriate sites of −136pSBP2-GUS construct, −92pSBP2-GUS construct and pCAMBIA1381Z. (B) Schematic illustration of CRD-A fragments fused to GUS. Sequential extensions of CRD-A (fragments II, III, IV, V) were isolated by PCR and individually fused to the 5′end of −92pSBP2-GUS. (C) SBP2-GFP fusion, under the control of GmSBP2 promoter. The positions of restriction enzyme sites used for cloning are indicated
To obtain a translational fusion of SBP2 and GFP under control of the SBP2 promoter, the 2-kb SBP2 promoter fragment was amplified with the appropriate primers (Table 1) and inserted into EcoRI/SmaI sites of pCAMBIA1381Z to generate pUFV674. A GFP (green fluorescent protein) cDNA was amplified from pK7FWG2 (Karimi et al. 2002) with the primers eGFPPstR (5′-GCATGCCTGCAGGTCACTGGATTTTGG-3′, coordinates 8 and 34 of pK7FWG2, PstI site underlined) and eGFPSalF (5′-GTGGTGGTCGACATGGTGAGCAAGGGC-3′, positions 978 to 951 of pK7FWG2, SalI site underlined), digested with SalI and PstI, inserted into pUC118 to give pUFV714. The GFP cDNA was then relieved from pUFV714 with SalI and PstI and inserted into the same sites of pUFV674, yielding pUFV760 (also designated −2000pSBP-GFP). Likewise, the SBP2 cDNA was obtained by PCR from pUFV30, (Pirovani et al. 2002), with the primers S64BamF (5′-AGAGGATCCCCGGGTACCGAGCTC-3′, coordinates 93 and 116, creating a BamHI site at position 96) and S64SalNSR (5′-CCTCCACACGTCGACCGCAACAGCGCG-3′, coordinates 1499 and 1473, creating a SalI site at position 1484), inserted into SalI and BamHI sites of pUC118, generating pUFV713 and, then, transferred to the same sites of pUFV760. The resulting clone pUFV763 (also designated −2000pSBP2:SPB2-GFP) contains the SBP2 cDNA fused in-frame to GFP cDNA, under the control of the S-64/SBP2 promoter.
Generation and selection of transgenic plants
The pCAMBIA-derived recombinant plasmids or pCAMBIA1381Z binary vector alone were used to transform Nicotiana tabacum L. cv. Havana plants by Agrobacterium tumefaciens-mediated leaf disc transformation (Alvim et al. 2001) and transformed plants were selected and regenerated on medium containing hygromycin (Buzeli et al. 2002). Most of the rooted plants were tested for the incorporation of the hygromycin (hptII) and SBP2 promoter-GUS fusion genes by PCR analysis and GUS activity, as previously described (Waclawovsky et al. 2006a). The transgenic lines were further selected by the transgene single-copy number criterion as judged by Southern blots. Detailed sectional analyses for tissue-specific expression were carried out on five independent pSBP2-GUS transgenic lines of each construct. One hygromycin-resistant plant for the pCAMBIA1381Z incorporated binary vector was used as a negative control.
Determination of GUS activity and histochemical in situ localization of GUS in tobacco organs
Protein extraction and fluorometric assay for GUS activity were performed as described by Buzeli et al. (2002) with methylumbelliferone (MU) as a standard. The histochemical analysis of β-glucuronidase activity was performed as previously described (McCabe et al. 1988). The tissues (roots, stems and leaves) were sectioned using a hand microtome. Tissue sections were embedded in the GUS assay buffer [100 mM NaH2PO4·H2O (pH 7.0), 0.5 mM K4Fe(CN)6·3H2O, 10 mM Na2EDTA·2H2O, 0.1% (v/v) Triton X-100] containing 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc) (McCabe et al. 1988) and incubated at 37°C in the dark for 4 h. Pigments were extracted from stained tissues with methanol:acetone (3:1). The micrographs were taken under an Olympus AX-70 microscope.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared from soybean and tobacco leaves and stems as previously described (Polanco et al. 2002). The integrity of the nuclear extracts was monitored by SDS-PAGE. Fragment II, encompassing sequences positions −1765 to −1485 on pgsS641.1 was amplified as described above, digested with HindIII and radiolabeled with [α-32P]dCTP and one unit of E. coli DNA polymerase (Klenow fragment). An aliquot of nuclear extract was incubated with the radiolabeled probe in the presence of 2 μg of sonicated salmon sperm DNA and the binding buffer (12 mM Hepes, pH 7.9, 4 mM Tris-Cl, pH 7.9, 60 mM KCl; 1 mM EDTA, 1 mM DTT, 12% (v/v) glycerol) in a final volume of 15 μl for 30 min at room temperature. The reaction was resolved by electrophoresis in 5% acrylamide gel at 35 mA, for 4 h with the running buffer (6.7 mM Tris-Cl, pH 7.9, 3.3 mM sodium acetate, 1 mM EDTA) under circulation. The gel was dried and revealed by autoradiography at room temperature for 6 h.
Real-time RT-PCR analysis
Total RNA was extracted from frozen tissues with TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions and was further purified through silica columns. The quality and integrity of the RNA was monitored by spectrophotometry and agarose gel electrophoresis, respectively. We extracted RNA from leaves, roots and stems at different developmental stages. For the quantitative RT-PCR, 3 μg of total RNA were treated with DNase (Promega, Madison, WI) and fractionated through RNA purification columns (Qiagen, Valencia, CA). Reverse transcription was carried out using M-MLV reverse transcriptase (Invitrogen) and oligo-dT (18, IDT, Coralville, IA) primers (according to the protocol of the manufacturer). We used a set of primers, which was designed to anneal specifically to SBP2 cDNA and another set of primers designed to anneal to all three isolated soybean SBP cDNAs (Table 2).
Real-time RT-PCR reactions were performed on an ABI7500 instrument (Applied Biosystems, Foster City, CA), using SYBR® Green PCR Master Mix (Applied Biosystems). The amplification reactions were performed as follows: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 94°C for 15 s and 60°C for 1 min. To confirm quality and primer specificity, we verified the size of amplification products after electrophoresis through a 1.5% agarose gel, and analyzed the T m (melting temperature) of amplification products in a dissociation curve, performed by the ABI7500 instrument. Gene expression was quantified using the \( 2^{{ - \Delta C_{{{\rm T}}} }} \) method. The RNA helicase was used as a control gene to normalize all values in the real-time RT-PCR assays.
Tissue localization of SBP2-GFP
Confocal imaging was performed using a Zeiss inverted LSM510 META laser scanning microscope with an argon laser and a 40× oil immersion objective. For imaging GFP, the excitation line 488 nm and the 505–530 nm band pass filter were used. The pinhole was usually set to give a 1–1.5 μm optical slice. Post-acquisition image processing was done using the LSM 5 Browser software (Zeiss). We examined samples from ten independently transformed −2000pSBP2:SBP-GFP tobacco lines.
Results
The GmSBP2 promoter directs accumulation of a SBP2-GFP fusion to the vascular tissues of N. tabacum
We have previously demonstrated that the GmSBP2 promoter directs vascular tissue-specific expression of reporter genes in transgenic tobacco (Contim et al. 2003). These results were based on the expression of pSBP2-reporter gene transcriptional fusions in transgenic plants and may not reflect the accumulation of SBP2 protein in soybean tissues due to the lack of transcriptional regulatory elements and post-transcriptional control. To address these possibilities we have expressed a SBP-GFP fusion under the control of the full-length GmSBP2 promoter in transgenic tobacco and analyzed the localization of the recombinant protein through confocal laser scanning microscopy in ten independently transformed lines. In the stem, intense GFP fluorescence was observed in the vascular tissue, both in the xylem tracheary elements and in the phloem fibers (Fig. 2A). In the shoot apex, the fusion protein accumulated in the procambium region (Pc), phloem (P) and xylem (X) (Fig. 2B). The phloem was clearly identified by the presence of the sieve plates between adjacent cells (see inset in Fig. 2B), whereas the xylem was distinguished by the annular and helical secondary wall thickenings. Collectively, these results indicate that the SBP2 protein is in fact located in the vascular system of vegetative organs which is consistent with the pattern of SBP2 promoter activity in transgenic lines as well as with the SBP2 involvement in sucrose translocation-dependent physiological processes.
Confocal images of SBP2-GFP stably expressed in shoot (A) and shoot apex (B) of tobacco plants. First and second columns show plants expressing SBP2-GFP and the last column shows wild type plants. Left images show the signal from the green channel. Right images display mixed green and transmission channels. X: xylem; P: phloem; Pc: procambium; SP: sieve plate. Size bars = 100 μm
The SBP2 promoter distal region (position −2000 to −700) is essential for vascular tissue-specific expression of reporter genes
The vascular tissue-specific activity of the SBP2 promoter in vegetative organs is confined to a distal region (−2000 to −700), designated CRD-A (cis-regulatory domain-A), which contains repressing sequences that prevent gene expression in tissues other than vascular tissues (Waclawovsky et al. 2006a). Having established that the tissue-specificity of the promoter-GUS transcriptional fusions reflected the accumulation of the protein, we confirmed the negative tissue-specific nature of CRD-A through gain-of-function experiments. The 1.3 kb CRD-A sequences were fused to the −136pSBP2-GUS construct, which has been shown to induce constitutive GUS expression in leaves (Fig. 1, CRD-A/−136pSBP2-GUS) and to the SBP2 minimal promoter−92pSBP2-GUS (CRD-A/−92pSBP2-GUS), which has been shown to direct high levels of constitutive GUS expression in all vegetative organs (Waclawovsky et al. 2006a). These constructs were introduced into a tobacco plants and the GUS activity was determined in at least three independent transformants for each construct. CRD-A causes a drastic reduction in both −136pSBP2- and −92pSBP2-mediated GUS expression (Fig. 3). This quantitative measurements of GUS activity confirmed that the distal region on the GmSBP2 promoter, CRD-A (−2000 to −700), possesses cis-regulatory modules capable of repressing gene expression.
Functional analysis of the SBP2 promoter in mature leaves of transgenic tobacco plants expressing SBP2-GUS fusion genes. Specific GUS activity was determined by fluorometric assays with total extracts from leaves and expressed as nmol of 4-methylumbelliferone μg protein−1 min−1. The bars represent average (±S.E.) of three independent measurements using extracts from independent transgenic lines. Control represents the promoter-less binary vector (pCAMBIA1381Z) transformed plants
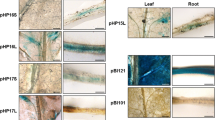
For tissue-specific GUS activity, we analyzed more than five independent primary transgenic plants expressing the SBP2 promoter-GUS fusion genes. We also included independent transgenic lines of the previously described −2000pSBP2-GUS, −136pSBP2-GUS and −92pSBP2-GUS constructs in the histochemical assays, which demonstrated the same expression pattern as described before (Contim et al. 2003; Waclawovsky et al. 2006a; Fig. 4). The fusion of the CRD-A sequences to the 5′ end of −136pSBP2-GUS caused a reduction in GUS activity in all organs analyzed. This reduction was clearly evident in the leaf surface, in which the GUS activity was restricted to the vascular tissue (Fig. 4, a4). In the stem, the GUS expression was also more pronounced in the vascular bundle, although the parenchymatic adjacent cells displayed a weak GUS staining (Fig. 4, compare b4 and b3). Likewise, in the shoot apex and roots, CRD-A reduced −136pSBP2-mediated GUS expression in all tissues except in vascular tissues (Fig. 4, c4 and d4, respectively).
Tissue-specific regulation of SBP2-GUS fusion gene expression in transgenic plants. Photographs of transgenic organs plants (as indicated) harboring the indicated SBP2-GUS fusion gene stained for GUS activity. Control corresponds to promoter-less pCAMBIA-transformed plants. Abbreviations: VT, Vascular Tissue; EP, Outer Phloem; IP, Inner Phloem; PA, Parenchyma; X, Xylem; LP, Leaf Primordia; SAM, Shoot Apical Meristem; E, Epidermis; VC, Vascular Cylinder; EZ, Elongation Zone; AM, Apical Meristem; RC, Root Cap. Bars in a1–a7 = 100 μm and in b1–b7, c1–c7, d1–d7 = 200 μm
The fusion of CRD-A to the 5′ end of −92pSBP2-GUS also reduced drastically GUS activity in all tissues analyzed (Fig. 4, a6, b6, c6, d6) and restored the tissue-specific expression pattern of the full promoter (Fig. 4, a2, b2, c2, d2), except in the shoot apex, in which the GUS staining was reduced but was not confined to the vascular tissue (Fig. 4, c6). In the leaf surface, a robust and restricted GUS staining was observed in the vascular bundle (Fig. 4, a6), while in the stem, GUS expression was restricted to the inner phloem (Fig. 4, b6). In roots, CRD-A abolished −92pSBP2-mediated GUS expression in the meristem (Fig. 4, compare d5 and d6) and confined the GUS staining to the vascular region (Fig. 4, d6). Taken together, these results confirmed the negative regulatory nature of CRD-A which promoted drastic reduction in the SBP2 promoter activity in the majority of the tissues analyzed.
The distal region CRD-A of the GmSBP2 promoter functions independently on the proximal TATA-containing region
The CRD-A fragment was also inserted in the pCAMBIA1381Z binary vector in the absence of a minimal promoter (Fig. 1). This DNA construct was initially designed to serve as a negative control for GUS expression, but rather it directed GUS expression in a tissue-specific manner similar to the full-length promoter-mediated expression pattern. These analyses were performed in six independently transformed lines. Apart from the shoot apex, in which CRD-A failed to direct minimal promoter independent GUS expression (Fig. 4, c7), CRD-A functioned independently on the proximal TATA-containing region in all other tissues analyzed. Both in leaves (Fig. 4, a7) and roots (Fig. 4, d7), GUS expression was restricted to the vascular tissue, whereas in stem, GUS staining was detected only in the inner phloem (Fig. 4, b7). These results indicate that CRD-A harbors cis-regulatory elements capable of promoting basal transcription and is sufficient to drive GUS expression to the vascular tissue in a context-independent manner (Fig. 4). Furthermore, they indicate that sequences downstream of −92 are important for expression in the shoot apex, as their absence abolished detectable expression in the region (Fig. 4, c7).
The tissue-specific expression pattern is controlled by distinct cis-regulatory modules on the distal region of the SBP2 promoter
Potential tissue-specific controlling elements were found on the distal region of the GmSBP2 promoter (Fig. 5). These include the GLUB1 sequence AACAAAC, which has been associated with the negative tissue-specific control of the rice glutelin gene (Yoshihara et al. 1996), both in the sense (positions −1197 to −1203) and reverse orientation (positions −1053 to −1059); three NtBBF1 binding sites ACTTTA (positions −1474 to −1479, −1586a −1591, −1713 to −1718) which have been implicated in tissue-specific expression of the rolB promoter in tobacco (Baumann et al. 1999); the GATA box (positions −855, −987 and −1142) that has been shown to be involved in tissue-specific expression and in light regulation of plant promoters (Gidoni et al. 1989; Lam and Chua 1989). Furthermore, several potential DOF binding sites (AAG core sequence) were located on CRD-A (Fig. 5A, red).
(A) Putative cis-regulatory elements on CRD-A region of SBP2 promoter. Numbers indicate the position relative to the translational start codon and the arrows delimit the sequential extension of the CRD-A fragments as Frag II (bold), Frag III (underlined) Frag IV (italic) and Frag V (regular letter). Direct and inverted CCAAT boxes are shown in green and potential TATA boxes in orange. Direct and inverted GLUB sequences are boxed. Putative NtBBF1 binding sites are shown in blue; potential DOF binding sites, in red and GATA boxes, in purple. (B) Alignment of CRD-G with Frag II and Frag IV of CRD-A. A highly conserved sequence is shown in red
To characterize functional regulatory cis-acting elements on the SBP2 promoter, we performed gain-of-function experiments for distinct modules of the distal region. These modules, designated Frag II (−1785/−1508), Frag III (−1507/−1237), Frag IV (−1236/−971) and Frag V (−971/−700), were directly fused to the −92pSBP2-GUS construct (Fig. 1B). All modules were capable of reducing the SBP2 promoter activity as their individual insertion at the 5′ end of −92pSBP2 altered its constitutive expression pattern (Fig. 6). However, the intensity of reduction varied to different extents for the four modules and according to the organ analyzed. While Frag V was the least effective in reducing −92pSBP2-mediated GUS expression, Frag IV restored the vascular tissue-specific expression of the full-promoter in leaves (Fig. 6, a6). Likewise, in stem, Frag III sequences caused the most drastic reduction in −92pSBP2-mediated GUS expression, which was restricted to the inner phloem mimicking the full-promoter-mediated GUS expression (Fig. 6, b5). Although the other modules, Frag II, IV and V, retained a prominent GUS staining in phloem, they reduced but did not abolish −92pSBP2 activity in the parenchymatic cells of stems (Fig. 6, b4, b6, b7). In the shoot apex, Frag II, III and IV abolished −92pSBP2-mediated GUS expression (Fig. 6, c4, c5, c6), whereas Frag V promoted a general reduction in expression with a predominant GUS staining in the vascular bundle (Fig. 6, c7). Finally, in roots, while all fragments were able to repress −92pSBP2-mediated GUS expression in the meristem (Fig. 6, d4, d5, d6, d7), Frag II and IV also abolished expression in the elongation zone (Fig. 6, d4 and d6). Furthermore, Frag II recovered the tissue-specific expression pattern of the full-length promoter, as Frag II/−92pSBP2-mediated GUS expression was confined to the root vascular tissue (Fig. 6, d4).
Histochemical analysis of tissue-specific regulation of SBP2-GUS fusion gene expression in transgenic plants. Photographs of transgenic organs plants (as indicated) harboring the indicated SBP2-GUS fusion gene stained for GUS activity. Control corresponds to promoter-less pCAMBIA-transformed plants. Abbreviations: VT, Vascular Tissue; EP, Outer Phloem; IP, Inner Phloem; PA, Parenchyma; X, Xylem; LP, Leaf Primordia; SAM, Shoot Apical Meristem; E, Epidermis; VC, Vascular Cylinder; EZ, Elongation Zone; AM, Apical Meristem; RC, Root Cap. Bars in a1–a7 = 100 μm and in b1–b7, c1–c7, d1–d7 = 200 μm
Negative cis-regulatory elements that prevent expression in the root elongation zone have been previously identified in the proximal promoter region, positions −136 to −92, designated cis-regulatory domain G, CRD-G (Waclawovsky et al. 2006a). To search for potential procambium expression-repressing determinants, the CDR-G sequence was compared with Frag II and Frag IV sequences (Fig. 5B). Sequences matching the 44 bp-CRD-B were detected in the Frag II and Frag IV (about 50% sequence identity). This conserved DNA segment harbors at the 3′ end a nearly identical nucleotide sequence that may function as a root-specific repressing element.
Soybean nuclear activities interact with distal cis-regulatory sequences of GmSBP2
Because the promoter analyses were performed in the tobacco heterologous system, it was of interest to evaluate whether the GmSBP2 promoter-driven expression pattern could reflect endogenous expression in soybean. To address this possibility, the accumulation of the SBP2 transcripts was assayed by qRT-PCR (see below) in the same soybean organs as those in which the SBP2 promoter activity was detected in tobacco. In addition, soybean nuclear activities were assayed for interactions with the distal region of the SBP2 promoter by EMSA. Frag II was radiolabeled and incubated in the presence of leaf and stem nuclear extracts from soybean and tobacco and used for gel shift assays. As expected from the GmSBP2 promoter analysis, tobacco nuclear extracts resulted in complex formation with Frag II (Fig. 7, lanes 4 and 5). Likewise, Frag II formed complexes with proteins of nuclear extracts prepared from soybean leaves (lane 2) and stems (lane 3), although with a different migration pattern from that found for tobacco extracts. The interactions were specific as the introduction of a ten-fold excess of unlabeled probe competed for binding (data not shown). Although these results clearly demonstrated that DNA binding activities from soybean nuclear extracts specifically recognized the GmSBP2 promoter region, the significance and identity of the resulting complexes remain to be determined.
Electrophoretic mobility shift assay. The radiolabeled −1785/−1508 fragment of the SBP2 promoter was incubated with nuclear extracts from soybean leaves (lane 2) and stem (lane 3) as well as from tobacco leaves (lane 4) and stems (lane 5) and the complexes formed were resolved by non-denaturing polyacrylamide gel electrophoresis. The arrow on the left indicates the position of free probe, whereas the arrows on the right the positions of the retarded complexes
GmSBP2 transcripts accumulate in soybean vegetative organs
The accumulation of SBP2 in vegetative organs was analyzed by qRT-PCR with soybean SBPs-general primers (Fig. 8, SBP) and GmSBP2-specific primers (Fig. 8, SBP2). Consistent with the expression pattern driven by the GmSBP2 promoter in tobacco, GmSBP2 transcripts were detected in all soybean vegetative organs analyzed. In general, GmSBP2 expression in mature plants was higher than in young plants. This result contrasts with the observation that SBP functions predominantly in the initial stages of plant development when the sink/source ratio is high (Waclawovsky et al. 2006b). Nevertheless, GmSBP2 expression is higher in sink organs, such as young leaves and roots, than in source mature leaves (Fig. 8), which is consistent with the involvement of SBP in determining sink strength.
SBP transcripts accumulate in soybean vegetative organs.SBP2 expression was verified in soybean plants during the reproductive phase (RP) and vegetative phase (VP). Total RNA was extracted from source leaves (SoL), sink leaves (SiL), stem (S), roots (R), leaves (L) and seedlings (Sd) and quantified by qRT-PCR using SBP2-specific primers (SPB2) and SBP family-general primers (SBP)
Discussion
In addition to its high activity in seeds, the GmSBP2 promoter has been shown to direct phloem-specific expression of a linked reporter gene in vegetative organs of transgenic tobacco (Contim et al. 2003; Waclawovsky et al. 2006a). While the vascular tissue-specific SBP2 promoter activity is consistent with the proposed involvement of GmSBP in sucrose transport-dependent physiological processes (Pedra et al. 2000; Waclawovsky et al. 2006b), the accumulation of the protein in vascular tissues of soybean vegetative organs is still a matter of debate. Although GmSBP had previously been immunolocalized in association with the plasma membrane of the sieve element–companion cell complexes of mature phloem (Grimes et al. 1992), these observations could not be reproduced using antibodies prepared against an E. coli-expressed truncated GmSBP1 (Elmer et al. 2003). Based on the specificity of the truncated SBP-antibody, the GmSBP family was found to accumulate exclusively in cotyledons and to be resolved in four pI-distinct cross-reacting polypeptides by immunoblotting assays of two-dimensional (2D) gels. In contrast, we demonstrated here that a GmSBP-GFP fusion protein accumulates in the vascular tissue of tobacco vegetative organs when the expression of the recombinant gene is driven by the full-length GmSBP2 promoter. We have also detected by qRT-PCR the accumulation of GmSBP transcripts in soybean vegetative organs (Fig. 8). Consistent with the proposed role for SBP in sink strength (Waclawovsky et al. 2006b), GmSBP2 transcripts were found to be predominantly expressed in sink organs, such as young leaves and roots. Taken together, these results confirm that the GmSBP2 gene is expressed in soybean vegetative organs and the protein accumulates in the vascular tissue.
We have also determined by gain-of-function experiments that the distal region (CDR-A fragment) of the SBP2 promoter harbors repressing elements that control the tissue-specific activity of the promoter. Fusion of CRD-A to −136pSBP2-GUS or −92pSBP2-GUS constructs diminished GUS expression and altered their constitutive pattern of expression. While the CRD-A/−92pSBP2-GUS restored the spatial pattern of the full-length promoter-mediated expression, fusion of CRD-A to −136pSBP2-GUS failed to confine the expression of the reporter gene to the inner phloem of stems and to the vascular tissue of roots (Fig. 4). This indicates that the region between −136 and −92 may harbor a positive element capable of attenuating but not abolishing the negative and tissue-specific effect of CDR-A. In fact, the region delimited by positions −136 to −92 contains a putative CCAAT box which has been associated with transcriptional activation and high levels of transcriptional activity of plant promoters (Kusnetsov et al. 1999; Buzeli et al. 2002).
Further characterization of the CRD-A fragment illustrates the silencer nature of distal elements on SBP2 promoter and suggests that the tissue-specific control of SBP2 gene expression requires a complex integration of multiple cis-acting regulatory elements, which are responsible for different levels of expression according to the organ analyzed. A direct comparison of the CRD-A/−92pSBP2-mediated GUS expression and the repressing effects of discrete modules from CRD-A allowed us to delimit the individual contribution of each cis-regulatory module to the general pattern of tissue-specific expression of SBP2 promoter in vegetative organs. Frag V (−970/−700) contains the cis-elements responsible for the expression pattern of GmSBP2 promoter in shoot apex, as when directly fused to the 5′ end of −92pSBP2-GUS it restored the expression pattern of the full-length promoter in shoot apex but not in the other organs analyzed. Likewise, in leaves, the GmSBP2 promoter-mediated vascular tissue-specific expression is contribution of Frag IV (−1236/−971), whereas, in roots, it is directed by cis-regulatory elements presents in Frag II (−1782/−1508). In stems, the inner phloem-specific expression of the full-length promoter is mediated by Frag III delimited by positions −1507 and −1237. These results further substantiate the notion that tissue-specificity may be mediated by distal regions of plant promoters. Although in the majority of plant promoters the determinants for tissue-specificity are often located on proximal upstream sequences (Stougaard et al. 1987; Zhao et al. 1994; Hamilton et al. 1998; Ruiz-Rívero and Prat 1998), distal tissue-specific elements have previously been mapped in the distal 5′ region of the soybean Msg promoter and Flaveria trinervia C4pppcA1 promoter (Stromvik et al. 1999; Gowik et al. 2004). Nevertheless, the negative nature of the SBP2 distal repressing modules differs from other plant promoters as the determinants for tissue-specificity in distal upstream regions often act in a positive regulatory manner.
The gain-of-function experiments for individual modules of CRD-A also mapped several negative cis-regulatory domains on SBP2 promoter distal regions. The sequences −1785 to −970 may contain shoot apex-specific expression-repressing elements, as all the three modules (Frag II, III and IV) were capable of abolishing GUS expression in this region. Likewise, strong silencers for root meristem expression are contained in the region −1785 to −700 because all modules repressed −92pSBP2-mediated root meristem expression. Frag II (−1785/−1508) tightly restricted the promoter activity to the vascular tissue of roots and may therefore harbor strong root expression silencers. Strong stem expression-repressing elements were located at positions −1507 to −1237, as Frag III confined −92pSBP2-mediated GUS expression to the inner phloem. Finally, the presence of any of the two modules (−1785/−1508 and −1236/−971) corresponding to Frag II and IV prevented GUS expression in the root elongation zone. Negative cis-regulatory elements that prevent expression in the root elongation zone have previously been identified in the proximal promoter region, positions −136 to −92, designated CRD-G (Waclawovsky et al. 2006a). A direct comparison among Frag II, Frag IV and CRD-G sequences identified the conserved consensus sequence CAGTTnCaAccACATTcCT present in all three cis-regulatory domains. This conserved sequence may represent a procabium expression-repressing element and hence a rational target for further functional promoter analysis in roots.
Among the four modules, Frag V (−970/−700) exhibited the most attenuated effects causing a slight decrease in GUS expression in the organs analyzed, except for the root meristem in which Frag V/−92pSBP2-mediated GUS expression was undetectable. The Frag V module harbors three TATA box-like sequences (positions −790, −783 and −761), in addition to an AT-rich region. We found that the CRD-A lacking the proximal 92-bp-minimal promoter mediates the similar tissue-specific expression pattern as the full-length promoter- and the CRD-A/−92pSBP2-mediated expression, although with weaker levels. The presence of putative TATA box-like sequences, correctly positioned in Frag V when directly fused to an ORF, may explain the capacity of the CRD-A to sustain tissue-specific transcription in the absence of a minimal promoter. Furthermore, Frag V contains AT-rich regions which have been identified in numerous promoters as enhancers of transcription (Bustos et al. 1989; Rieping and Schoffl 1992). Fortuitous AT-rich regions on the distal region of promoters that can take over the function of TATA boxes have already been reported from other soybean genes (Stomvik et al. 1999). The capacity of the CRD-A to sustain basal transcription in the absence of an external minimal promoter, clearly demonstrated that the vascular tissue-specific determinants of the GmSBP2 promoter are indeed contained in this region and function in a context-independent manner.
In summary, a detailed examination of the expression patterns of the SBP2:GUS constructs revealed that the vascular tissue-specific expression of the full promoter is conferred by the distal region, CRD-A, which is organized into repressing modules that act independently of one another to confer the vascular tissue-specific expression in distinct organs. The promoter activity reflected the accumulation of a SBP-GFP fusion protein into the vascular system of vegetative organs, which is consistent with the proposed involvement of GmSBP in sucrose transport-dependent physiological process. Our results also led to the identification of relevant cis-regulatory elements on SBP2 promoter that suppress expression in tissues other than vascular tissues of vegetative organs.
Abbreviations
- GmSBP2:
-
Glycine max sucrose-binding protein 2
- VfSBPL:
-
Vicia faba sucrose-binding protein-like protein
- PCR:
-
polymerase chain reaction
- CRD:
-
cis-regulatory domain
- GUS:
-
β-glucuronidase
References
Alvim FC, Carolino SMB, Cascardo JCM, Nunes CC, Martinez CA, Otoni WC, Fontes EPB (2001) Enhanced accumulation of BiP in transgenic plants confers tolerance to water stress. Plant Physiol 126:1042–1054
Baumann K, De Paolis A, Constantino P, Gualberti G (1999) The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell 11:323–334
Bustos M, Guiltinan M, Jordano J, Bergum D, Kalkan F, Hall T (1989) Regulation of B-glucuronidase expression in transgenic tobacco plants by an A/T-rich cis-acting sequence found upstream of a French bean B-phaseolin gene. Plant Cell 1:839–853
Buzeli RAA, Cascardo JCM, Rodrigues LAZ, Andrade MO, Almeida RS, Loureiro ME, Otoni WC, Fontes EPB (2002) Tissue-specific regulation of BiP genes: a cis-acting regulatory domain is required for BiP promoter activity in plant meristems. Plant Mol Biol 50:757–771
Castillo J, Rodrigo MI, Marques JA, Zuanigat AA, Franco L (2000) A pea nuclear protein that is induced by dehydration belongs to the vicilin superfamily. Eur J Biochem 267:2156–2165
Contim LAS, Waclawovsky AJ, Delú-Filho N, Pirovani CP, Clarindo WR, Loureiro ME, Carvalho CR, Fontes EPB (2003) The soybean sucrose binding protein gene family: genomic organization, gene copy number and tissue-specific expression of the SBP2 promoter. J Exp Bot 54:2643–2653
Delú-Filho N, Pirovani CP, Pedra JHF, Matrangolo FSV, Macedo JNA, Otoni WC, Fontes EPB (2000) A sucrose binding protein homologue from soybean affects sucrose uptake in transgenic tobacco suspension-cultured cells. Plant Physiol Biochem 38:353–361
Dunwell JM, Khuri S, Gane PJ (2000) Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol Mol Biol Rev 64:153–179
Dunwell JM, Purvis A, Khuri S (2004) Cupins: the most functionally diverse protein superfamily. Phytochemistry 65:7–17
Elmer A, Chao W, Grimes H (2003) Protein sorting and expression of a unique soybean cotyledon protein, GmSBP, destined for the protein storage vacuole. Plant Mol Biol 52:1089–1106
Gidoni D, Brosio P, Bond-Nutter D, Bedbrook J, Dunsmuir P (1989) Novel cis-acting elements in Petunia Cab gene promoters. Mol Gen Genet 215:337–344
Gowik U, Burscheidt J, Akyildiz M, Schule U, Koczor M, Streubel M, Westhoff P (2004) Cis-regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell 16:1077–1090
Grimes HD, Overvoorde PJ (1996) Functional characterization of sucrose binding protein-mediated sucrose uptake in yeast. J Exp Bot 47:1217–1222
Grimes HD, Overvoorde PJ, Ripp KG, Franceschi VR, Hitz WD (1992) A 62-kDa sucrose binding protein is expressed and localizes in tissues actively engaged in sucrose transport. Plant Cell 4:1561–1574
Hamilton DA, Schwarz YH, Mascarenhas JP (1998) A monocot pollen-specific promoter contains separable pollen-specific and quantitative elements. Plant Mol Biol 38:663–669
Heim U, Wang Q, Kurz T, Borisjuk L, Golombek S, Neubohn B, Adler K, Gahrtz M, Sauer N, Weber H, Wobus U (2001) Expression patterns and subcellular localization of a 52 kDa sucrose binding protein homologue of Vicia faba (VfSBPL) suggest different functions during development. Plant Mol Biol 47:461–474
Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7:193–195
Kühn C, Quick WP, Schultz A, Sonnewald U, Frommer WB (1996) Companion cell-specific inhibition of the potato sucrose transporter SUT1. Plant Cell Environ 19:1115–1123
Kusnetsov V, Landsberger M, Meurer J, Oelmuller R (1999) The assembly of the CAAT-box binding complex at a photosynthesis gene promoter is regulated by light, cytokinin, and the stage of the plastids. J Biol Chem 274:36009–36014
Lam E, Chua NH (1989) ASF-2: a factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in cab promoters. Plant Cell 1:1147–1156
McCabe DE, Swain WF, Martinell BJ, Christou P (1988) Stable transformation of soybean (Glycine max) by particle bombardment. Biotechnology 6:923–926
Overvoorde PJ, Frommer WB, Spencer D (1996) A soybean sucrose binding protein independently mediates nonsaturable sucrose uptake in yeast. Plant Cell 8:271–280
Overvoorde PJ, Chao WS, Grimes HD (1997) A plasma membrane sucrose-binding protein that mediates sucrose uptake shares structural and sequence similarity with seed storage proteins but remains functionally distinct. J Biol Chem 272:15898–15904
Pedra JHF, Delú-Filho N, Pirovani CP, Contim LAS, Dewey RE, Otoni WC, Fontes EPB (2000) Antisense and sense expression of a sucrose binding protein homologue gene from soybean in transgenic tobacco affects plant growth and carbohydrate partitioning in leaves. Plant Sci 152:87–98
Pirovani CP, Macedo JNA, Contim LAS, Matrangolo FSV, Loureiro ME, Fontes EPB (2002) A sucrose binding protein homologue from soybean exhibits GTP-binding activity that functions independently of sucrose transport activity. Eur J Biochem 269:3998–4008
Polanco R, Lobos S, Vicuna R (2002) Binding of nuclear proteins to the promoter region of the lactase gene Cs-lcs1 from the basidiomycete Ceriporiopsis subvermispora. Enzyme Microb Technol 30:525–528
Rieping M, Schoffl F (1992) Synergistic effect of upstream sequences, CCAAT box elements, and HSE for enhances expression of chimeric heat shock genes in transgenic tobacco. Mol Gen Genet 231:226–232
Riesmeir JW, Willmitzer L, Frommer WB (1994) Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J 13:1–7
Ripp KG, Viitanen PV, Hitz WD, Fransceschi VR (1988) Identification of a membrane protein associated with sucrose transport into cells of developing soybean cotyledons. Plant Physiol 88:1435–1445
Rocha CS, Luz DF, Oliveira ML, Baracat-Pereira MC, Medrano FJ, Fontes EPB (2007) Expression of the sucrose binding protein from soybean: renaturation and stability of the recombinant protein. Phytochemistry 68:802–810
Ruiz-Rivero OJ, Prat S (1998) A -308 deletion of the tomato LAP promoters is able to direct flower-specific and MeJA-induced expression in transgenic plants. Plant Mol Biol 36:639–648
Stougaard J, Sandal NN, Gron A, Khule A, Marcker KA (1987) 5′ analysis of the soybean leghaemoglobin lbc(3) gene: regulatory elements required for promoter activity and organ specificity. EMBO J 6:3565–3569
Stromvik MV, Sundeararaman VP, Vodkin LO (1999) A novel promoter from soybean that is active in a complex development pattern with and without its proximal 650 base pairs. Plant Mol Biol 41:217–231
Waclawovsky AJ, Freitas RL, Rocha CS, Contim LAS, Fontes EPB (2006a) Combinatorial regulation modules on GmSBP2 promoter: a distal cis-regulatory domain confines the SBP2 promoter to the vascular tissue in vegetative organs. Biochim Biophys Acta 1759:89–98
Waclawovsky AJ, Loureiro ME, Freitas RL, Rocha CS, Cano MAO, Fontes EPB (2006b) Evidence for the sucrose-binding protein role in carbohydrate metabolism and transport at early development stage. Physiol Plant 128:391–404
Warmbrodt RD, Buckhout TJ, Hitz WD (1989) Localization of a protein, immunologically similar to a sucrose-binding protein from developing soybean cotyledons, on the plasma membrane of sieve-tube members of spinach leaves. Planta 180:105–115
Warmbrodt RD, Vanderwoude WJ, Hitz WD (1991) Studies on the localization of a protein, immunologically similar to a 62-kilodalton sucrose-binding protein isolated from developing soybean cotyledons, in the shoot and root of spinach. New Phytol 118:501–511
Yoshihara T, Washida H, Takaiwa F (1996) 45-bp proximal region containing AACA and GCN4 motif is sufficient to confer endosperm-specific expression of the rice storage protein glutein gene, GluA-3. FEBS Lett 383:213–218
Zhao Y, Leisy DJ, Okita TW (1994) Tissue-specific expression and temporal regulation of the rice glutein Gt3 gene are conferred by at least two spatially separated cis-regulatory elements. Plant Mol Biol 25:429–436
Acknowledgements
We are grateful to Dr. Simon L. Elliot for critically reading the manuscript. This research was supported by the Brazilian Government Agencies CNPq grant 50.6119/2004-1 and 470878/2006-1 (to E.P.B.F.) and FAPEMIG grant EDT 560/05 and EDT 523/07 (to E.P.B.F.). R.L.F. was supported by a graduate fellowship from the Brazilian Government Agency CAPES. C.M.C. is a FAPEMIG postdoctoral fellow (CBB 00112/07).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Freitas, R.L., Carvalho, C.M., Fietto, L.G. et al. Distinct repressing modules on the distal region of the SBP2 promoter contribute to its vascular tissue-specific expression in different vegetative organs. Plant Mol Biol 65, 603–614 (2007). https://doi.org/10.1007/s11103-007-9225-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-007-9225-0