Abstract
The area between the upper part of the leaf sheath and the basal portion of the leaf blade contains several specialized organs, such as the laminar joint, auricle and ligule. Here we report the identification of T-DNA insertional mutant lines that lack all of these organs. The gene knocked out in the mutant lines encodes a protein that contains a SBP (SQUAMOSA promoter Binding Protein)-domain and is highly homologous to the maize LIGULELESS1 (LG1) gene. At the amino acid sequence level, the OsLG1 protein is 69% identical to maize LG1 and 78% identical to barley LG1. We named the rice gene OsLIGULELESS1 (OsLG1). Transient expression of an OsLG1:RFP (Red Fluorescent Protein) fusion protein indicated that the protein is localized to the nucleus. Transgenic plants harboring the OsLG1 promoter:GUS (β-glucuronidase) reporter gene construct display preferential expression in developing laminar joint regions and meristemic regions. The gene is also weakly expressed in the ligule, auricles, and leaf sheaths at the basal region. These results indicate that OsLG1 is a transcriptional factor that plays an important role in building the laminar joint between leaf blade and leaf sheath boundary, thereby controlling ligule and auricle development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A rice leaf consists of a leaf sheath, a leaf blade, and a laminar joint that contains a pair of auricles and the ligules. The auricle is a hairy and sickle-shaped organ that surrounds the culm, thus keeping the leaf sheath and culm together. The ligule is a thin and tongue-like white membrane that keeps the leaf sheath closed tightly against the auricles to prevent rainwater or dust from entering. The laminar joint is mechanical tissue that bends the leaf blade away from the vertical axis toward the abaxial side when the leaf blade and leaf sheath are fully elongated (Hoshikawa 1989). Since the laminar joint angle increases proportionally to brassinosteroid level, a joint bending assay has been used for screening brassinosteroid-related mutants (Wada et al. 1981; Yamamuro et al. 2000; Hong et al. 2004).
Morphological features of the ligule, auricles, and laminar joint, as well as their presence or absence, are used as key identification features of agricultural and horticultural crop varieties. For example, barnyard grasses (Echinochloa crusgalli) do not have a ligule and goose grasses (Eleusine indica) are absent in auricles (Miller 1999). Although most rice varieties contain all three organs, some rice cultivars (Oryza sativa L. Jal lahri) lack the auricles.
Development of the laminar joint region has been characterized in maize at genetic and molecular levels using liguleless1 (lg1) and liguleless2 (lg2) mutants (Becraft et al. 1990; Harper and Freeling 1996; Moreno et al. 1997; Walsh et al. 1998). Loss of function of the lg1 gene prevented the proper formation of ligules and auricles at the blade-sheath boundary. Ligules were completely absent from about the first 10 leaves; however, on the upper leaves a rudimentary ligule developed without accompanying auricles. In contrast, the lg2 mutant showed an age-dependent phenotype (Harper and Freeling 1996; Walsh and Freeling 1999). The first and sometimes the second leaf completely lacked the ligule and auricle, but the organs were developed at the margin of the third leaf. Successive leaves gradually developed more ligules and auricles, until they looked almost normal. Therefore, the two genes appear to play different roles in ligule–auricle development. LG1 encodes a putative transcription factor containing an internal domain of 77 amino acids with significant similarity to the conserved domain present in SBP (SQUAMOSA Promoter Binding Protein) DNA binding proteins (Moreno et al. 1997). LG1 mRNA exists predominantly in the ligular region of very young leaves but is also present weakly in the sheath and blade (Moreno et al. 1997). The LG2 gene encodes a bZIP (basic-leucine zipper) transcription factor and is ubiquitously expressed throughout leaf primordia (Walsh et al. 1998). bZIP transcription factors regulate pathogen defense, light and stress signaling, seed maturation, and the development of various organs such as leaves and flowers (Jakoby et al. 2002). Double mutant analysis suggests that LG1 and LG2 act in the same pathway (Harper and Freeling 1996). It was proposed that LG2 specifies the precise location of ligule and auricle development and interacts with LG1 to control the development of ligule and auricle. Comparative genetic studies using the LG1 gene identified putative candidates for barley (Rossini et al. 2006) and sorghum (Zwick et al. 1998).
In rice, little is known about molecular mechanism that controls the development of the laminar joint region and the ligule. Ligule development is started by periclinal division of the adaxial side in leaf primordium. Some genes which expressed in the ligular region were identified. The GL2-type homeobox gene, ROC1 (Rice outermost cell-specific gene1) is specifically expressed in the epidermis including the outmost cells of the ligule (Ito et al. 2002). The rice SCARECROW (OsSCR) gene is expressed not only in the endodermal cell layer of the root, but also in ligular cells (Kamiya et al. 2003).
Among the 2,000 traits have been reported in rice (Kinoshita 1998; Kurata et al. 2005), three mutants, auricleless (aur), liguleless (lg), and collarless (col) affect the leaf blade-sheath boundary (Maekawa 1988; Sanchez and Khush 1998). However, the mutants have not been studied at the molecular level. Here, we report the molecular characterization of rice LIGULELESS1 (OsLG1), a gene that plays a key role in development of laminar joint and the organs developed from the region.
Materials and methods
Plant growth
Surface-sterilized T2 seeds of oslg1 and wild-type (Oryza sativa cv. Japonica Dongjin) were germinated in the Murashige and Skoog (MS) medium comprising 3% sucrose, 0.2% phytagel and 0.55 mM myo-inositol. Plants were grown for 7 days at 27°C in the continuous light conditions and then transplanted into soil in the greenhouse until maturity
Screening of T-DNA insertional allele in the OsLG1 gene from the DNA pools
Systematic reverse genetics screening was performed as described previously (Lee et al. 2003). PCR was performed using the OsLG1-specific primers (5′-TCCACCCGAACCCTAGATAGATGA-3′ and 5′-ATGAACACTGACCTGCTGCACTG-3′), and the T-DNA right-border primer (5′-ATCCAGACTGAATGCCCACAGGC-3′). The PCR product from the superpool DNA was separated by agarose gel and the blot was hybridized with the OsLG1-specific probe. After identification of an individual line, T-DNA insertion was confirmed by sequencing the PCR product.
Genotyping the mutant plants
PCRs for genotyping were performed in 20 μl of a mixture containing 20 ng of plant DNA, 10× Taq buffer, 0.2× band doctor, 0.2 mM dNTP, 0.2 unit of Taq polymerase (Solgent, Daejeon, Korea), and 1 μM of the primers. The protocol included 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 120 s. Primers for oslg1-1 genotyping were OsLG1–1 primers a (5′-ATGCGTGCATGTTTCTAGGT-3′), OsLG1-1 primers b (5′-ACTGTTCCATCCCAGAGACA-3′, and for a T-DNA right-border primer c (5′-ATCCAGACTGAATGCCCACAGGC-3′). Primers for oslg1-2 genotyping were OsLG1-2 primers a (5′-TCCACCCGAACCCTAGATAGATGA-3′), OsLG1-2 primer b (5′-CTCAAACATGCGAAGGAGAAGTCA-3′), and for a T-DNA right-border primer c (5′-ATCCAGACTGAATGCCCACAGGC-3′).
RT-PCR analysis
Total RNA was extracted from various organs using Tri-reagent (MRC Inc., Cincinnati, Ohio, USA) according to the manufacturer’s instruction manual. For first-strand cDNA synthesis, 2 μg of total RNA was reverse-transcribed in a volume of 25 μl containing 10 ng of oligo (dT) 12–18 primer, 10 mM dNTPs, and 1 units of M-MLV reverse transcriptase (Promega, Madison, WI, USA). The PCR reaction was performed in a 20 μl solution containing a 2 μl aliquot of the cDNA reaction, 0.2 μM gene specific primers, 10 mM dNTPs, 1 unit of Taq DNA polymerase (Solgent, Daejeon, Korea), 5× band doctor, and 10× reaction buffer. The reaction included an initial 5-min denaturation at 94°C, followed by 23–32 cycles of PCR (94°C, 30 s; 58–60°C, 30 s; and 72°C, 1–1.5 min), and a final 10 min at 72°C. Afterward, 10 μl of the reaction mixture was analyzed on a 1.2% (w/v) agarose gel. OsLG1 specific primers (forward: 5′-TCCACCCGAACCCTAGATAGATGA-3′ and reverse: 5′-GCAGCTCTTCTTGGCATCGT-3′) and OsActin primers (forward: 5′-CCTCTTCCAGCCTTCCTTCAT-3′ and reverse: 5′-ACGGCGATAACAGCTCCTCTT-3′) were used.
Quantitative real-time RT-PCR
Real-time PCR was performed using a Roche Lightcyler with cDNA prepared from the 4th (early emerged) and 5th (newly emerged) leaves of 30-day-old plants. cDNA was synthesized using total RNA isolated from the region of the sheath 3–8 mm directly below the laminar joint, 2 mm of the laminar joint region containing sheath and blade, and 3–8 mm of the leaf blade directly above the laminar joint. Real-time RT-PCR procedure was performed as described previously (Han et al. 2006). The mRNA level of ubiquitin was used to normalize the expression ratio of each gene. The primers for OsLG1 were 5′-CCAAGAAGAGCTGCAGGAAG-3′ and 5′-CCATGGCTTGGTCTTGATCT-3′. The primers for the rice ubiquitin gene were 5′-TGAAGACCCTGACTGGGAAG-3′ and 5′-CACGGTTCAACAACATCCAG-3′. The changes in gene expression were calculated using the ΔCt method.
Transient expression of the OsLG1-RFP fusion
To investigate the cellular localization, we constructed an OsLG1-RFP fusion molecule placed under the control of the CaMV 35S promoter. We replaced the stop codon of OsLG1 with a SalI restriction site via PCR, using the forward primer (5′-TCTAGAGCGAGGATGATGAACGTTCCAT-3′) and the reverse primer (5′-GTCGACCAGTGATCGAAGTCGAGATCAA-3′). The amplified fragment was inserted between XbaI and SalI of the pCaMV35S-RFP vector (Sohn et al. 2003; Jang et al. 2004), generating an in-frame fusion between the OsLG1 and RFP genes. As a positive control, the OsSNB coding region was ligated in-frame to the N terminus of the soluble modified green fluorescent protein (smGFP) under the control of the CaMV 35S promoter (Jung et al. 2002; Jang et al. 2004; Lee et al. 2006). Protoplast isolation and the electroporation procedure are as described previously (Kyozuka and Shimamoto 1991) with some modifications. Rice seeds were germinated on MS medium under continuous light at 26°C. Leaves of 10-day-old seedlings were cut into approximately 0.5 mm strips and placed in 20 ml enzyme solution containing 0.6 M mannitol, 10 mM MES, 1 mM CaCl2, 0.1% BSA, 1.5% cellulase R-10 (Yakult Honsha), 0.1% pectolyase, and 0.5% macerozyme R-10 (Yakult Honsha). After vacuum-infiltration for 10 min, samples were digested at 25°C with gentle shaking at 40 rpm for 5 h. The same volume of KMC solution (117 mM KCl, 82 mM MgCl2, and 85 mM CaCl2, pH 5.6) was added to the enzyme solution and filtered by using a 20 μm nylon mesh. Protoplasts were collected by centrifuging at 1,500 rpm for 90 s at 4°C. Pellets were resuspended in EP3 solution containing 70 mM KCl, 5 mM MgCl2, 0.1% MES, and 0.4 mM mannitol. Protoplasts were counted and 2 × 106 protoplasts were mixed with 20 μg of the plasmid DNA. Electroporation was performed in ice using the Gene Pulser Xcell Electroporation System (Bio-Rad) at 300 V cm−1 with a 450 μF capacitor. The protoplasts were transferred to incubation solution (0.6 mM mannitol, 4 mM MES, and 4 mM KCl, pH 5.6) and kept in dark for 12–18 h at 28°C. Expression of the fusion construct was monitored by a fluorescence microscopy (Zeiss, Jena, Germany) and the images were captured with a cooled charge-coupled device camera (Zeiss, Jena, Germany).
Construction of the OsLG1 promoter:GUS fusion
The 1.8-kb promoter region of OsLG1 was obtained by PCR using forward primer (5′-AAGCTTTTGCCCAAATAATGACACGATATA-3′) and reverse primer (5′-GGATCCCCTCGCGGCCTGTCTCCTCCTC-3′). The PCR product was inserted into the pGA1230 vector containing the GUS reporter gene (Jeon et al. 2000b). Transgenic plants carrying the construct were obtained by the stable transformation method as previously reported (Lee et al. 1999). Histochemical GUS staining was performed as described by Jefferson et al. (1987) and Dai et al. (1996). For light microscope analysis, the tissues were fixed in a solution containing 50% ethanol, 5% acetic acid, and 3.7% formaldehyde, and then embedded in Technovit 8100 Resin (Kulzer & Co., Wehrheim, Germany). The samples were sectioned to 4- to 9-μm thickness with a microtome (Leica, Nussloch, Germany), and observed under a microscope (Nikon, Kanagawa, Japan) using bright- and dark-field illumination.
Sequences alignments of SBP domain protein
Multiple sequence alignments were performed using the CLUSTAL X program (Thompson et al. 1997) and BioEdit program (Version 7.0.5, http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The following SBP-containing proteins were used as queries: AtSPL3 (At2 g33810), AtSPL4 (At1 g53160), AtSPL5 (At3 g15270), AtSPL8 (At1 g02065), and AtSPL14 (At1 g20980) from Arabidopsis, ZmLG1 (U89496) from maize, HvLG1 (AM117950) from barley and OsLG1 (Os04 g56170) from rice.
Scanning electron microscope analysis
The laminar joint region of 10-day-old seedling shoots and 6th leaves (newly emerged) from 70-day-old plants was used for SEM analysis. Samples were pre-fixed with 3% glutaraldehyde-sodium phosphate buffer (0.1 M) for 3 h at room temperature and rinsed with 0.1 M sodium phosphate buffer. Post-fixation was performed with 2% osmium tetroxide (OsO4), followed by rinsing with 0.1 M sodium phosphate buffer. After fixation, the samples were dehydrated through an ethanol series, and infiltrated through an isoamyl acetate series. The samples were critical point-dried, then sputter-coated with palladium. The mounted specimens were observed with a scanning electron microscope (Leo S1450VP, Carl Zeiss, Germany).
Results
Isolation of a liguleless mutant from rice T-DNA tagging population
We have previously reported the generation of T-DNA tagging lines in rice (Oryza sativa var. japonica) using the Agrobacterium-mediated co-cultivation method (Jeon et al. 2000a; Jeong et al. 2002). From the population, we have identified a liguleless mutant line, 3A-12312.
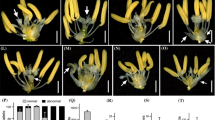
Wild-type rice plant leaves consist of three parts: the blade, sheath, and laminar joint (Fig. 1A–E). The leaf blade is a broad, flat structure located at the upper part of the leaf. The sheath is located between leaf blade and a node, and warps around the stem internode above this node. The laminar joint region is a belt-like structure present in the boundary between blade and sheath (Fig. 1A, C–E). A pair of auricles and a ligule are present at the sides of the lamina joint region (Fig. 1B–E). The ligule is a thin, white, tongue-like organ and is said to be the degenerated tip of the leaf sheath (Fig. 1B–F) (Hoshikawa 1989). Auricles are horn-like tissues that surround the culm, thus keeping the leaf sheath and the culm together (Fig. 1C–E).
Phenotypes of the wild-type and oslg1 plants. Comparison of leaves in the abaxial side (A) and adaxial side (B) among wild-type plant (left) and OsLG1 knockout plants (10-day-old seedlings). (C) Comparison of 2nd leaves in the wild-type plant (left) and oslg1-1 mutant plant (right) at 70-day-old plants. Comparison of 5th mature leaves in the abaxidal side (D) and in the adaxial side (E) at 70-day-old plants. (F) Comparison of flag leaves in the wild-type plant (left) and oslg1-1 mutant plant (right) at flowering stage, and (G) Comparison of wild-type plant (left) and oslg1-1 mutant plant (right) at maturity. li, ligule; au, auricle; lj, laminar joint; bl, leaf blade; sh, leaf sheath. Bar, 100 μm
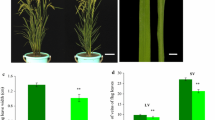
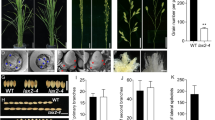
In the liguleless mutant that we have isolated, leaf sheaths were directly connected to blades without a laminar joint region (Fig. 1A). In addition, the ligule and auricles were not present at the region (Fig. 1B). At maturity, homozygotic mutant plants showed upright attitudes (Fig. 1G), and the ligules and auricles were absent in all the leaves including the flag leaves (Fig. 1C–F). We named this mutant allele Oryza saliva liguleless1-1 (Oslg1-1).
Scanning electron microscopy analyses
From SEM analyses, the epidermal tissue on the abaxial side of the wild-type leaf blades was found to consist of stomata, silica cells with a wart-like protuberances, and trichomes (Fig. 2B). The epidermis of the sheath was basically similar to that of the leaf blade, containing stomata, dumbbell-shaped silica cells, and trichomes (Fig. 2D). The epidermis of the laminar joint region had smooth surfaces lacking silica cells or trichomes (Fig. 2C). The laminar joint region exhibited progressive thickening from the margin toward the midrib (Fig. 2A). In oslg1-1 mutant plants, the epidermis of the leaf blade and sheath was identical to that of wild-type plants, containing stomata and silica cells (Fig. 2F and H). However, cellular structures characteristic of the laminar joint region were absent from of oslg1-1 mutant plants (Fig. 2E and G).
Scanning electron micrographs of the abaxial surfaces of wild-type and oslg1-1 leaves. The epidermis structures of the laminar joint region of 10-day-old wild-type plant (A) were compared to those of the oslg1-1 plant at the same stage (E). The boxed areas b, c, d, f, g, and h were enlarged to Figs. B, C, D, F, G, and H, respectively. B and F, leaf blade; C and G, laminar joint; D and H, leaf sheath. s, silical cell; st, stomata
In the adaxial view, the leaf blade (Fig. 3B and F) and the sheath (Fig. 3D and H) of 70-day-old oslg1-1 and segregating wild-type plants showed no difference in the surface structure. The epidermis of laminar joint in the wild-type consisted of wrinkled, unshaped cells containing the no trichomes and silica cell (Fig. 3C). In oslg1-1 mutants, the epidermis of laminar joint region had no obvious boundary (Fig. 3G, Supplementary Fig. 1). Histological analysis showed the difference in the cellular structure between wild-type and oslg1-1 plants did not observe in the 10-day-old plants (Supplementary Fig. 2).
Scanning electron micrographs of the adaxial surfaces of wild-type and oslg1-1 leaves. The epidermis structures of the laminar joint regions of 70-day-old wild-type plant (A) were compared to those of the oslg1-1 plant at the same stage (E). The boxed areas b, c, d, f, g, and h were enlarged to Figs. B, C, D, F, G, and H, respectively. B and F, leaf blade; C and G, laminar joint; D and H, leaf sheath. lj, laminar joint; sh, leaf sheath; bl, leaf blade. Ab, abaxial side view
Isolation of the T-DNA-tagged gene
The DNA sequence flanking the inserted T-DNA was isolated from the oslg1-1 mutant line using the inverse PCR method (Jeong et al. 2006). T-DNA was found inserted in the first intron of the gene LOC_Os04g56170 (The Institute for Genomic Research, http://www.tigr.org/tdb/e2k1/osa1/LocusNameSearch.shtml), located on chromosome 4 (Fig. 4A, Supplementary Fig. 3). The full-length cDNA sequence of the gene is present in KOME (AK068104), and it encodes a predicted protein of 416 amino acids (Fig. 4B).
Genomic structure of the OsLG1 gene and insertion positions of T-DNA. (A) Scheme of the OsLG1 gene and relative insertion positions of T-DNA. Black boxes represent exons and intervening lines represent introns. Insertion positions of T-DNA are indicated at top. Number 1 indicates the translational initiation site and 3441 indicates the end of translation. An arrow below the T-DNA indicates the direction of the GUS reporter gene in the tagging vector. a, b, and c are primers used for genotyping. SBP, SQUAMOSA promoter binding protein domain. NLS, nuclear localization signal. Bar, 0.5 kb. (B) Alignment of SBP domains. Shade box indicates the eight conserved cysteine and histidine residues. Zn-1 and Zn-2 indicate two Zn-coordinating structures. The triangle (▼) above the sequences refers to the position of an intron found in the SBP domain. * complete conservation; : indicates strong conservation; ., weak conservation. The accession numbers of the SBP genes are listed in “Materials and methods” section. (C) RT-PCR analysis of the OsLG1 transcripts in wild-type, oslg1-1, and oslg1-2. RT-PCR was performed using the P1 and P2 primers shown in Fig. 4A. OsActin was used as a control to demonstrate equal RNA loading
Analysis of the OsLG1 protein sequences using the NCBI Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) identified a conserved SBP DNA binding domain, which is present only in plants (Fig. 4B). The SBP domain consists of 76 highly conserved amino acids that overlap with a nuclear localization signal (NLS). The SBP proteins bind specifically to the GTAC core motif in the Antirrhinum majus SQUAMOSA promoter and Arabidopsis thaliana APETALA1 promoter (Klein et al. 1996; Birkenbihl et al. 2005). Structural analysis of the SBP domain showed two zinc finger-like structures (Zn-1 and Zn-2) formed by eight conserved cysteine and histidine residues, a Cys3HisCys2HisCys or Cys6HisCys sequence motif (Fig. 4B) (Yamasaki et al. 2004). A NLS sequence (KKSCRKRLADHNRRRRK) is found within the highly conserved SBP DNA binding domain (aa 242–259) (Fig. 4B). Cardon et al. (1999) have shown that the Arabidopsis SBP box consists of two exons interrupted by an intron, and that the position of this intron is conserved. OsLG1 also has a conserved intron within the SBP box (Fig. 4A)
A homology search using the BLASTP algorithm (BLOSUM 62 matrix) showed that the predicted protein is most homologous to maize LG1 (ZmLG1) with 69% identity (Supplementary Fig. 4). The next homologous protein among the maize SBP proteins is ZmSBP3, which is 27% identical to OsLG1 (Supplementary Fig. 5). In barley, OsLG1 showed 56% identity with HvLG1. The most homologous protein in Arabidopsis is AtSPL8 (encoded by At1G02065), which is 51% identical to OsLG1.
Comparative genetics approaches showed that the region on rice chromosome 4L, where the OsLG1 gene is located, is syntenous to maize chromosome 2S, where LG1 is present (Becraft et al. 1990; Kaplinsky et al. 2002). Likewise, the rice chromosomal region is syntenous to barley chromosome 2H, where the barley HvLG1 gene is located (Rossini et al. 2006). These results indicate that the OsLG1 gene identified by our T-DNA insertional mutagenesis is an ortholog of the maize LG1 gene and barley HvLG1.
Identification of an additional allele by systematic reverse genetics screening
To verify that the liguleless phenotype observed from the oslg1-1 mutant was indeed due to T-DNA insertion, we conducted co-segregation analysis between the phenotype and genotype. We obtained T2 progeny from the original mutant plant and their genotypes were analyzed. Among the 80 plants analyzed, 14 were liguleless mutants. Genotyping of these plants showed that all of the mutants were homozygous (oslg1/oslg1) to the T-DNA insertion, whereas the remaining 66 were either wild-type or heterozygous (OsLG1/oslg1) plants (Supplementary Fig. 6). In T3 generation, among the 40 plants examined, 13 were liguleless mutants and all of them were oslg1/oslg1 (data not shown). Genomic DNA gel-blot analysis using the 5′ region of GUS probe, which is located in T-DNA, showed that one copy of T-DNA was inserted in Line 3A-12312 (Supplementary Fig. 7).
To further confirm that the liguleless mutant phenotype is due to the mutation in the OsLG1 gene, we obtained an additional insertional allele (oslg1-2) from the T-DNA insertional populations via the systematic reverse genetics screening method (Lee et al. 2003). In Line 1B-15010, T-DNA was inserted 5′ UTR region (−1 bp) from the translation initiation site (+1) of the OsLG1 gene (Fig. 4A, Supplementary Fig. 3). The oslg1-2 mutant plants showed the same mutant phenotypes as observed from the oslg1-1 plants (Fig. 1A and B), confirming that the mutant phenotypes were caused by a loss-of-function mutation of the OsLG1 gene. Semi-quantitative PCR experiments showed that the OsLG1 mRNA is absent in both oslg1-1 and oslg1-2 mutants (Fig. 4C).
OsLG1 protein is targeted to the nucleus
Previous studies indicate that the ZmLG1 protein and SBP domain-containing proteins are localized into the nucleus (Moreno et al. 1997; Birkenbihl et al. 2005). The presence of the NLS sequence in the OsLG1 protein and a high similarity among these proteins suggest that the OsLG1 protein would be also localized to the nucleus. In order to determine its cellular location, the OsLG1 coding region was fused to the red fluorescent protein (RFP) gene under control of the 35S cauliflower mosaic virus (CaMV) promoter (Fig. 5A) (Sohn et al. 2003; Jang et al. 2004), and the construct was transiently expressed in mesophyll protoplasts isolated from rice seedling leaves. As a positive control, a previously characterized nuclear targeted protein, OsSNB:GFP, was co-expressed in the protoplasts (Lee et al. 2006). Figure 5B illustrates that the RFP signal was found in the nucleus and coincided with the GFP signal, indicating that OsLG1 is a nuclear protein and may function as a transcriptional regulator.
Nuclear localization of OsLG1:RFP protein. (A) Schematic diagrams of fusion constructs. 35S Pro, CaMV 35S promoter; Tnos, nopaline synthase gene terminator. (B) Rice mesophyll protoplasts were co-transformed with 35S pro:OsLG1:RFP and 35S pro:OsSNB:GFP. Images were obtained by a confocal laser scanning microscope (Zeiss) at either GFP channel (upper left) or RFP channel (upper middle). The merged image between two channels is presented (lower middle). The protoplast visualized by a bright microscope is also shown (lower right). As a control, the Ubi:GFP construct was independently transformed to protoplasts (upper right)
OsLG1 is preferentially expressed in the laminar region
To study the expression pattern of OsLG1, we constructed a binary vector carrying a fusion molecule between the OsLG1 promoter and the GUS reporter gene. The vector was introduced into rice by the Agrobacterium-mediated transformation method (Lee et al. 1999). Among five independently transformed T1 transgenic plants, four showed GUS positive in the same expression pattern. We observed an intensive GUS signal in the laminar joint regions and in the vein of the sheath but not in roots (Fig. 6A–C) (Xie et al. 2006). GUS expression was detected in the laminar joint region of the newly emerged leaves (Fig. 6A and D). GUS expression was preferentially detected in the initiation site of the laminar joint region from the edge to midrib in leaves at younger stages of development (Fig. 6G, H, and I). Cross sections of the tissues showed preferential GUS staining in the vascular bundle tissues of the sub-apical region (Fig. 6J and K), laminar joints (Fig 6O), leaf sheaths (Fig. 6K–M, Supplementary Fig. 8) especially in the region directly below the laminar joint (Fig. 6N), and ligules and auricles (Fig. 6P). Whereas a low level of expression was observed in newly emerging leaves (Fig. 6L and M), strong expression was detected in the laminar joints (Fig. 6O).
Histochemical localization of GUS activity directed by the OsLG1 promoter:GUS fusion in transgenic rice plants. (A–C) Expression pattern of the OsLG1:GUS transgenic plants at six-leaf stage. GUS images at the laminar joint regions at four-leaf stage (D–F) and six-leaf stage (G–I). Bar, 1 mm. (J–P) Cross sections of OsLG1:GUS T1 transgenic plants. Photographs were taken with a dark-field microscope, where GUS activity appeared as pink color. (J–M) Cross section of sub-apical region. (N) Cross section of the lower part of the laminar joint region. (O) Laminar joint region. (P) Upper part of the laminar joint region. Pink color can be seen in the vascular bundle of the ligule and auricles (white arrowhead). au, auricle; cl, central lacuna; lg, ligule; vb, vascular bundle; lj, laminar joint; nl, newly emerged leaf. The boxed areas b, c, e, f, h, and i were enlarged to Figs. B, C, E, F, H, and I, respectively
To examine whether the GUS assay accurately represented the expression pattern of OsLG1, we conducted a semi-quantitative RT-PCR analysis using total RNA prepared from leaf blades directly above the laminar joint, laminar joint regions, and sheaths directly below the joint. The result in Fig. 7A shows that the level of OsLG1 transcript expression is higher in the laminar joint regions (Samples 2 and 5) and a low level of the transcript was detected in sheaths (Samples 1 and 4). A similar preferential expression pattern was observed from both the 5th and 4th leaves. Real-time PCR analyses also support the observation that the OsLG1 transcript level was higher in the laminar joint regions (Fig. 7B). Semi-quantitative RT-PCR analysis showed OsLG1 expressed in the ligule and auricles (Supplementary Fig. 9). These results are consistent with the GUS expression pattern, which shows preferential activity at the laminar joint and sub-apical regions and a low activity in sheaths, ligules and auricles. Therefore, we could conclude that the GUS assay showed a good correlation with the OsLG1 expression pattern.
RT-PCR analysis of the OsLG1 expression pattern. PCR was performed using the OsLG1-specific primers the P1 and P2 primers shown in Fig. 4A. Actin was used as a control to demonstrate equal RNA loading. (A) Samples were obtained from 4th (1–3) and 5th leaves (4–6) of 30-day-old plants. 1 and 4, 3–8 mm of sheath directly below the laminar joint; 2 and 5, laminar joint region containing the 2 mm of sheath and blade region; 3 and 6, 3–8 mm of leaf blade directly above the laminar joint. (B) Quantitative real-time RT-PCR analysis with the RNA samples prepared from the three independent samples shown in Fig. 7A. Error bars indicate the propagation of the standard deviation of three biological replicates. Ubiquitin served as a reference for normalization
Discussion
In this study, we identified T-DNA insertional mutants in the OsLG1 gene. The knockout mutants were defective in the ligule, auricles, and lamina joint. We have characterized the mutants in detail and studied expression patterns of the gene using the GUS reporter.
The OsLG1 encodes a SBP DNA binding protein containing a highly conserved region of 76 amino acids that overlaps with a NLS. SBP domain proteins were originally identified from Antirrhinum majus and bind to the promoter region of SQUAMOSA (Klein et al. 1996). Recently, 19 SBP (OsSPL) genes were identified in the rice genome. They can be classified into six subgroups (S1–S6), and OsLG1 is OsSPL8, which a member of the subgroup S5 containing the maize LIGULELESS1. Among the rice SBP genes, OsSPL2, OsSPL12, OsSPL13, OsSPL14, OsSPL16, and OsSPL18 are likely regulated by OsmiR156 (Xiong et al. 2005; Xie et al. 2006). To date, there is no report on functional characterization of the OsSPL genes. OsLG1 (OsSPL8) is the first characterized SBP gene in rice.
The oslg1 mutants showed defects in the laminar joint during leaf development. In particular, OsLG1 was required for the proper development of the laminar joint at the leaf blade-sheath boundary. In addition, the mutations also affected auricle and ligule formation. In maize, two recessive mutations, lg1 and lg2, have been reported to affect ligular region development (Becraft et al. 1990; Harper and Freeling 1996). A lg1 mutation did not influence the overall growth pattern in leaf development but altered the anticlinal division pattern in the preligular region, causing the removal of the ligule and auricle. However, the presence of some rudimentary ligules in upper leaves suggests that the lg1 mutation is leaky (Becraft et al. 1990; Sylvester et al. 1990; Moreno et al. 1997). The second mutant lg2 also affected correct initiation and positioning of the ligule and auricle (Harper and Freeling. 1996; Walsh et al. 1998). A synergistic phenotype was observed in the upper leaves of double mutants of lg1 and lg2, indicating that ZmLG1 and ZmLG2 function in the same pathway (Harper and Freeling. 1996). Another recessive mutation, extended auricle1 (eta1) caused the outgrowth of auricles and diffusion of the leaf blade-sheath boundary. In double mutants of eta1 and zmlg1 or zmlg2, synergistic phenotypes were also found as observed from the zmlg1 zmlg2 double mutant. The eta1 zmlg1 mutants do not form the ligule and auricles in both the lower and upper leaves, suggesting that eta1 is also involved in the formation of ligules and auricles (Osmont et al. 2003).
Phylogenetic and comparative genetic analysis indicated that the OsLG1 gene is orthologous to ZmLG1 (Kaplinsky et al. 2002; Xie et al. 2006). However, while a single mutation of OsLG1 resulted in complete loss of the ligules, auricles, and laminar joints throughout the plant, the maize lg1 mutant displayed a leaky phenotype and required an additional mutant to suppress organ formation in the upper leaves. Harper and Freeling (1996) have proposed two possible explanations for the production of rudimentary ligules in adult leaves. One proposal was “adult ligule elaboration program”, in which rudimentary ligule is produced in an age-dependent manner, and the other proposed explanation was gene duplication. A single OsLG1 mutation caused a complete loss of the ligule development in rice. Similarly, a mutation in barley Liguleless (HvLG1) gene, which is collinear with the Oslg1 gene and maize lg1 gene also generated complete liguleless phenotype on all leaves (Pratchett and Laurie 1994; Rossini et al. 2006). Therefore, the first possibility can be eliminated if the same molecular mechanism is under operation in both rice and maize. The reason that maize lg1 and lg2 mutants are leaky is probably due to the recent genome duplication (Ahn et al. 1993; Swigonova et al. 2004). A homologue (LOC_Os01g64020) of ZmLG2 is present in the rice genome. We did not observe the liguleless phenotypes in the T-DNA knockout mutants in the oslg2 homologous gene, perhaps due to genome duplication in rice as well (unpublished data).
Prior to this study, three mutants that affect the leaf sheath-blade boundary have been reported in rice (Kurata et al. 2005). The auricleless (aur) mutant lacks the auricle and rudimentary ligule, and the lg mutant is defective in the ligule, auricle, and laminar joint (Maekawa 1988). The collarless (col) mutant lacks the laminar joint region (Sanchez and Khush 1998). Among these three mutants, the lg mutant phenotypes bear the strongest resemblance to our oslg1 mutant. Sanchez and Khush (1998) have proposed that the lg mutant phenotypes were caused by double mutations in two independent genes: collarless and liguleless. Maekawa (1988) has isolated another liguleless mutant that showed defective development of all three organs. He demonstrated that the liguleless mutant is allelic to lg examined by Sanchez and Khush (1998). The results from our study support Maekawa’s observation that a single mutation in the LG gene is the source of alteration in all three organs. One possible explanation for this discrepancy is that the lg mutation examined by Sanchez and Khush could be leaky, therefore resulting in occasional development of the organs in mutant plants. It is also possible that the OsLG1 gene we studied is different from the LG gene previously reported, although they are on the same chromosome 4.
In Arabidopsis, the most homologous gene to OsLG1 is AtSPL8 (51% identity at protein level). However, its function is quite diverged from that of rice. Mutation in the Arabidopsis gene affects pollen sac development and causes the reduction of fertility (Unte et al. 2003). Since auricles and ligules are not present in Arabidopsis, the divergence is expected.
In addition to LG1 and LG2, other genes are also involved in ligular region development in maize. The kn1 (knotted 1) mutations cause the formation of ectopic knots (Vollbrecht et al. 1991), and the lg3 (Liguleless 3) mutation transforms blades to sheaths (Muehlbauer et al. 1997). RS1 (Rough sheath 1) is a dominant mutation that causes rough sheath (Schneeberger et al. 1995), and gnarley1 (gn1) produces altered ligules and sheaths (Foster et al. 1999). Therefore, these genes likely affect ligular region development.
Although the expression pattern of the maize LG1 gene had been investigated by the RT-PCR method (Moreno et al. 1997), its precise expression pattern has not been investigated. They reported that the gene was expressed in the ligular region of newly emerged leaves and meristems containing primordia regions in young seedlings. To study the expression pattern of the OsLG1 gene in detail, we generated a fusion molecule between the OsLG1 promoter and the GUS reporter gene. When the molecule was introduced to rice, the reporter gene expression was preferentially detected in the laminar joint region of newly emerged leaves. In the laminar joint region, GUS was expressed more strongly in the edge. The gene was preferentially expressed in the vascular bundle. This result indicates that the lack of the ligule and auricle in the oslg1 mutant is likely due to the failure of laminar joint development.
References
Ahn S, Tanksley SD (1993) Comparative linkage maps of the rice and maize genomes. Proc Natl Acad Sci USA 90:7980–7984
Becraft PW, Bongard-Pierce DK, Sylvester AW, Poethig RS, Freeling M (1990) The liguleless-1 gene acts tissue specifically in maize leaf development. Dev Biol 141:220–232
Birkenbihl RP, Jach G, Saedler H, Huijser P (2005) Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J Mol Biol 352:585–596
Cardon G, Hohmann S, Klein J, Nettesheim K, Saedler H, Huijser P (1999) Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237:91–104
Dai Z, Gao J, An K, Lee JM, Edwards GE, An G (1996) Promoter elements controlling developmental and environmental regulation of a tobacco ribosomal protein gene L34. Plant Mol Biol 32:1055–1065
Foster T, Veit B, Hake S (1999) Mosaic analysis of the dominant mutant, Gnarley1-R, reveals distinct lateral and transverse signaling pathways during maize leaf development. Development 126:305–313
Han MJ, Jung KH, Yi G, Lee DY, An G (2006) Rice Immature Pollen 1 (RIP1) is a Regulator of Late Pollen Development. Plant Cell Physiol 47:1457–1472
Harper L, Freeling M (1996) Interactions of liguleless1 and liguleless2 function during ligule induction in maize. Genetics 144:1871–1882
Hong Z, Ueguchi-Tanaka M, Matsuika M (2004) Brassinosteroids and rice architecture. J Pestic Sci 29:184–188
Hoshikawa K (1989) The growing rice plant—an anatomical monograph. Nobunkyo Press, Tokyo
Ito M, Sentoku N, Nishimura A, Hong SK, Sato Y, Matsuoka M (2002) Position dependent expression of GL2-type homeobox gene, Roc1: significance for protoderm differentiation and radial pattern formation in early rice embryogenesis. Plant J 29:497–507
Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111
Jang S, Hur J, Kim SJ, Han MJ, Kim SR, An G (2004) Ectopic expression of OsYAB1 causes extra stamens and carpels in rice. Plant Mol Biol 56:133–143
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions:β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. Embo J 6:3901–3907
Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Yang K, Nam J, An K, Han MJ, Sung RJ, Choi HS, Yu JH, Choi JH, Cho SY, Cha SS, Kim SI, An G (2000a) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22:561–570
Jeon JS, Lee S, Jung KH, Jun SH, Kim C, An G (2000b) Tissue-preferential expression of a rice α-tubulin gene, OsTubA1, mediated by the first intron. Plant Physiol 123:1005–1014
Jeong DH, An S, Kang HG, Moon S, Han JJ, Park S, Lee HS, An K, An G (2002) T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol 130:1636–1644
Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR, Kim J, Shin M, Jung M, An G (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45:123–132
Jung JY, Kim YW, Kwak JM, Hwang JU, Young J, Schroeder JI, Hwang I, Lee Y (2002) Phosphatidylinositol 3- and 4-phosphate are required for normal stomatal movements. Plant Cell 14:2399–2412
Kamiya N, Itoh J, Morikami A, Nagato Y, Matsuoka M (2003) The SCARECROW gene’s role in asymmetric cell divisions in rice plants. Plant J 36:45–54
Kaplinsky NJ, Braun DM, Penterman J, Goff SA, Freeling M (2002) Utility and distribution of conserved noncoding sequences in the grasses. Proc Natl Acad Sci USA 99:6147–6151
Kinoshita T (1998) Linkage mapping using mutant genes in rice. Rice Genet Newslett 15:13–74
Klein J, Saedler H, Huijser P (1996) A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol Gen Genet 250:7–16
Kurata N, Miyoshi K, Nonomura K, Yamazaki Y, Ito Y (2005) Rice mutants and genes related to organ development, morphogenesis and physiological traits. Plant Cell Physiol 46:48–62
Kyozuka J, Shimamoto K (1991) Transformation and regeneration of rice protoplasts. In: Lindsey K (ed) Plant tissue culture mannual B1. Kluwer Academic Publisher, Dordrecht, pp 1–17
Lee DY, Lee J, Moon S, Park SY, An G (2006) The rice heterochronic gene/SUPERNUMERARY BRACT/regulates transition from spikelet meristem to floral meristem. Plant J 49:64–78
Lee S, Jeon JS, Jung KH, An G (1999) Binary vectors for efficient transformation of rice. J Plant Biol 42:310–316
Lee S, Kim J, Son JS, Nam J, Jeong DH, Lee K, Jang S, Yoo J, Lee J, Lee DY, Kang HG, An G (2003) Systematic reverse genetic screening of T-DNA tagged genes in rice for functional genomic analyses: MADS-box genes as a test case. Plant Cell Physiol 44:1403–1411
Maekawa M (1988) A new allele at the lg locus conferring short ligules. Rice Genet Newslett 5:87–88
Miller T (1999) Puzzled by Poaceae?—a grass identification workshop. Washington State Weed Conference, November 3
Moreno MA, Harper LC, Krueger RW, Dellaporta SL, Freeling M (1997) liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev 11:616–628
Muehlbauer GJ, Fowler JE, Freeling M (1997) Sectors expressing the homeobox gene liguleless3 implicate a time-dependent mechanism for cell fate acquisition along the proximal-distal axis of the maize leaf. Development 124:5097–5106
Osmont KS, Jesaitis LA, Freeling M (2003) The extended auricle1 (eta1) gene is essential for the genetic network controlling postinitiation maize leaf development. Genetics 165:1507–1519
Pratchett N, Laurie DA (1994) Genetic map location of the barley developmental mutant liguleless in relation to RFLP markers. Hereditus 120:35–39
Rossini L, Vecchietti A, Nicoloso L, Stein N, Franzago S, Salamini F, Pozzi C. (2006) Candidate genes for barley mutants involved in plant architecture: an in silico approach. Theor Appl Genet 112:1073–1085
Sanchez AC, Khush GS (1998) A gene for collarless phenotype in rice. Rice Genet Newslett 15:99–100
Schneeberger RG, Becraft PW, Hake S, Freeling M (1995) Ectopic expression of the knox homeo box gene rough sheath1 alters cell fate in the maize leaf. Genes Dev 9:2292–2304
Sohn EJ, Kim ES, Zhao M, Kim SJ, Kim H, Kim YW, Lee YJ, Hillmer S, Sohn U, Jiang L, Hwang I (2003) Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 15:1057–1570
Swigonova Z, Lai J, Ma J, Ramakrishna W, Llaca V, Bennetzen JL, Messing J (2004) On the tetraploid origin of the maize genome. Comp Funct Genom 5:281–284
Sylvester AW, Cande WZ, Freeling M (1990) Division and differentiation during normal and liguleless-1 maize leaf development. Development 110:985–1000
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Unte US, Sorensen AM, Pesaresi P, Gandikota M, Leister D, Saedler H, Huijser P (2003) SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell 15:1009–1019
Vollbrecht E, Veit B, Sinha N, Hake S (1991) The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350:241–243
Wada K, Marumo S, Ikekawa N, Morisaki M, Mori K (1981) Brassinolide and homobrassinolide promotion of lamina inclination of rice seedlings. Plant Cell Physiol 22:323–325
Walsh J, Waters CA, Freeling M (1998) The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade-sheath boundary. Genes Dev 12:208–218
Walsh J, Freeling M (1999) The liguleless2 gene of maize functions during the transition from the vegetative to the reproductive shoot apex. Plant J 19:489–495
Xie K, Wu C, Xiong L (2006) Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol 142:280–293
Xiong Y, Liu T, Tian C, Sun S, Li J, Chen M (2005) Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Mol Biol 59:191–203
Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12:1591–1606
Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Nunokawa E, Ishizuka Y, Terada T, Shirouzu M, Osanai T, Tanaka A, Seki M, Shinozaki K, Yokoyama S (2004) A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J Mol Biol 337:49–63
Zwick MS, Islam-Faridi MN, Czeschin DG Jr, Wing RA, Hart GE, Stelly DM, Price HJ. (1998) Physical mapping of the liguleless linkage group in Sorghum bicolor using rice RFLP-selected sorghum BACs. Genetics 148:1983–1992
Acknowledgements
We thank Jong-seong Jeon, Sichul Lee, Shinyoung Lee, Sunok Moon, Choong-Hwan Ryu, and Dong-Yean Lee for helpful discussion, Young-Ock Kim and Ja Kyung Yi for assistance in protoplast cell culturing, Min-Jung Han for technical instruction the real-time PCR analysis, In-soon Park for generating transgenic GUS plants. We also express our thanks to Chahm An for critical reading of the manuscript. This work was funded in part by grants from the Crop Functional Genomic Center, the 21st Century Frontier Program (CG-1111); from the Biogreen 21 Program, Rural Development Administration; and from the Korea Science and Engineering Foundation (KOSEF) through the National Research Laboratory Program funded by the Ministry of Science and Technology (M10600000270–06J0000–27010).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lee, J., Park, JJ., Kim, S.L. et al. Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol Biol 65, 487–499 (2007). https://doi.org/10.1007/s11103-007-9196-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-007-9196-1