ABSTRACT
Purpose
We describe a nucleation-based method which allows for the generation of monodisperse lipid nanoparticles over a range of diameters. Using a set of novel zwitterionic lipids and inverse phosphocholine lipids with pKas ranging from 2 to 5, we showed how the hydrodynamic diameter of lipid nanoparticles can be systematically manipulated over a 60 nm to 500 nm size range.
Method
Lipid nanoparticles were prepared by adding an anti-solvent, such as water, to the organic phase containing the lipid components. This led to super-saturation and the spontaneous formation of particles.
Results
The growth and final particle size was controlled by the ratio of the components in the ternary system: lipid, organic solvent and aqueous phase. Particles with diameter below 125 nm were formed under conditions where the super-saturation coefficient was between 2.3 and 20. PEG-lipid served as an efficient growth inhibitor except at very high and low lipid concentrations. Encapsulation efficiency of siRNA into lipid nanoparticles was shown to be pH-dependent and requires the protonation of the anionic carboxylate groups of the zwitterionic lipids, emphasizing the importance of electrostatic forces.
Conclusion
This process enables high encapsulation efficiency of nucleic acids and allows the size of lipid nanoparticles to be controlled.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
INTRODUCTION
The systemic delivery of siRNA is a multi-step process in which several barriers must be overcome before the target cells are reached (1). Because siRNA is negatively charged and subject to degradation in the bloodstream, carriers are developed to transport the siRNA to the site of action. Among the vectors developed for siRNA delivery, lipid based delivery systems such as liposomes, lipoplexes and lipid nanoparticles have proved to be effective carriers and are currently assessed as clinical candidates for the treatment of liver cancer, liver-related diseases, and solid tumors (2,3).
siRNA/lipid nanoparticles must be within a specific range of sizes in order to be effectively delivered to the site of action after systemic administration. Certain organ and tumor tissues are best reached with particles smaller than 150 nm (4,5). Access to organ and tissue cells is restricted by the extracellular matrix and connective tissue, which represent a dense network with only small gaps (6). For example, transendothelial openings in fenestrated capillaries of tumor tissues with diameters of 40–80 nm put a limit on the size of particles which can be effectively delivered to the site of disease. Similarly, access to hepatocytes in the liver is restricted by the fenestrated endothelium with gaps of 100–150 nm in diameter (2,7).
Existing lipid nanoparticle preparation methods include the T-mixing method to reproducibly prepare cationic lipid-DNA complexes (8), the ‘ethanol-destabilized preformed liposomes’ process (9), and the ‘stepwise ethanol dilution method’ (10). For the ethanol-destabilized liposome approach, the sizes of the preformed and extruded liposomes are very sensitive to the nucleic acid addition step. Thus, under certain conditions particles can increase in size in an uncontrolled manner or irreversibly aggregate. Similar to the ethanol-destabilized liposome process, small particles can be obtained with the stepwise ethanol method. Using this procedure, lipids are dissolved in ethanol and mixed with a buffer solution containing nucleic acid. A second dilution step with buffer is required to obtain high encaspulation efficiency and stable particles. However, a mechanism-based rationale has not yet been described to explain how these nanoparticles form, how particle size can be controlled, or how aggregation can be circumvented.
In this paper, we present a nucleation based method, which allows for the generation of monodisperse lipid nanoparticles over a broad size range (60–500 nm). The nucleation of lipid nanoparticles is caused by super-saturation. This is initiated by the addition of an anti-solvent to the organic solvent phase which contains the lipid components (11,12). An aqueous buffer system is used as the anti-solvent, while the organic solvent phase can be any solvent, that is miscible with water and in which the lipid components are soluble. This includes methanol, ethanol, tetrahydrofuran, etc. The final size of the particles is dictated by the nucleation and growth kinetics. Both can be controlled by altering parameters in the super-saturation method (13). The hydrodynamic diameters of the particles can be regulated and shifted to smaller or larger size ranges through manipulation of the following process parameters: (1) lipid composition and concentration, (2) solvent to anti-solvent ratio, (3) lipid solubility in the final mixture. These parameters influence the systems super-saturation coefficient. A fourth way is the use of additives such as PEG to provide a steric barrier to prevent coalescence.
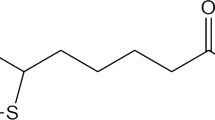
Zwitterionic lipids (ZL) and inverse phosphocholine (iPC) lipids with pKas ranging from 2 to 5 were used to generate lipid/siRNA nanoparticles via controlled nucleation (14, 15). The ZL consist of anionic protonable carboxylate groups and tertiary or quarternary amine groups (Fig. 1); the iPC lipids also contain quarternary amines, but have a phosphate as the protonable anionic group (Fig. 1). By constructing a ternary phase diagram we demonstrate systematically how parameters of a multicomponent nanoparticle system (lipid components, siRNA, organic phase, and water phase ratio) can be used to control the hydrodynamic diameter of the particles and the encapsulation of siRNA in the particles.
MATERIALS AND METHOD
Materials
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioctadecanoyl-sn-glycero-3-phosphocholine (DSPC), Cholesterol, N-palmitoyl-sphingosine-1-{succinyl [methoxy (polyethylene glycol)750] (C16 PEG750), and N-palmitoyl-sphingosine-1-{succinyl [methoxy(polyethylene glycol)2000] (C16 PEG2000) were obtained from Avanti Polar Lipids, Inc. (Alabaster, AL, USA). PEG-DMG was purchased from NOF-corporation (Tokyo, Japan). Zwitterionic lipids (ZL) and iPC lipids were synthesized as described in (14,15). Slide-A-Lyzer Dialysis cassettes, 10 K MWCO, were purchased from Thermo Scientific. The anti-luciferase siRNA: 5’-GAUUAUGUCCGGUUAUGUA-3’ (cat. No. D-002050-01-20) was purchased from Dharmacon. Quant-iT™ RiboGreen® RNA Reagent was purchased from Invitrogen (Carlsbad, USA). All other chemicals were purchased from Sigma Aldrich.
Nanoparticle Preparation Using the Controlled Nucleation Method
Lipid nanoparticles were prepared using a batch mixing process at 25 °C. The lipid or lipid mixture was dissolved in a water-miscible organic solvent (methanol) and sonicated at 25 °C for 5 min. The lipid/organic solvent solution was subsequently injected into a magnetically stirred 2.5 ml vial, which contained siRNA dissolved in an aqueous buffer system. For reproducible injection speed, a KD Scientific pump holding a syringe with a 22 G × 1 1/2 needle was used to inject the methanol lipid solution into the aqueous phase. The injection rate was 5 ml/min unless specified otherwise. The lipid suspension was stirred for 10 min at 350 rpm. Methanol was either removed under reduced pressure using a rotavapor for 1–4 h or by dialysis against PBS without Ca2+ and Mg2+. The amount of lipid, organic phase and aqueous phase was adjusted to obtain the desired mass fractions (mf) in the three-component system. These components were expressed as mass fraction of lipid (mflipid), mass fraction of methanol (mfMeOH) and mass fraction of water (mfH2O), where mflipid + mfMeOH + mfH2O = 1. A typical lipid nanoparticle formulation was prepared as follows: 0.5 g of aqueous phase (50 mM citric buffer, pH 3) containing 150 μg of siRNA was placed into a 2.5 ml glass reaction vial containing a magnetic stirring bar. Then 3 mg of lipid mixture was dissolved in 0.5 g of methanol and injected into the aqueous buffer system under constant stirring.
Extrusion
To investigate the effect of extrusion on particle size, lipid nanoparticles were extruded in the presence of methanol or after removal of methanol through 80 nm polycarbonate membrane (Avestin, Ottawa, CA) 5 times at room temperature.
3D Surface Plot: Three Component System
A surface plot was constructed for two different lipid nanoparticle compositions. Formulation 1 was composed of DLinPAT/DOPE/PEG-DMG at a molar ratio of 60:30:10. Formulation 2 was composed of DLinPAT/DSPC/Chol/PEG-DMG at a molar ratio of 40:10:40:10. The lipid/siRNA ratio used was 20:1 (wt:wt). The lipid nanoparticles were prepared using the controlled nucleation method and consist of three components: aqueous phase, organic phase and lipids. The ratios between these 3 components determine nucleation and the final particle size.
In the surface plot, the hydrodynamic diameters of the lipid nanoparticles were plotted onto the y-axis, the mass fraction of methanol on the x-axis, and the mass fractions of lipids onto the z-axis. The mass fraction of the aqueous phase (aqueous buffer), can be determined by subtracting the mass fraction of methanol and the corresponding mass fraction of lipid from 1 (mfH2O = 1 − mfMeOH − mflipid). For the DLinPAT/DOPE/PEG-DMG formulation seven mass fractions of methanol (0.1, 0.2, 0.3, 0.4, 0.5, to 0.7) each combined with eight mass fractions of lipid (0.0001, 0.0003, 0.001, 0.003, 0.01, 0.015, 0.02, and 0.05) were tested. For formulation 2 (DLinPAT/DSPC/Chol/PEG-DMG) five mass fractions of lipid (0.0001, 0.0003, 0.001, 0.003, and 0.01), each combined with five mass fractions of methanol (0.1, 0.2, 0.3, 0.4, 0.5) were investigated. Higher mass fractions of lipid could not be tested due to limited solubility of DLinPAT/DSPC/Chol/PEG-DMG in methanol. The hydrodynamic diameters were measured after methanol was removed by dialysis. Lipid nanoparticles were not subjected to extrusion. Results are given as mean values ± sd of three independent experiments.
Determination of Super-Saturation
In precipitations generated by the addition of an anti-solvent to a dissolved solute, the super-saturation is reported as the ratio of the total mass of solute added divided by the final solution volume, c, to the bulk solubility, c∞, in the final solvent mixture: Super-saturation : S = c/c∞ (11). The super-saturation of the DLinPAT/DOPE/PEG-DMG lipid mixture was determined at multiple solvent/anti-solvent mixtures and lipid concentrations (mflipid 0.05, 0.01, 0.0001). Since DLinPAT and DOPE are the major contributors to the super-saturation of the DLinPAT/DOPE/PEG-DMG (60/30/10) lipid formulation in the resulting solvent mixture, the super-saturation is presented as combined values of DOPE and DLinPAT. The solubility of the lipid mixture in different aqueous buffer/methanol compositions was used to determine super-saturation. Three different lipid mass fractions (0.05, 0.01, 0.0001) of DLinPAT/DOPE/PEG-DSPE were dissolved in MeOH and added at the indicated mass fraction ratios to aqueous buffer solution. After 15 min, the liposomal mixtures were centrifuged for 60 min at 100,000 rpm at 4 °C (TLA 100.2 rotor, Beckman TL-100, Beckman Coulter, Fullerton, CA). The supernatant was then separated from the pellet and the amount of lipid in the pellet and supernatant was quantified by reverse phase HPLC. HPLC was performed using an analytical reverse phase C8 column (Agilent Zorbax Eclipse XDB-C8 (4.6 × 150 mm; 5 μm)) with a mobile phase composed of a mixture of water and methanol containing 0.1 % trifluoroacetic acid. DLinPAT and DOPE were eluted with a 15 min MeOH gradient (90-99 %) using a flow rate of 1.25 ml/min, a column temperature of 45 °C and peak monitoring at 205 nm. The injection volume was 50 μl. The elution times for DLinPAT and DOPE were 5.339 and 7.125 min, respectively. Figure 3c is representative of a sample chromatogram. Lipid standard curves in the range from 0.1 mM to 1 mM were generated for both DLinPAT and DOPE, and used to determine the lipid concentration in each sample run. Experiments were repeated twice.
Particle Size of Lipid Nanoparticles
Hydrodynamic diameter and particle size distributions were measured by dynamic light scattering using a Malvern Zetasizer NanoZS (Malvern, UK). The z-average (Z-ave) diameters of at least ten measurements were reported. All measurements are given as mean values ± sd of three independent runs, each performed in triplicate
Phase Contrast Images of Lipid Nanoparticle Suspension
For phase contrast images, lipid nanoparticle suspensions were placed onto a glass slide and covered with a cover glass. Phase contrast microscopy was performed on a Nikon Eclipse TS 100 using a Plan Flour ELWD 20×/0.45 objective. Images were recorded with a RT Monochrome Diagnostic instruments camera and the SPOT advanced software.
Effect of PEG on Particle Growth
To investigate the effect of different concentrations of PEG on particle size and the growth of lipid nanoparticles, increasing amounts of PEG-lipid (C16 PEG750 and C16 PEG2000) were incoporated into the DLinPAT/DSPC/Chol/PEG-lipid. The DSPC concentrations were adjusted to account for the decreasing amounts of PEG-content (10 %, 5 %, 1.5 %, and 1 %). The mole ratio of the lipid components were as follows: DLinPAT/DSPC/Chol/PEG-lipid (40/10 + (10-x)/40/x), where x is the mole % of PEG-lipid. Results are given as mean values ± sd of three independent experiments.
Encapsulation of siRNA
siRNA encapsulation efficiency was determined using the Ribogreen® assay. Ribogreen is a membrane-impermeable dye and increases its fluorescence upon binding to siRNA. Encaspulation efficiency of siRNA was quantified by measuring the fluorescence signal upon addition of Ribogreen to aliquots of lipid nanoparticle formulations in the presence or absence of 0.4 % Triton-X. Fluorescence was measured using the Fluostar fluorescence plate reader (BMG) at excitation and emission wavelengths of 485 nm and 520 nm respectively. siRNA encapsulation efficiency was quantified as follows: \( \% = \left( {{{\hbox{F}}_{\rm{T}}} - {{\hbox{F}}_{\rm{B}}}} \right)/{{\hbox{F}}_{\rm{T}}} * {1}00 \), where FB represents the fluorescence of liposomal formulation before treatment with Triton, and FT represents the fluorescence of liposomal formulation after treatment with Triton. siRNA was quantified by a calibration curve ranging from 50 ng/ml to 1 μg/ml siRNA. Results are given as means of triplicate measurements ± sd and are representative of three independent experiments.
Effect of pH and pKa on Encapsulation Efficiency of siRNA
To investigate the influence of pH on siRNA encapsulation efficiency, the following buffers were used: 50 mM citric acid buffer, pH 3; 50 mM citric acid buffer, pH 4; 50 mM sodium aceate buffer, pH 5; 50 mM sodium phosphate buffer, pH 7.4; and 50 mM Tris, pH 8.5. DOBAQ/DOPE/PEG-DMG (molar ratio 60/30/10) at a 20/1 lipid/siRNA wt/wt ratio was used as a representative lipid formulation. The effect of lipid pKa on siRNA encapsulation was investigated using a set of lipids with pKas ranging from 2 to 5: DOBAQ, DLinPAT, DOMPAQ, DOAAQ, DOCP, and DOCPe. Lipid nanoparticles were prepared in a 50 mM citric acid buffer, pH 3. Experiments were repeated twice and results are given as means of triplicate measurements ± sd.
RESULTS
Hydrodynamic Diameter: A Function of Solvent, Anti-Solvent and Lipid Ratio
Different therapeutic applications require specific physicochemical properties of particles (i.e. size, charge) for efficient delivery. Thus methods to fine-tune these parameters become increasingly important. The optimal ratio of solvent/anti-solvent to lipid for generating particles in a certain size range is specific for each lipid composition. Therefore, this needs to be adjusted accordingly. The hydrodynamic diameter of DLinPAT/DOPE/PEG-DMG lipid nanoparticles can be controlled by adjusting the mass fractions of the ternary system: mflipid, mfH2O, mfMeOH. As illustrated in the 3D surface plot in Fig. 2a, small particles in the size range of 50–100 nm are obtained with methanol mass fractions from 0.1 to 0.5 in certain combinations with lipid mass fractions from 0.0003 to 0.05. Regions with large particles are found in regions of low methanol mass fractions (0.1 & 0.2) and high lipid mass fractions (0.015, 0.02 and 0.05). Highly diluted lipid solutions can also result in the formation of large particles (up to 800 nm). This is the case when a low mass fraction of lipid (mflipid 0.0001) is dissolved in a high mass fraction of methanol (mfMeOH 0.7). A growth in particle size of 40–500 nm is observed for all lipid mass fractions when the methanol mass fraction is increased from 0.5 to 0.7.
A simplified 2D graph (Fig. 2b), which details the relationship between mflipid, mfMeOH and particle diameter reveals that intermediate particle sizes of 175 nm to 275 nm are generated at a low lipid mass fraction (mf lipid = 0.0001) with methanol mass fractions ranging from 0.1 to 0.5. For the 2 highest lipid mass fractions (0.02 and 0.05), small particles (60–100 nm) were obtained at methanol mass fractions from 0.3 to 0.5. Both, decreasing the mfMeOH to 0.1 or increasing it to 0.7 resulted in the formation of larger particles. For lower lipid mass fractions (0.001 and 0.003) particles in the range of 50–100 nm are generated with methanol mass fractions from 0.1 to 0.4. Lipid mass fractions of 0.015 and 0.01 displayed the widest range of particle sizes in the desired spectrum of 50–100 nm. When increasing the methanol mass fraction from 0.5 to 0.7, a trend of increasing sizes for all lipid mass fractions was observed.
(a) 3D surface plot showing the hydrodynamic diameter of DLinPAT/DOPE/PEG-DMG (60/30/10) formulation as a function of mfMeOH and mflipid. (b) Stable nanoparticle region: plot shows the effects of methanol mass fraction (mfMeOH) and lipid mass fraction (mflipid) on hydrodynamic diameter of lipid nanoparticles with sizes <300 nm.
Effect of Super-Saturation on Particle Size
As described by Addio and Prud’homme (11), super-saturation can be controlled through the concentration of the solute in the system and the solubility of the solute in the resulting mixed solvent system (in this case aqueous buffer/methanol). The super-saturation of the DLinPAT/DOPE/PEG-DMG lipid mixture in different aqueous buffer/methanol compositions was determined by mixing a lipid methanol solution at certain mass fractions with the aqueous phase. mfMeOH 0.1 to 1 corresponds to methanol concentrations of 10–100 %. Super-saturation was determined for 3 different mflipid (0.0001, 0.01, 0.05) by reverse phase HPLC (Fig. 3c). In Fig. 3a, the super-saturation coefficients of DLinPAT and DOPE, combined, are presented as a function of mfMeOH and mflipid. Super-saturation increased with decreasing percentage of methanol (decreasing mfMeOH) and increased with increasing mflipid (Fig. 3a). The highest super-saturation was obtained for the highest lipid concentration (mflipid 0.05). Low super-saturation in the range of 1.1 to 1.7 was observed for mflipid 0.0001. In Fig. 3b, the super-saturation is correlated with the hydrodynamic diameter. Particle sizes smaller than 125 nm were obtained with super-saturation from 2.3 to 20. This corresponds to mflipid 0.01 at mfMeOH 0.1 to 0.5, and mflipid 0.05 combined with mfMeOH 0.3 to 0.7. Lower super-saturation in the range of 1.7 yielded particle sizes larger than 125 nm (177 to 220 nm). At the lowest super-saturation of 1.1 (mflipid 0.0001 combined with mfMeOH 0.7) particle sizes increased to 784 nm. Super-saturation ~40 resulted in particle sizes of 885 nm.
Influence of Lipid Composition on Particle Size
In order to investigate how lipid composition affects size distribution, the helper lipids in the first formulation were replaced with DSPC/Chol/PEG-DMG while keeping DLinPAT as the lead lipid. Cholesterol and DSPC were chosen because they both display different molecular geometry compared to DOPE (16). The difference in lipid geometry can influence packing of the lipids in the bilayer and hence particle sizes. For DLinPAT/DSPC/Chol/PEG-DMG, the optimal region for generating lipid nanoparticles <100 nm was obtained with mflipid 0.0001 to 0.003 at mfMeOH below 0.3. Large, aggregated particles >500 nm were found at mfMeOH above 0.3. This corresponds to a methanol concentration of >35 % MeOH in the final solution (Fig. 4a, b). An expanded graph depicting the particle sizes in the smaller diameter ranges shows that particles <100 nm were generated at methanol mass fractions of 0.2 to 0.3 and lipid mass fractions of 0.0001 to 0.003 (Fig. 4c). Decreasing the volume and mass fraction of methanol to 0.1 or increasing the lipid mass fraction to 0.01 both resulted in larger particles.
Phase contrast images of lipid nanoparticles generated with high mass fractions of methanol were taken (mfMeOH > 0.3) (Fig. 5). The formation of cholesterol needles >10 μm was observed. This shows that at high methanol concentrations, two main processes were competing with another: nucleation to lipid nanoparticles and the formation of cholesterol needles.
a 3D surface plot showing the hydrodynamic diameter of DLinPAT/DSPC/Chol/PEG-DMG (40/10/40/10) formulation as a function of mfMeOH and mflipid. b 2D plot shows the effects of mfMeOH and mflipid on hydrodynamic diameter of lipid nanoparticles (c) Plot shows the distribution of hydrodynamic diameter of DLinPAT/DSPC/Chol/PEG-DMG lipid (40/10/40/10) nanoparticles in the size range of interest (60–140 nm).
Speed of Injection and its Influence on Hydrodynamic Diameter
Particle formation is mainly controlled by the local super-saturation at the boundary surface of the solvent and anti-solvent while mixing. Thus, the influence of speed of injection of the methanol lipid phase into the aqueous solution or vice versa was investigated using the lipid composition DLinPAT/DOPE/PEG-DMG. A significant increase in size of 12–15 nm occurred when the injection speed was decreased from 5 ml/min to 0.5 ml/min or 0.1 ml/min (Fig. 6a). A further increase in particle size can be obtained when injecting the siRNA/buffer solution into the lipid/methanol mixture (Fig. 6b). At an injection speed of 5 ml/min particles were 100 nm. However, particle sizes increased to 200 nm when the injection speed was decreased to 0.1 ml/min.
Effect of PEG on Particle Growth
The colloidal stability of particle formulations is an important aspect for therapeutic delivery as well as long-term storage. Uncontrolled growth and irreversible aggregation of particles due to Van der Waals interactions often complicate the process of nanoparticle preparation. Since PEG coatings are known to stabilize particles in various polymeric and lipidic nanoparticle systems, we investigated its effect on the formation of DLinPAT/DSPC/Chol/PEG-lipid nanoparticles. Two PEG-lipids with different PEG chain lengths (750 Da and 2000 Da) were incorporated into the lipid nanoparticles in different mol concentrations (0–10 %). As shown in Fig. 7, sizes increased with decreasing PEG concentrations as well as with shorter PEG chain length. At the same mole concentrations, the PEG 2000 Da leads to smaller sizes than PEG 750 Da. Uncontrolled particle growth due to precipitation and aggregation of particles occurs when no PEG-lipid was added.
Extrusion of Lipid Nanoparticles Before Solvent Removal Allows Size Reduction
The particle sizes reported in Figs. 2–7 were all obtained through the optimal choices of mfMeOH, mflipid and mfH2O, without any further post-processing of the particles. The ability to manipulate particle diameter by extrusion may provide additional benefit for generating particles in the size range of interest. DLinPAT/DOPE/PEG-DMG and DLinPAT/DSPC/Chol/PEG-DMG lipid nanoparticles were either extruded before or after removal of methanol through 80 nm polycarbonate membranes. As shown in Fig. 8a, b, the hydrodynamic diameter of lipid nanoparticles can be further reduced by at least 40 nm (p < 0.01 for DLinPAT/DOPE/PEG-DMG and p < 0.001 for DLinPAT/DSPC/Chol/PEG-DMG), without affecting the encapsulation efficiency of siRNA, when extrusion is performed before the removal of methanol. However, if lipid nanoparticles were extruded after methanol removal, no significant reduction in size was observed.
Effect of pKa and pH on Encapsulation Efficiency of siRNA
A thorough understanding of the factors influencing the encapsulation efficiency of nucleic acids into lipid nanoparticles is just as important as the optimization of particle size. Encapsulation of nucleic acids into lipid nanoparticles can occur by a passive process (17,18) or through electrostatic interactions. To investigate the conditions of optimal siRNA encapsulation, various zwitterionic lipids with pKas ranging from 2 to 5 were used for the preparation of nanoparticles. Lipid nanoparticles were formed with all tested lipids. However, encapsulation efficiency of siRNA was only achieved when the pH of the buffer used was below the pKa of the lipids. For DOBAQ/DOPE/PEG-DMG lipid nanoparticles at pH 7.4 and pH 8.5, the percentage of siRNA encapsulated was negligible at 4–5 %. This could be further increased to 39 ± 5 % at pH 5 and 85 ± 7 % at pH 3 (Table Ia). When keeping the pH at 3, but using zwitterionic lipids with different pKas, only the one with a pKa value of 5 (DOBAQ, DLinPAT) showed high encapsulation efficiency of siRNA with 75 ± 5 to 83 ± 7. In contrast, lipids with pKa < 3, such as DOAAQ, DOCP, and DOCPe, showed no siRNA encapsulation at pH 3 (Table Ib). No statistically significant decrease in siRNA activity was observed, when exposing the siRNA to acidic pH 3 (data not shown). This is in line with studies showing that less than 3 % of the siRNA is degraded at low pH (9,19).
Effect of Process Parameters on Encapsulation Efficiency of siRNA
The ratios between methanol, water, and lipid not only affect hydrodynamic diameter, but also encapsulation efficiency of siRNA. As can be seen in Fig. 9a, encapsulation efficiency of siRNA for all mass fractions of lipids (DLinPAT/DOPE/PEG-DMG) is enhanced with increasing mass fractions of MeOH. This corresponds to a higher percentage of methanol in the final lipid formulations. Encapsulation efficiency of siRNA seems to be more affected by the mass fraction of MeOH when using high mass fractions of lipids. The encapsulation efficiency of siRNA was enhanced by 40 % for higher lipid mass fractions (0.01, 0.015, 0.02) when the methanol mass fraction was increased from 0.1 to 0.5. Although higher methanol mass fractions also lead to higher siRNA encapsulation when using low mass fractions of lipids (0.001 & 0.003) the effect was less pronounced and an increase of 20 % was achieved. This phenomenon is depicted in Fig. 9b: the steepest slopes were observed for the high lipid mass fractions (0.015 and 0.02) and decreased when lipid mass fractions were lowered to 0.01, 0.003 and 0.001.
DISCUSSION
In this paper, we have presented evidence showing that particle sizes of lipid nanoparticles and encapsulation efficiency of nucleic acids can be regulated using the controlled nucleation method. As the rate of super-saturation ultimately determines nucleation and particle growth (11,12), changing mass fractions of solvents, anti-solvents, and lipids will affect precipitation and particle formation. For the hydrophobic lipid components, nucleation is initiated by changing from the organic media, in which the lipids are highly soluble, to a poor solvent mixture by adding aqueous media. Although, this procedure allows for the use of all organic solvents, which are miscible with water and are able to dissolve the lipids (i.e. ethanol, tetradydrofuran, etc.), we chose methanol in these experiments. With its low boiling temperature, methanol can be removed not only by dialysis but also by rotary evaporation at reduced pressure.
Because super-saturation can be controlled by the lipid concentration in the system and its solubility in the solvent/anti-solvent mixture (11), we investigated the size distributions of 2 different lipid compositions as a function of different lipid concentrations and solvent mixtures. In both formulations, the zwitterionic lipid DLinPAT was used as a lead lipid, but formulated with different helper lipid compositions (DLinPAT/DOPE/PEG-DMG and DLinPAT/Chol/DSPC/PEG-DMG). The 3D surface plots showed that the optimal range of methanol, water and lipid mass fractions in which particle diameters smaller than 100 nm are obtained, is different for each lipid composition. This may be due to different solubilities of the helper lipids in methanol or molecular shapes that could result in different membrane packing (16,20). For DLinPAT/DOPE/PEG-DMG, at high super-saturation (c/c∞ = 40) which is the case at low methanol mass fractions (mfMeOH 0.1) combined with high lipid mass fractions (mflipid 0.05), particles aggregated (Figs. 2a, 3). This may be attributed to a local increase in the number of lipid molecules, which can participate in the formation of particles and aggregation. Higher lipid concentrations also enhances the probability of collision between the nanoparticles, and hence increase aggregation (21). An increase in particle size of ~50 nm was also observed for DLinPAT/DSPC/Chol/PEG-DMG at high lipid concentrations (mfMeOH 0.2 & 0.3 and mflipid 0.01) compared to those generated at lower mass fractions of lipid (Fig. 4b, c). Higher lipid mass fractions, at which the effect of concentration may be more pronounced, could not be investigated for DLinPAT/DSPC/Chol/PEG-DMG lipid nanoparticles due to the limited solubility of cholesterol in methanol (22).
As shown in Fig. 3a, b, colloidally stable particles with large hydrodynamic diameters (500–800 nm) were found for DLinPAT/DOPE/PEG-DMG lipid nanoparticles at low mflipid 0.0001 in a high mfMeOH 0.7. This corresponds to a low super-saturation (c/c∞ = 1.1) in the final mixture. One explanation could be that PEG depletes at the interface of the particles due to its high solubility in water. This could cause a loss of steric stabilization and accelerated particle growth, resulting in particles with large diameters and multi-lamellarity. An alternative explanation why larger particles are formed at the same lipid mass fraction but higher methanol concentrations (mfMeOH 0.7) could be that the lipids are more soluble at high methanol concentrations. Hence, the initial numbers of nucleating particles formed are lower at high methanol concentrations than at lower methanol concentrations. Upon slow removal of methanol by dialysis, the lipid molecules that remained in solution, precipitates onto the fewer nucleation sites that were initially formed (21). Thus larger particle sizes are generated. This corroborates the findings by Chen et al. who showed that peptide nanoparticles and ß-carotene particles were larger when the super-saturation rate was lower (23,24).
For the DLinPAT/DSPC/Chol/PEG-DMG formulation, particles smaller than 140 nm were obtained at all mass lipid fractions as long as the mass fraction of methanol did not exceed 0.3. With higher amounts of methanol (mfMeOH = 0.4 and mfMeOH = 0.5) particles grew in an uncontrolled manner leading to irreversible aggregation (Fig. 4a). Phase contrast images showed the formation of large cholesterol needles >10 μm (Fig. 5). Since a common method to crystallize cholesterol from organic solvents involves adding water, two main processes were competing with one another at high methanol concentrations: nucleation to lipid nanoparticles and the formation of cholesterol needles.
Because the particle formation takes place at the boundary surface between the solvent and anti-solvent upon mixing, changes in the meso-mixing and micro-mixing mode of these two solvents significantly affect nucleation kinetics and particle growth (12). As shown in Fig. 6, particle size does not only depend on the mass fraction ratios of the ternary system, but also on the mixing mode and mixing speed. Although the same amount of lipid, methanol, and water was used for all DLinPAT/DOPE/PEG-DMG formulations; the speed of mixing (5 ml/min, 0.5 ml/min, 0.1 ml/min) and the mode of mixing (organic phase into aqueous phase and vice versa) result in different initial super-saturation rates of the lipid in the solvent mixture. According to theoretical deductions by Cao, final particle sizes depend on the concentration of a solute in a solvent (25). High initial super-saturation of a solute in a solvent favors the formation of a large number of nuclei. Since the number of available molecules at a given concentration of solute is the same, a larger number of nuclei favor smaller particle sizes. The highest initial super-saturation resulting in smallest particle sizes was obtained when the lipid/methanol solution was injected at high speed into the aqueous solution. Slower injection resulted in smaller amounts of lipid and lower super-saturation at the very beginning. The number of nucleation sites formed was smaller and the nucleation sites continued to grow by collecting still dissolved or newly added lipid molecules. Lower super-saturation leading to slightly larger particles (100 nm vs. 60 nm) also occurs with high initial amounts of solvents and low concentrations of the anti-solvent water. This is the case when the aqueous phase is added to the methanol/lipid phase (21). The effect is even stronger when the injection speed was reduced from 5 ml/min to 0.1 ml/min. Particle sizes increased to 200 nm, indicating a reduction in the number of particles. Changes in particle diameter were also observed when altering the stirring from 120 rpm to 350 rpm and 1200 rpm. For DLinPAT/DSPC/Chol/PEG-DMG, particle sizes of 82 nm ± 7 nm were obtained at both 350 rpm and 1200 rpm. However, at the low stirring speed (120 rpm) particle sizes slightly increased to 100 nm ± 6 nm.
Particle size was not only controlled by adjusting the super-saturation of the lipid compositions in the solvent and anti-solvent mixture (Fig. 3), but also using a PEG-coating. As shown in Fig. 7, particle sizes decreased with the mole % of PEG and PEG chain lengths. Lipid nanoparticles irreversibly aggregated when no PEG-lipid was included in the formulations. This indicates that PEG serves as an effective steric barrier and is important for suspension stability (26). The PEG coating acts as a growth inhibitor and prevents flocculation by reducing the number of collisions and preventing the particles from fusing (27,28). Thus, longer PEG-chain lengths are more effective as particle growth inhibitors than shorter ones.
A 3D-surface plot is convenient for visualizing how three components can impact hydrodynamic diameter of particles. It is also helpful to determine process parameters that yield particles in the size range of interest. However, such a process requires that data be collected over a large range of independent variable values. This can be time consuming and requires a large amount of lipid and siRNA. Thus, we have shown that extrusion is a viable method for refining particle sizes without having to investigate all the process parameters. We found that the size of the lipid nanoparticles can be reduced by at least 40 nm when the extrusion was performed before the removal of the solvent (Fig. 8). Furthermore, extrusion did not affect encapsulation efficiency of siRNA (data not shown). Particle sizes could not be decreased when extrusion was done after solvent removal. One explanation for this could be that lipid nanoparticles are more fluidic in the presence of the organic solvent. During the extrusion process they are more likely to deform, re-organize and adapt to the size of the membrane pores (9). In contrast, after removal of the organic solvent, the solubility of the lipid nanoparticles in the final aqueous solution is reduced. This makes them stiffer and less likely to form smaller structures.
The encapsulation efficiency of siRNA depends on several factors: (1) the siRNA to lipid ratios; (2) the pKa of the lipids and the pH of the buffer used for particle preparation; and (3) the percentage (mass fraction) of the organic solvent in the final solution. As can be seen in Table Ia and b, encapsulation of siRNA was pKa and pH dependent. Electrostatic interactions with the siRNA, and thus the encapsulation of the siRNA, can only occur at pH values where the zwitterionic lipids are in a protonated state and therefore positively charged (9). Increasing the pH above the pKa of the lipids decreased or completely inhibited siRNA encapsulation. ZL with a pKa of 5 required a pH of 3 for substantial siRNA encapsulation. However, pH 3 was not sufficient for lipids with even lower pKa (<3), such as DOAAQ or DOCP, and DOCPe. At the optimal pH, the mass fraction of methanol and lipid are also important factors which affect the encapsulation efficiency. As shown in Fig. 9a, b, encapsulation efficiency of siRNA increased in a linear manner with increasing concentrations of methanol. Enhancement of up to 50 % encapsulation efficiency was observed when mfMeOH increased from 0.1 to 0.5. Strikingly, the effect of methanol concentration on siRNA encapsulation efficiency was more noticeable when using high mass fractions of lipid (see increasing slope factors for regression lines in Fig. 9b). One of the reasons could be that, at low methanol mass fractions, lipids may nucleate too fast at the interface after mixing. With low methanol concentration, the vesicles are also less deformable and may not be able to rearrange during the nucleation process to encapsulate siRNA that is further away from the initial interface.
Based on these results we suggest a model to explain the lipid nanoparticle formation generated by the controlled nucleation method (Fig. 10). (Step 1) Upon mixing of the lipid/methanol solution with an aqueous buffer, precipitation of lipid molecules at the interfaces between the organic and aqueous phase occurs due to local super-saturation. Super-saturation is achieved by adding the lipid/methanol solution into an anti-solvent, in this case aqueous buffer solution. Lipid molecules orient themselves with the hydrophilic headgroups pointing in the direction of the aqueous phase and the hydrophobic tails pointing towards the organic phase. (Step 2) As the methanol is diluted by mixing of the aqueous and organic phase, lipid bilayer fragments are formed. (Step 3) If lipid bilayer fragments are positively charged, negatively charged nucleic acids in the aqueous solution will serve as nucleation sites for the precipitation of lipid molecules into vesicles. (Step 4) Finally, to reduce the overall energy of the system bilayer fragments assemble into vesicles to form lipid nanoparticles (29,30).
Since nucleation of the lipid nanoparticles occurs at the interfaces between the organic phase and aqueous phase, the mixing mode plays a critical role in the formation of, and thus the size of, the lipid nanoparticle. For batch mixing processes, which are presented here, lipid concentration, lipid composition, and the mixing mode (i.e. injection speed) control particle sizes. For microfluidic devices, the flow conditions in the channels determine the mixing mode and the interfaces between two solutions (31–35). Thus, the distribution of lipid nanoparticle sizes generated by microfluidics are a function of the microchannels’ architecture, the flow rate and mixing speed of the two solutions (33). Microfluidic devices can be designed to mix solutions under laminar flow conditions or by chaotic advection (31). The latter enables shorter mixing times and higher flow-rates by using microchannels with staggered herringbone, zig-zag shaped, or serpentine structures (36,37). This can result in a more homogeneous nucleation mixing environment capable of producing smaller particle sizes (38). Thus, microfluidic devices would seem to be the preferred technologies to exploit super-saturation conditions formed by solvent dilution methodologies to produce liposomes, emulsions, and other nanoparticles of defined diameters.
CONCLUSION
In this study we demonstrated how lipid nanoparticles can be generated based on the controlled nucleation process. Using a mechanism-based approach we showed how sizes of lipid nanoparticles and encapsulation efficiency of nucleic acid can be manipulated and systematically controlled by altering process conditions. Since the final particle size depends on factors such as super-saturation, lipid composition, and amounts of growth inhibitors (PEG), experimental conditions need to be optimized for each lipid nanoparticle formulation. For DLinPAT/DOPE/PEG-DMG particles with hydrodynamic diameters below 125 nm are obtained in regions of intermediate super-saturation. Both, low and high super-saturations of lipids lead to larger particles. Further investigations are required to learn whether this is a general phenomenon for other liposomal formulations. By adjusting the ratios of lipid, organic solvent, and aqueous phase, lipid nanoparticles with size ranges tailored to specific biological and therapeutic applications can be generated.
REFERENCES
Nguyen J, Szoka FC. Nucleic acid delivery: the missing pieces of the puzzle? Accounts of Chemical Research. 2012.
Akinc A, Goldberg M, Qin J, Dorkin JR, Gamba-Vitalo C, Maier M, et al. Development of lipidoid-siRNA formulations for systemic delivery to the liver. Mol Ther. 2009;17(5):872–9.
Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28(2):172–6.
Gaumet M, Vargas A, Gurny R, Delie F. Nanoparticles for drug delivery: the need for precision in reporting particle size parameters. Eur J Pharm Biopharm. 2008;69(1):1–9.
Nagayasu A, Uchiyama K, Kiwada H. The size of liposomes: a factor which affects their targeting efficiency to tumors and therapeutic activity of liposomal antitumor drugs. Adv Drug Deliv Rev. 1999;40(1–2):75–87.
Li W, Szoka Jr FC. Lipid-based nanoparticles for nucleic acid delivery. Pharm Res. 2007;24(3):438–49.
Wisse E, Jacobs F, Topal B, Frederik P, De Geest B. The size of endothelial fenestrae in human liver sinusoids: implications for hepatocyte-directed gene transfer. Gene Ther. 2008;15(17):1193–9.
Hirota S, de Ilarduya CT, Barron LG, Szoka Jr FC. Simple mixing device to reproducibly prepare cationic lipid-DNA complexes (lipoplexes). Biotechniques. 1999;27(2):286–90.
Semple SC, Klimuk SK, Harasym TO, Dos Santos N, Ansell SM, Wong KF, et al. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: formation of novel small multilamellar vesicle structures. Biochim Biophys Acta. 2001;1510(1–2):152–66.
Jeffs LB, Palmer LR, Ambegia EG, Giesbrecht C, Ewanick S, MacLachlan I. A scalable, extrusion-free method for efficient liposomal encapsulation of plasmid DNA. Pharm Res. 2005;22(3):362–72.
D’Addio SM, Prud’homme RK. Controlling drug nanoparticle formation by rapid precipitation. Adv Drug Deliv Rev. 2011;63(6):417–26.
Horn D, Rieger J. Organic nanoparticles in the aqueous phase-theory, experiment, and use. Angew Chem Int Ed Engl. 2001;40(23):4330–61.
Mahajan A, Kirwan D. Nucleation and growth-kinetics of biochemicals measured at high supersaturations. J Chryst Growth. 1994;144:281–90.
Walsh CL, Nguyen J, Szoka FC. Synthesis and characterization of novel zwitterionic lipids with pH-responsive biophysical properties. Chem Commun, in press.
Perttu EK, Kohli AG, Szoka FC. Inverse-phosphocholine lipids: a remix of a common phospholipid. J Am Chem Soc. 2012;134(10):4485–8.
Kumar VV. Complementary molecular shapes and additivity of the packing parameter of lipids. Proc Natl Acad Sci U S A. 1991;88(2):444–8.
Shew RL, Deamer DW. A novel method for encapsulation of macromolecules in liposomes. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1985;816(1):1–8.
Szoka Jr F, Papahadjopoulos D. Procedure for preparation of liposomes with large internal aqueous space and high capture by reverse-phase evaporation. Proc Natl Acad Sci U S A. 1978;75(9):4194–8.
Zhang N, Tan C, Cai P, Jiang Y, Zhang P, Zhao Y. Synthesis and properties of morpholino chimeric oligonucleotides. Tetrahedon Lett. 2008;49(22):3570–3.
Schmid P, Hunter E. Extraction and purification of lipids. I. Solubility of lipids in biologically important solvents. Physiol Chem Phys. 1971;3:98–102.
Aubry J, Ganachaud F, Cohen Addad JP, Cabane B. Nanoprecipitation of polymethylmethacrylate by solvent shifting: 1. Boundaries. Langmuir. 2009;25(4):1970–9.
Bar LK, Garti N, Sarig S, Bar R. Solubilities of cholesterol, sitosterol, and cholesteryl acetate in polar organic solvents. J Chem Eng Data. 1984;29(4):440–3.
Chen T, D’Addio SM, Kennedy MT, Swietlow A, Kevrekidis IG, Panagiotopoulos AZ, et al. Protected peptide nanoparticles: experiments and brownian dynamics simulations of the energetics of assembly. Nano Lett. 2009;9(6):2218–22.
Liu Y, Kathan K, Saad W, Prud’homme RK. Ostwald ripening of beta-carotene nanoparticles. Phys Rev Lett. 2007;98(3):036102.
Cao G. Nanostructures & nanomaterials: synthesis, properties & applications. 2004.
Lasic DD, Ceh B, Stuart MC, Guo L, Frederik PM, Barenholz Y. Transmembrane gradient driven phase transitions within vesicles: lessons for drug delivery. Biochim Biophys Acta. 1995;1239(2):145–56.
Guo X, Szoka Jr FC. Steric stabilization of fusogenic liposomes by a low-pH sensitive PEG–diortho ester–lipid conjugate. Bioconjug Chem. 2001;12(2):291–300.
Holland JW, Hui C, Cullis PR, Madden TD. Poly(ethylene glycol)–lipid conjugates regulate the calcium-induced fusion of liposomes composed of phosphatidylethanolamine and phosphatidylserine. Biochemistry. 1996;35(8):2618–24.
Guida V. Thermodynamics and kinetics of vesicles formation processes. Adv Colloid Interface Sci. 2010;161(1–2):77–88.
Lasic DD, Joannic R, Keller BC, Frederik PM, Auvray L. Spontaneous vesiculation. Adv Colloid Interface Sci. 2001;89–90:337–49.
DeMello AJ. Control and detection of chemical reactions in microfluidic systems. Nature. 2006;442(7101):394–402.
Huang X, Caddell R, Yu B, Xu S, Theobald B, Lee LJ, et al. Ultrasound-enhanced microfluidic synthesis of liposomes. Anticancer Res. 2010;30(2):463–6.
Jahn A, Stavis SM, Hong JS, Vreeland WN, DeVoe DL, Gaitan M. Microfluidic mixing and the formation of nanoscale lipid vesicles. ACS Nano. 2010;4(4):2077–87.
Jahn A, Vreeland WN, DeVoe DL, Locascio LE, Gaitan M. Microfluidic directed formation of liposomes of controlled size. Langmuir. 2007;23(11):6289–93.
Jahn A, Vreeland WN, Gaitan M, Locascio LE. Controlled vesicle self-assembly in microfluidic channels with hydrodynamic focusing. J Am Chem Soc. 2004;126(9):2674–5.
Mengeaud V, Josserand J, Girault HH. Mixing processes in a zigzag microchannel: finite element simulations and optical study. Anal Chem. 2002;74(16):4279–86.
Stroock AD, Dertinger SK, Ajdari A, Mezic I, Stone HA, Whitesides GM. Chaotic mixer for microchannels. Science. 2002;295(5555):647–51.
Valencia PM, Basto PA, Zhang L, Rhee M, Langer R, Farokhzad OC, et al. Single-step assembly of homogenous lipid-polymeric and lipid-quantum dot nanoparticles enabled by microfluidic rapid mixing. ACS Nano. 2010;4(3):1671–9.
ACKNOWLEDGMENTS & DISCLOSURES
We are grateful for the financial support from the NIH (2R01EB003008-08) and Pfizer QB3. Juliane Nguyen is supported by the postdoctoral research fellowship from the Deutsche Forschungsgemeinschaft (DFG). Colin Walsh and Emily Perttu are supported by the NSF pre-doctoral fellowship. Michael Motion is supported by a fellowship of the NIH NIGMS. We acknowledge Charles Noble and Mark Hayes for their technical help with the HPLC.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nguyen, J., Walsh, C.L., Motion, J.P.M. et al. Controlled Nucleation of Lipid Nanoparticles. Pharm Res 29, 2236–2248 (2012). https://doi.org/10.1007/s11095-012-0752-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-012-0752-2