ABSTRACT
Purpose
Peroxisome proliferator-activated receptor α (PPARα) is an important transcriptional factor that regulates genes encoding endo/xenobiotic enzymes and lipid metabolizing enzymes. In this study, we investigated whether microRNAs (miRNAs) are involved in the regulation of PPARα in human liver.
Methods
Precursor or antisense oligonucleotide for miR-21 or miR-27b was transfected into HuH7 cells; expression of PPARα and acyl-CoA synthetase M2B was determined by Western blot and real-time RT-PCR. Luciferase assay was performed to identify the functional miRNA recognition element (MRE). Expression levels of PPARα, miR-21, and miR-27b in a panel of 24 human livers were determined.
Results
The overexpression and inhibition of miR-21 or miR-27b in HuH7 cells significantly decreased and increased the PPARα protein level, respectively, but not PPARα mRNA level. The miRNA-dependent regulation of PPARα affected the expression of its downstream gene. Luciferase assay identified a functional MRE for miR-21 in the 3′-untranslated region of PPARα. In human livers, the PPARα protein levels were not correlated with PPARα mRNA, but inversely correlated with the miR-21 levels, suggesting a substantial impact of miR-21, although the contribution of miR-27b could not be ruled out.
Conclusions
We found that PPARα in human liver is regulated by miRNAs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Peroxisome proliferator-activated receptor α (PPARα), which is mainly expressed in the liver, regulates the expression of many genes involved in fatty acid transport, catabolism and energy homeostasis (1). The binding of ligands such as fatty acids, eicosanoids and fibrate hypolipidemics activates PPARα to form a heterodimer with retinoid X receptor (RXR), followed by the binding to PPARα response elements in the promoter region of target genes (2) such as fatty acid transport protein (3), acyl-CoA synthetase (ACS) (4), cytochrome P450 4A11 (5) UDP-glucuronosyltransferase 1A9 and 2B4 (6,7).
MicroRNAs (miRNAs) are endogenous ~22-nucleotide noncoding RNAs. The miRNAs play important roles in the regulation of target genes by recognizing the complementary region in mainly the 3′-untranslated region (UTR) and causing translational repression or mRNA degradation (8). There is accumulating evidence that miRNAs are involved in diverse biological processes including development, cell proliferation, differentiation, apoptosis, and cancer initiation or progression (9–11). The human genome contains ~1000 miRNAs, and 60% of human mRNAs are predicted to be targets of miRNAs (12). Recently, we found that some nuclear receptors such as pregnane X receptor (13), vitamin D receptor (14), and hepatocyte nuclear factor 4α (15) were regulated by miRNA. In addition, PPARγ (16), glucocorticoid receptor (17), and estrogen receptor (18) have been reported to be targets of miRNA. Employing an on-line search, we found that some miRNAs may possibly bind to human PPARα mRNA. In this study, we examined whether miRNAs might be involved in the regulation of PPARα in human liver.
MATERIALS AND METHODS
Chemicals and Reagents
Bezafibarate was purchased from Sigma-Aldrich (St. Louis, MO). The pGL3-promoter vector, phRL-TK vector, Tfx-20, and dual luciferase reporter assay system were purchased from Promega (Madison, WI). LipofectAMINE 2000, LipofectAMINE Reagent, and LipofectAMINE RNAiMAX were from Invitrogen (Carlsbad, CA). Pre-miR miRNA Precursors for miR-21, -22, -24, -27b, -181a, let-7a and negative control #1 were from Ambion (Austin, WI). Antisense locked nucleic acid (LNA)/DNA oligonucleotides (AsO) for miR-21 (5′-ACAGTTCTTCAACTGGCAGCTT -3′; LNA is indicated by the underline), for miR-27b (5′-GCAGAACTTAGCCACTGTGAA-3′), and for negative control (5′-AGACUAGCGGUAUCUUAAACC-3′) were commercially synthesized at NIPPON EGT (Toyama, Japan). RNAiso, random hexamer, and SYBR Premix Ex Taq were from Takara (Shiga, Japan). ROX was purchased from Stratagene (La Jolla, CA). ReverTra Ace was obtained from Toyobo (Osaka, Japan). Restriction enzymes were from Takara, Toyobo and New England Biolabs (Ipswich, MA). All primers and oligonucleotides were commercially synthesized at Hokkaido System Sciences (Sapporo, Japan). Rabbit anti-human PPARα polyclonal antibody (H-98), rabbit anti-human GAPDH polyclonal antibodies (S-20), and IRDye 680 goat anti-rabbit IgG were from Santa Cruz Biotechnology (Santa Cruz, CA), IMAGENEX (San Diego, CA), and LI-COR Biosciences (Lincoln, NE), respectively. All other chemicals and solvents were of the highest grade commercially available.
Human Livers and Preparation of Homogenates and Total RNA
Human liver samples from 16 donors were obtained from the Human and Animal Bridging (HAB) Research Organization (Chiba, Japan), which is in partnership with the National Disease Research Interchange (NDRI, Philadelphia, PA), and those from 8 of the donors were obtained from autopsy materials that were discarded after pathological investigation. The use of the human livers was approved by the Ethics Committees of Kanazawa University (Kanazawa, Japan) and Iwate Medical University (Morioka, Japan). Total cell homogenates were prepared from 24 human liver samples by homogenization with lysis buffer (50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40) containing protease inhibitors (0.5 mM (p-amidinophenyl) methanesulfonyl fluoride, 2 μg/ml aprotinin, 2 μg/ml leupeptin). The protein concentration was determined using Bradford protein assay reagent (Bio-Rad, Hercules, CA) with γ-globulin as a standard. Total RNA was prepared using RNAiso according to the manufacturer’s protocols, and the integrity was assessed by estimating the ratio of the band density of 28S and 18S rRNA.
Cell Culture
The human hepatocellular carcinoma cell lines HuH7 and HepG2 were obtained from Riken Gene Bank (Tsukuba, Japan). The human embryonic kidney cell line HEK293 and the human uterus carcinoma cell line HeLa were obtained from the American Type Culture Collection (Manassas, VA). HuH7 and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Nissui Pharmaceutical, Tokyo, Japan) supplemented with 10% fetal bovine serum (FBS) (Invitrogen). HepG2 cells were cultured in DMEM supplemented with 0.1 mM nonessential amino acid (Invitrogen) and 10% FBS. HEK293 cells were cultured in DMEM supplemented with 4.5 g/l glucose, 10 mM HEPES, and 10% FBS. These cells were maintained at 37°C under an atmosphere of 5% CO2-95% air.
Transfection of Precursor or AsO for miRNAs into HuH7 or HepG2 Cells and Preparation of Cell Homogenates and Total RNA
The cells were seeded into 6-well plates and transfected with 50 nM precursor or AsO for miRNAs or control using LipofectAMINE RNAiMAX. After incubation for 72 h, the cells were harvested, suspended in a small amount of TGE buffer (10 mM Tris–HCl, 20% glycerol, 1 mM EDTA (pH 7.4)), disrupted by freeze-thawing three times and homogenized. Total RNA was also prepared as described above. In some cases, the HuH7 cells transfected with precursor or AsO for miRNA or control were treated with 100 μM bezafibrate 48 h after the transfection. After incubation for 24 h, total RNA was prepared.
SDS-PAGE and Western Blot Analysis of PPARα
Total cell homogenates from the human liver samples (3 μg), HuH7 cells (10 μg), and HepG2 cells (30 μg) were separated in 10% SDS-PAGE and transferred to Immobilon-P transfer membrane (Millipore, Billerica, MA). The membranes were probed with rabbit anti-human PPARα or rabbit anti-human GAPDH antibodies and the corresponding fluorescent dye-conjugated second antibodies. The PPARα protein level was normalized with the GAPDH protein level.
Real-Time RT-PCR for PPARα and Acyl-CoA Synthetase M2B (ACSM2B)
The cDNA was synthesized from total RNA using ReverTra Ace. The real-time RT-PCR was performed using Mx3000PTM (Stratagene). The forward and reverse primers for PPARα mRNA were 5′-ACG GAA AGC CCA CTC TGC CCC CTC TC-3′ and 5′-CTT GTC CCC GCA GAT TCT ACA TTC G-3′, respectively. The forward and reverse primers for ACSM2B mRNA were 5′- AGT GAA AAC AGC CAG GA-3′ and 5′-GAC TTC ATC CCC AGC AAC-3′, respectively. A 1-μl portion of the reverse-transcribed mixture was added to a PCR mixture containing 10 pmol of each primer, 12.5 μl of SYBR Premix Ex Taq solution and 75 nM ROX in a final volume of 25 μl. The PCR conditions of PPARα and ACSM2B were as follows: after an initial denaturation at 95°C for 30 s, the amplification was performed by denaturation at 94°C for 4 s and 20 s, annealing and extension at 64°C for 20 s for 45 cycles. The PPARα and ACSM2B mRNA levels were normalized with GAPDH mRNA as described previously (19).
Real-Time RT-PCR for Mature miR-21 and miR-27b
Polyadenylation and reverse transcription were performed using Ncode miRNA first strand cDNA synthesis kit (Invitrogen) according to the manufacturer’s protocol. The forward primers for miR-21 and miR-27b were 5′-TAG CTT ATC AGA CTG ATG TTG A-3′ and 5′-TTA ACA GTG GCT AAG TTC T -3′, respectively, and the reverse primer was the supplemented universal qPCR primer. The PCR analyses were performed as follows: after an initial denaturation at 95°C for 30 s, the amplification for miR-21 and miR-27b was performed by denaturation at 95°C for 4 s and 10 s, annealing and extension at 64°C and 56°C for 20 s and 10 s for 40 cycles, respectively. The mature miR-21 and miR-27b levels were normalized with U6 snRNA determined by real-time RT-PCR as described previously (19).
Construction of Reporter Plasmids
Various fragments of the human PPARα mRNA were inserted into the Xba I site, downstream of the luciferase gene in the pGL3-promoter vector. As the positive controls, a fragment containing the perfectly matching sequences of the mature miR-21 and miR-27b, 5′-CTA GAT AGC TTA TCA GAC TGA TGT TGA T-3′ and 5′-CTA GAC AGA ACT TAG CCA CTG TGA AT-3′ (the matching sequences of miR-21 and miR-27b are italicized), was also inserted, and the products were termed pGL3/c-miR-21 and pGL3/c-miR-27b, respectively. DNA sequencing analysis was performed to confirm the nucleotide sequences of these constructed plasmids using Long-Read Tower DNA sequencer (GE Healthcare Bio-Sciences, Piscataway, NJ).
Luciferase Assay
Various pGL3 luciferase reporter plasmids were transiently transfected with the phRL-TK vector into HEK293, HepG2, or HeLa cells. Briefly, the day before transfection, the cells were seeded into 24-well plates and cultured for 24 h. To the HEK293 cells, 170 ng of pGL3 plasmid, 30 ng of phRL-TK vector and the precursors for miRNA or control were co-transfected using LipofectAMINE 2000. To the HepG2 and HeLa cells, 170 ng of pGL3 plasimd, 30 ng of phRL-TK vector and the AsO for miR-21 or control were transfected using Tfx-20 and LipofectAMINE Reagent, respectively. After incubation for 48 h, the cells were lysed with a passive lysis buffer, and then the luciferase activity was measured with a luminometer (Wallac, Turku, Finland) using the dual-luciferase reporter assay system.
Statistical Analyses
Statistical significance was determined by unpaired, two-tailed student’s t-test or ANOVA followed by Tukey’s method test. Correlation analyses were performed by Pearson’s product-moment method. A value of P < 0.05 was considered statistically significant.
RESULTS
miR-21 and miR-27b Decreased the PPARα Protein Level Affecting the Induction of its Target Gene in Human Liver-Derived Cell Lines
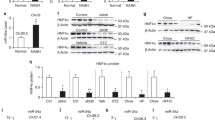
The length of the 3′-UTR of human PPARα is about 8.4 kb. Computational prediction using Target scan (http://www.targetscan.org/) indicated that 28 miRNAs share complementarity with the sequence in the 3′-UTR. Meanwhile, when PicTar (http://pictar.mdc-berlin.de/) was used, 9 miRNAs were found to share complementarity. Among these miRNAs, we selected liver-enriched miRNAs (miR-21, miR-22, miR-24, miR-27b, miR-181a, and let-7a) according to miRanda (http://www.microrna.org/microrna/getMirnaForm.do). We sought to investigate whether these miRNAs can down-regulate PPARα by transfection of the precursors for the miRNAs. First, we determined the PPARα protein and mRNA levels in HuH7, HepG2, HEK293, and HeLa cells by Western blot and real-time RT-PCR analyses, respectively (Fig. 1a). HuH7 cells showed the highest PPARα protein level, which corresponds to the levels in human liver samples. HepG2 cells showed the highest PPARα mRNA level. Among the four cell lines, no positive correlation was observed between the PPARα protein and mRNA levels, suggesting the involvement of post-transcriptional regulation. When we transfected the precursors for the miRNAs into the HuH7 cells, the PPARα protein levels were significantly decreased by the overexpressions of miR-21 (P < 0.001, 47% of control) and miR-27b (P < 0.01, 43% of control) (Fig. 1b), but not by those of miR-22, miR-24, miR-181a and let-7a. When miR-21 and miR-27b were inhibited by the transfection of AsO, a significant increase of PPARα protein was observed (Fig. 1c). In contrast, the PPARα mRNA level was not affected by the overexpression or inhibition of miR-21 and miR-27b (Fig. 1b and c), suggesting that the mechanism of the down-regulation would be translational repression. We confirmed that the mature miRNA levels were significantly changed by the transfection of the precursor or AsO for miRNA (Fig. 1d). When we performed this experiment using HepG2 cells that showed relatively lower PPARα protein level (Fig. 1a), similar changes were observed (Fig. 1b). Thus, we found that PPARα in human liver-derived cells was negatively regulated by miR-21 and miR-27b. The miR-21 levels in the intact HuH7 and HepG2 cells and AsO for miR-21-treated HuH7 were within the range of the miR-21 levels in human livers, which were described in the latter section. The miR-27b levels in the intact HuH7 and HepG2 cells and precursor or AsO for miR-27b-treated HuH7 cell were within the range of the miR-27b levels in human livers. The fact suggests that the regulation by miR-21 and miR-27b for PPARα would be feasible in human liver.
Effects of miR-21 and miR-27b on the PPARα protein, mRNA and ACSM2B mRNA levels. (a) The expression levels of PPARα protein in four cell lines were determined by Western blot and normalized with GAPDH protein levels. A sample of human liver homogenate showing average level of PPARα protein was also applied. The expression levels of PPARα mRNA were determined by real-time RT-PCR and normalized with GAPDH mRNA levels. ND: Not detected. (b, c and d) HuH7 (or HepG2) cells were transfected with 50 nM precursor or AsO for miRNAs or control. After 72 h, total RNA and total cell homogenates were prepared. The expression levels of PPARα protein were determined by Western blot and normalized with GAPDH protein levels. The expression levels of PPARα mRNA, mature miR-21, and mature miR-27b were determined by real-time RT-PCR and normalized with GAPDH mRNA and U6 snRNA levels, respectively. The data are relative to that with the control. Each column represents the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, compared with control. NT: No transfection. (e) Forty eight hours after the transfection of precursor or AsO for miRNAs, HuH7 cells were treated with 100 μM bezafibrate for 24 h. The expression levels of ACSM2B mRNA were determined by real-time RT-PCR and normalized with the GAPDH mRNA level. Each column represents the mean ± SD of three independent experiments. **P < 0.01, ***P < 0.001, compared with bezafibrate (−). ††† P < 0.001, compared with precursor or AsO for control.
Next, we investigated the effect of the miR-21- and miR-27b-dependent regulation of PPARα on the expression of target genes. Treatment of HuH7 cells with 100 μM bezafibrate resulted in a significant induction (1.3-fold) of ACSM2B mRNA, a target of PPARα. When miR-21 and miR-27b were overexpressed, no significant induction was observed, whereas the inhibition of miR-21 and miR-27b resulted in the increased induction (1.9-fold and 1.4-fold, respectively) of ACSM2B mRNA (Fig. 1e). Moreover, overexpression of the miRNAs significantly (P < 0.001) decreased the basal level (52% and 46% of control, respectively), suggesting that PPARα would play an important role in the constitutive expression of ACSM2B. In contrast, the inhibition of the miRNAs did not increase the basal level. This would probably be due to the relatively mild changes of PPARα protein (Fig. 1c) as well as miRNAs (Fig. 1d) by AsO transfection. Collectively, it was suggested that miR-21 and miR-27b appear to affect the expression of genes downstream of PPARα.
Identification of Functional MRE in PPARα mRNA
By the computational search of miRNA recognition elements (MREs) for miR-21 and miR-27b in the 3′-UTR of the PPARα mRNA, two sites for miR-21 (termed MRE21_1 and MRE21_2) and six sites for miR-27b (MRE27b_1, MRE27b_2, MRE27b_3, MRE27b_4 MRE27b_5, and MRE27b_6) were predicted (Fig. 2a). The alignments of these MREs with miR-21 and miR-27b based on RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/welcome.html) are also shown in Fig. 2a. To identify functional MREs, we performed luciferase assays with the overexpression or inhibition of miR-21 and miR-27b. We selected HEK293 and HepG2 cells for the overexpression and inhibition studies, respectively, because these cell lines showed the lowest and highest expression levels of miR-21 and miR-27b (Fig. 2b). When the precursor for miR-21 was transfected into HEK293 cells, the luciferase activity of pGL3/c-miR-21 was significantly decreased (P < 0.001, 12% of control), suggesting that the overexpressed miR-21 was actually functional. The luciferase activity of the plasmid containing MRE21_1 was significantly decreased (P < 0.01, 60% of control) by the overexpression of miR-21, but that of the plasmid containing MRE21_2 was not (Fig. 2c). When the AsO for miR-21 was transfected into HepG2 cells, the increase of the luciferase activity of the plasmid pGL3/c-miR-21 was not observed (data not shown), because the repression of miR-21 level might be insufficient. Then, we used HeLa cells showing relatively lower miR-21 level. When the AsO for miR-21 was transfected into HeLa cells, the luciferase activity of pGL3/c-miR-21 was significantly increased (P < 0.05, 7.0-fold). The luciferase activity of the plasmid containing MRE21_1, but not the plasmid containing MRE21_2, was significantly increased (P < 0.01, 1.7-fold). Taken together, it was suggested that MRE21_1 is functional.
Luciferase assay using various reporter plasmids containing MRE21 and MRE27b in the human PPARα mRNA. (a) Schematic representation of human PPARα mRNA and the predicted target sequences of miR-21 and miR-27b in human PPARα mRNA. The numbering refers to the 5′ end of mRNA as 1, and the coding region is from +183 to +1589. Bold letters, seed sequence. (b) The expression levels of mature miR-21 and mature miR-27b in cell lines were determined by real time RT-PCR and normalized with the U6 snRNA levels. Each column represents the mean of two independent experiments. (c, d) The reporter plasmids (170 ng) were transiently transfected with phRL-TK plasmid (30 ng) and 50 nM precursor or AsO for miRNAs or control into HEK293 or HeLa cells, respectively. The firefly luciferase activity for each construct was normalized with the Renilla luciferase activities. Values are expressed as percentages of the relative luciferase activity of pGL3-p plasmid. Each column represents the mean of two or mean ± SD of three independent experiments, respectively. *P < 0.05, **P < 0.01, ***P < 0.001, compared with the precursor or AsO for control.
When the precursor for miR-27b was transfected into HEK293 cells, the luciferase activity of pGL3/c-miR-27b was significantly decreased (P < 0.001, 9% of control), suggesting that the overexpressed miR-27b was functional (Fig. 2d). However, the luciferase activities of five plasmids containing MRE27b_1 ~ MRE27b_6 were not affected by the overexpression of miR-27b. Next, we searched additional MREs for miR-27b in the coding region and 5′-UTR of PPARα mRNA and found in each one a potential site, termed MRE27b_7 and MRE27b_8, respectively. The luciferase activities of the plasmids containing these MREs were not affected by the overexpression of miR-27b (Fig. 2d). Thus, we could not identify a functional MRE for miR-27b. The inhibition study using AsO for miR-27b was not performed, since the significant decrease of the luciferase activities of the plasmids containing PPARα fragment were not observed by the overexpression study.
Relationship between the Expression Levels of PPARα Protein, PPARα mRNA, miR-21, and miR-27b in Human Livers
To investigate the significance of the miRNA-dependent regulation of PPARα in human livers, we sought correlations between the expression levels of PPARα protein, PPARα mRNA, miR-21, and miR-27b using 24 human livers. The PPARα protein levels showing 7-fold variability were not correlated with the PPARα mRNA levels showing 11-fold variability (Fig. 3a), supporting the involvement of post-transcriptional regulation. Interestingly, the miR-21 levels (5-fold variability) showed a significant inverse correlation with the protein levels (r = −0.56, P < 0.01) (Fig. 3b) and the translational efficiency of PPARα (protein/mRNA ratio) (r = −0.44, P < 0.05) (Fig. 3c). On the other hand, the miR-27b levels showing large interindividual variability (70-fold) were not correlated with the protein levels (Fig. 3d) and translational efficiency of PPARα (Fig. 3e). Thus, it was considered that miR-21, rather than miR-27b, would have a substantial impact on the PPARα expression in human livers.
Relationship between the miR-21, miR-27b, PPARα protein levels and translational efficiency in human livers. Relationship between the PPARα mRNA and protein levels (a), the mature miR-21 and PPARα protein levels (b), the mature miR-21 levels and translational efficiency of PPARα (PPARα protein/mRNA) (c), the mature miR-27b and PPARα protein levels (d), and the mature miR-27b levels and translational efficiency of PPARα (e). Expression levels of mature miR-21, mature miR-27b, and PPARα mRNA in a panel of 24 human livers were determined by real-time RT-PCR and normalized with the U6 snRNA and GAPDH mRNA levels, respectively. The PPARα protein level was determined by Western blot analysis and normalized with the GAPDH protein level. The values represent the levels relative to that of the lowest sample. Data are the mean of two independent experiments.
DISCUSSION
Earlier studies demonstrated that rat PPARα is activated by protein kinase C (PKC) signaling pathway (20), p38 pathway (21), and glucocorticoids (22). Human PPARα is also activated by PKC signaling pathway (23) and is up-regulated by insulin through the mitogen-activated protein kinase pathway (24). Thus, the mechanisms or factors regulating human PPARα expression are very limited. In this study, we examined the possibility of the post-transcriptional regulation of human PPARα by miRNAs.
We found that the PPARα protein level in human liver-derived cell lines was decreased by the overexpression of miR-21 and miR-27b, but not by that of miR-22, miR-24, miR-181a and let-7a (Fig. 1b). A possible concern is that the decrease of the target expression might be an artifact of the overexpression. This possibility can be excluded because we found that the PPARα protein level was increased by the inhibition of endogenous miR-21 and miR-27b (Fig. 1c). Furthermore, we found that the miR-21- and miR-27b-dependent down-regulation of PPARα protein affected the expression level or the induction of its target gene (Fig. 1e). Our findings revealed a significant role for miR-21 and miR-27b in the regulation of PPARα in human liver-derived cells. Previously, Iliopoulos et al. (25) reported that human PPARα protein was down- and up-regulated by the overexpression and inhibition of miR-22 in normal and osteoarthritic chondrocytes, respectively. However, this finding was not reproducible in the present study using human liver-derived cells. Possible explanations for this discrepancy are largely speculative but may include the possibility that other targets take precedence over PPARα for miR-22 in the liver. In addition, Zheng et al. (26) recently reported that PPARα was regulated by miR-10b using human hepatocyte cell line (L02) cultured with high concentrations of free fatty acid (as a non-alcoholic fatty acid liver disease model). Actually, miR-10b was predicted to be a potential regulator of PPARα by in silico analysis, but we did not focus on miR-10b because its expression level was low (as 137-fold low as the miR-21 level) in human liver (27). Zheng et al. (26) revealed that miR-10b is most highly up-regulated (5.4-fold) by treatment with high concentrations of free fatty acid. Therefore, it is speculated that the role of miR-10b in the PPARα regulation might be specific for some aberrant conditions. The significance of miR-21 in regulating the constitutive expression levels of PPARα in human livers was supported by our correlation analysis showing an inverse correlation between the PPARα protein and miR-21 levels (Fig. 3).
It has been reported that the expression of miR-21, which is an intergenic miRNA, is induced by interleukin-6 (IL-6) via the activation of the signal transducer and activator of transcription 3 (Stat3) (28). IL-6, a major mediator of the acute phase response in liver, induces fibrinogen-β, an acute phase protein gene (29), but PPARα inhibits the IL-6-dependent activation of fibrinogen-β promoter by its interaction with glucocorticoid receptor-interacting protein 1 (30). The present study suggests additional pathways by which the inhibitory effects of PPARα on fibrinogen-β expression may be diminished by miR-21. Taken together, miR-21 and PPARα are likely involved in the acute inflammation response. The sequences of miR-21 are common in human, mouse and rat. The seed sequence of the functional MRE for miR-21 we identified (MRE21_1) is conserved between human and mouse (Fig. 2a), but not in rat. Therefore, mouse may be a good model to investigate the significance of the miR-21-dependent regulation of PPARα in vivo.
Cell-based experiments clearly demonstrated that miR-27b down-regulates PPARα. Since we could not identify a functional MRE27b, we cannot conclude whether the regulation might be through a direct mechanism or indirect mechanism via some factor(s) involved in a post-transcriptional process. We also failed to obtain a clue from the analysis of correlations between the miR-27b and PPARα protein levels or translational efficiency using a panel of human liver samples. However, we consider that the absence of an inverse correlation would not be sufficient to rule out the involvement of miR-27b. Another concern is the highly variable expression levels of miR-27b (70-fold) in comparison with the variability in miR-21 (5-fold) and PPARα protein (7-fold) levels. The miR-27b is an intronic miRNA in the C9orf3 gene located on chromosome 9. So far, there is no information about how miR-27b or C9orf3 expression is regulated. Recent studies have described that miR-27b regulates PPARγ (16,31) or RXR (32), a heterodimer partner of PPARα, PPARγ, and other nuclear receptors. Thus, the involvement of miR-27b in adipogenesis and energy metabolism is implied. Therefore, elucidation of the causes of the large interindividual variability in the miR-27b levels would be valuable for understanding the physiological significance of miR-27b.
CONCLUSION
We found that miR-21 and miR-27b negatively regulate the expression of PPARα in human liver, affecting the expression of its downstream gene. Since PPARα is an important regulator of fatty acid catabolism, miR-21 and miR-27b would be one of the factors controlling lipid metabolism. It would be of interest to investigate in the future whether the expression of miR-21 and/or miR-27b might be changed in response to metabolic factors, insulin, glucose, glucocorticoid, or fasting. This study provides new insight into the regulation of PPARα in human liver.
Abbreviations
- ACS:
-
acyl-CoA synthetase
- AsO:
-
antisense LNA/DNA oligonucleotide
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- FBS:
-
fetal bovine serum
- IL-6:
-
interleukin-6
- LNA:
-
locked nucleic acid
- miRNA:
-
microRNA
- MRE:
-
miRNA recognition element
- PPAR:
-
peroxisome proliferator-activated receptor
- RXR:
-
retinoid X receptor
- Stat3:
-
signal transducer and activator of transcription 3
REFERENCES
Shah YM, Morimura K, Yang Q, Tanabe T, Takagi M, Gonzalez FJ. Peroxisome proliferator-activated receptor α regulates a microRNA-mediated signaling cascade responsible for hepatocellular proliferation. Mol Cell Biol. 2007;27(12):4238–47.
Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405(6785):421–4.
van Raalte DH, Li M, Pritchard PH, Wasan KM. Peroxisome proliferator-activated receptor (PPAR)-α: a pharmacological target with a promising future. Pharm Res. 2004;21(9):1531–8.
Martin G, Schoonjans K, Lefebvre AM, Staels B, Auwerx J. Coordinate regulation of the expression of the fatty acid transport protein and acyl-CoA synthetase genes by PPARα and PPARγ activators. J Biol Chem. 1997;272(45):28210–7.
Raucy JL, Lasker J, Ozaki K, Zoleta V. Regulation of CYP2E1 by ethanol and palmitic acid and CYP4A11 by clofibrate in primary cultures of human hepatocytes. Toxicol Sci. 2004;79(2):233–41.
Barbier O, Villeneuve L, Bocher V, Fontaine C, Pineda-Torra I, Duhem C, et al. The UDP-glucuronosyltransferase 1A9 enzyme is a peroxisome proliferator-activated receptor α and γ target gene. J Biol Chem. 2003;278(16):13975–83.
Barbier O, Duran-Sandoval D, Pineda-Torra I, Kosykh V, Fruchart JC, Staels B. Peroxisome proliferator-activated receptor α induces hepatic expression of the human bile acid glucuronidating UDP-glucuronosyltransferase 2B4 enzyme. J Biol Chem. 2003;278(35):32852–60.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97.
Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5.
Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101(9):2999–3004.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8.
Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian miRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105.
Takagi S, Nakajima M, Mohri T, Yokoi T. Post-transcriptional regulation of human pregnane X receptor by microRNA affects the expression of cytochrome P450 3A4. J Biol Chem. 2008;283(15):9674–80.
Mohri T, Nakajima M, Takagi S, Komagata S, Yokoi T. MicroRNA regulates human vitamin D receptor. Int J Cancer. 2009;125(6):1328–33.
Takagi S, Nakajima M, Kida K, Yamaura Y, Fukami T, Yokoi T. MicroRNAs regulate human hepatocyte nuclear factor 4α, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem. 2010;285(7):4415–22.
Jennewein C, Knethen AV, Schmid T, Brune B. MicroRNA-27b contributes to lipopolysaccharide-mediated peroxisome proliferator-activated receptor γ (PPARγ) mRNA destabilization. J Biol Chem. 2010;285(16):11846–53.
Vreugdenhil E, Verissimo CS, Mariman R, Kamporst JT, Barbosa JS, Zweers T, et al. MicroRNA 18 and 124a down-regulate the glucocorticoid receptor: implications for glucocorticoid responsiveness in the brain. Endocrinology. 2009;150(5):2220–8.
Al-Nakhle H, Burns PA, Cummings M, Hanby AM, Hughes TA, Satheesha S, et al. Estrogen receptor β1 expression is regulated by miR-92 in breast cancer. Cancer Res. 2010;70(11):4778–84.
Tsuchiya Y, Nakajima M, Kyo S, Kanaya T, Inoue M, Yokoi T. Human CYP1B1 is regulated by estradiol via estrogen receptor. Cancer Res. 2004;64(9):3119–25.
Yaacob NS, Norazmi MN, Gibson GG, Kass GE. The transcription of the Peroxisome proliferator-activated receptor α is regulated by protein kinase C. Toxicol Lett. 2001;125(1–3):133–41.
Barger PM, Browning AC, Garner AN, Kelly DP. p38 mitogen-activated protein kinase activates peroxisome proliferator-activated receptor α. J Biol Chem. 2001;276(48):44495–501.
Lemberger T, Saladin R, Vazquez M, Assimacopoulos F, Staels B, Desvergne B, et al. Expression of the peroxisome proliferator-activated receptor α gene is stimulated by stress and follows a diurnal rhythm. J Biol Chem. 1996;271(3):1764–9.
Blanquart C, Mansouri R, Paumelle R, Fruchart JC, Staels B, Glineur C. The protein kinase C signaling pathway regulates a molecular switch between transactivation and transrepression activity of the peroxisome proliferator-activated receptor α. Mol Endocrinol. 2004;18(8):1906–18.
Juge-Aubry CE, Hammar E, Siegrist-Kaiser C, Pernin A, Takeshita A, Chin WW, et al. Regulation of the transcriptional activity of the peroxisome proliferator-activated receptor α by phosphorylation of a ligand-independent trans-activating domein. J Biol Chem. 1999;274(15):10505–10.
Iliopoulos D, Malizos KN, Oikonomou P, Tsezou A. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS ONE. 2008;3(11):e3740.
Zheng L, Lv GC, Sheng J, Yang YD. Effect of miRNA-10b in regulating cellular steatosis level by targeting PPAR-α expression, a novel mechanism for the pathogenesis of NAFLD. J Gastroenterol Hepatol. 2010;25(1):156–63.
Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14(12):2486–94.
Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, et al. Interleukin-6-dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110(4):1330–3.
Wegenka UM, Buschmann J, Lutticken C, Heinrich PC, Horn F. Acute-phase response factor, a nuclear factor binding to acute-phase response elements, is rapidly activated by interleukin-6 at the posttranslational level. Mol Cell Biol. 1993;13(1):276–88.
Gervois P, Vu-Dac N, Kleemann R, Kockx M, Dubois G, Laine B, et al. Negative regulation of human fibrinogen gene expression by peroxisome proliferator-activated receptor α agonists via inhibition of CCAAT box/enhancer-binding protein β. J Biol Chem. 2001;276(36):33471–7.
Lin Q, Gao Z, Alarcon RM, Ye J, Yun Z. A role of miR-27 in the regulation of adipogenesis. FEBS J. 2009;276(8):2348–58.
Ji J, Zhang J, Huang G, Qian J, Wang X, Mei S. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 2009;583(4):759–66.
ACKNOWLEDGMENTS
This work was supported in part by Grant-in-Aid for Scientific Research (B) from Japan Society for the Promotion of Science. We acknowledge Mr. Brent Bell for reviewing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kida, K., Nakajima, M., Mohri, T. et al. PPARα Is Regulated by miR-21 and miR-27b in Human Liver. Pharm Res 28, 2467–2476 (2011). https://doi.org/10.1007/s11095-011-0473-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-011-0473-y