Abstract
Introduction
Glioblastoma, the most common and mortal primary brain tumor, accompanied with a dismal clinical outcome in adults. The oncogenic functions of long non-coding RNAs (lncRNAs) in glioblastoma have not been completely illuminated. In the present study, we aimed to investigate the potential role of lncRNA LINC00152 in glioblastoma.
Methods
We used bioinformatic method in public databases to select lncRNA LINC00152 and investigate its clinical value and potential mechanism in glioblastoma. CCK-8, transwell assay, colony formation and wound healing assays were used to explore the role of LINC00152 in glioblastoma malignant behaviors. PCR, western blot, immunofluorescence, reporter assays and nude mouse tumor intracranial model were employed to further verify the regulatory mechanism of LINC00152 in glioblastoma.
Results
LINC00152 was closely associated with glioma WHO classification and poor prognosis, and indicated a poor prognosis in glioblastoma patients. Tumor growth and invasion were suppressed both in vitro and vivo after LINC00152 was blocked. Moreover, LINC00152 modulated GBM malignant progression and proneural–mesenchymal transition through the miR-612 dependent AKT2/NF-κB pathway.
Conclusions
LINC00152 acted as a tumor oncogene with prognostic value for patients with glioblastoma through LINC00152/miR-612/AKT2/NF-κB axis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma (GBM), the most common and mortal primary brain tumor, accompanied with an unfavorable clinical outcome in adults [1]. Despite treatment involving surgery, following chemotherapy and radiation, patients with GBMs only have an average survival of no more than 15 months at the time of the initial diagnosis [2]. The proliferative, invasive and drug resistant hallmarks of GBM account for its poor prognosis [3]. Therefore, an urgent comprehending of the mechanisms of molecular biology involved in GBM malignant behaviors is needed.
AKT/NF-kB signaling plays a critical role in glioblastoma progression. NF-κB could be activated via an AKT-dependent pathway [4]. MK-2206 is a potent allosteric inhibitor of AKT, with anti-proliferative and anti-invasion activity in GBM [5]. The constitutive activation of NF-κB has been detected widespread in human cancers. Migration of GBM cells into the surrounding brain parenchyma is one of the primary reasons for tumor recurrence, and NF-kB signaling significantly contributes to this property of GBM cells [6]. Knockdown of p65 induces cytotoxicity in GBM cells, and expression of a p65 shRNA leads to a decrease in GBM xenograft growth [7].
Coding genes and their coding proteins have been thought to be the primary functional effectors of cells [8]. Until recent years, when the roles of noncoding RNAs (ncRNAs), began to be appreciated for their roles in most biological processes [9]. Long non-coding RNAs (lncRNAs) are a class of ncRNAs, which are greater than 200 bp in length and do not code any proteins [10]. LncRNA-mediated biology has been implicated in a wide variety of cellular processes, including histone modification, post-transcriptional processing inactivation and competing endogenous-RNA (ceRNA) [11]. Up to now, lncRNAs like HOTAIR [12], H19 [13], and HOXA11-AS [14], are dysregulated in diseases, especially in GBM. Uncovering GBM-associated lncRNAs would disclose a new level of the mechanism of brain tumor progression.
Although various classification programs have been done, the mesenchymal (MES) and proneural (PN) subtypes are unshakable [15]. Patients in the MES subtype exhibit relatively worse outcome than PN subtype [16]. More importantly, PN subtype has been proved to tend to MES recurrences, indicative of a proneural–mesenchymal transition during glioma progression [17, 18]. Previous studies have verified the occurrence of PN–MES shift in GBM and several major transcription factors, including STAT3 and NF-κB [19].
In this study, we aimed to explore how LINC00152 regulated the proneural–mesenchymal transition. We revealed that LINC00152 facilitated transition through miR-612/AKT2/NF-κB pathway.
Materials and methods
Clinical samples
The mRNA profiles and relevant clinical parameters were acquired from the Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org) and the Cancer Genome Atlas (TCGA, https://genome-cancer.ucsc.edu/). Human glioma samples were obtained from the Department of Neurosurgery, the Second Affiliated Hospital of Harbin Medical University. The grade of human glioma samples used in the present study was confirmed by the pathologist according to WHO standard [15].
Cell culture
LN229 and U87-MG cells were grown in Dulbecco’s Modified Eagle’s medium (DMEM, Gibco) separately containing 5 and 10% of fetal bovine serum (FBS, BI). N9 which was a patient-derived cell was grown in DMEM/F12 (1:1) (Gibco) and supplemented with 10% FBS and 1% penicillin–streptomycin described in our previous research [20]. All cells were maintained at 37 °C under a humidified atmosphere containing 5% CO2.
Transfection
Lentivirus containing LINC00152 RNAi segments, LINC00152 full length and luciferase lentivirus were obtained from Genechem (Shanghai, China). Chemosynthetic siRNA1, 2 and 3 (the siRNA sequences are 5′-GCGGAGCATGGAACTCGACAGTTAA-3′, 5′-CGGAGCATGGAACTCGACAGTTAAA-3′, 5′-GGGCAATAGGCGATACGATGCTTTA-3′) were obtained from Genechem (Shanghai, China). The miR-612 mimic, inhibitor and negative control (Genechem, Shanghai, China) were used in this study.
Cell proliferation, colony formation, invasion and wound healing assays
Cell proliferation was performed using the Cell-Counting Kit 8 (CCK-8; Dojindo Laboratories) according to the manufacturer’s instructions. For colony formation, cells were planted in six-well plates at 200/well for 14 days. Cell colony was fixed with 4% paraformaldehyde and observed by staining with 0.1% crystal violet for 30 s. Invasion assays were performed using Boyden chambers (Corning, NY) and 8-µm-sized pore membranes coated with matrigel. Chambers and plates were incubated at 37 °C with 5% CO2 for 36 h. Transfected GBM cells were seeded in six-well plates. After the confluence reached ~ 90%, the cell layers were scratched with a tip of a 1 mL pipette. After culture for 24 h, the wound width was measured to evaluate the wound healing ability of the tested cells. Each experiment was performed at least three times.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA from frozen normal brain and glioma specimens and cells was extracted using the Trizol reagent (Sigma). qRT-PCR was performed with CFX Connect™ (Bio-Rad) and used to analyze the expression of mRNA in human glioma samples and cell lines. The GAPDH and U6 were used as internal control for mRNAs and miRNAs. LINC00152 primers are 5′-ATGTGTAGGAGAGTCGGCCT-3′, 5′-ATGCCGTTTTAGGGGGACAG-3′. PDGFRA primers are 5′-CCTCTCGGGTCTCAGTTGAA-3′, 5′-CCACTGTCCGGCTTCTGAC-3′, Olig2 primers are 5′-TCGCATCCAGATTTTCGGGT-3′, 5′-AAAAGGTCATCGGGCTCTGG-3′. Vimentin primers are 5′-GGACCAGCTAACCAACGACA-3′, 5′-AAGGTCAAGACGTGCCAGAG-3′. CD44 primers are 5′-CACACCCTCCCCTCATTCAC-3′, 5′-CACACCCTCCCCTCATTCAC-3′.
Protein extraction and western blot
RIPA buffer Solarbio (Beijing, China) was used to extract total protein according to the manufacturer’s protocol. Nuclear and cytoplasmic protein extraction kit brought from Beyotime Biotechnology (Shanghai, China) was used to extract the different position of the protein in cells. Primary antibodies were P65 (rabbit, 1:1000, CST), phoshorylated-P65 (rabbit, 1:1000, CST), Olig2 (rabbit, 1:1000, Abcam), PDGFRA (rabbit, 1:1000, Abcam), Vimentin (rabbit, 1:1000, CST), CD44 (mouse, 1:1000, CST), AKT2 (rabbit, 1:1000, CST), IKKα (rabbit, 1:1000, CST), p-IKKα (rabbit, 1:1000, CST), p-AKT2 (rabbit, 1:1000, CST), H3 (mouse, 1:1000, CST) and GAPDH (mouse, 1:1000, Proteintech).
Bioinformatics prediction and luciferase reporter assay
The putative miR-612 target binding sequences in LINC00152, AKT2 and its mutant of the binding sites were synthesized and cloned downstream of the luciferase gene in the pmirGLO luciferase vector (Promega). The target gene of miR-612 was predicted with the support of computeraided algorithms: TargetScan. PGL4.32 (luc2P/NFκB-RE/Hypro Vector), which was purchased from Promega containing NF-κB response element. The plasmids were transfected into cells, and vector Renilla luciferase (Promega) served as an internal control. After 48 h, cells were lysed and tested by the Dual-Luciferase Reporter Assay System (Promega).
Immunofluorescence and microscopy
Immunofluorescence staining was performed using primary antibodies against P65 (1:100 dilutions, Cell Signaling Technology), followed by 1 h of incubation with 594 goat-anti-rabbit IgG (H + L) (Life Technology) at room temperature. All fluorescent results were visualized using the FV-1200 laser scanning confocal microscope.
Intracranial model build, hematoxylin-eosin staining (HE) and immunohistochemistry (IHC)
U87-MG cells were co-transfected with lenti-LINC00152 RNAi and luciferase lentivirus in vitro for 48 h. A total of 3.0 × 105 U87-MG cells infected with the virus were implanted stereotactically to establish intracranial GBM using cranial crews as the previous description [21]. After 10 and 20 days, tumors were monitored by fluorescent images of whole mice using the Living Imaging system. Paraffin sections (4 µm) were used for HE and IHC staining. Paraffin sections were dewaxed and incubated with the primary antibodies Ki67 (rabbit, 1:50 Santa Cruz), P65 (rabbit, 1:100, CST) and MMP9 (rabbit, 1:100, CST) at 4 °C for 12 h, followed by incubation with biotinylated secondary antibody (Zsbio) for 40 min at room temperature. After washing with PBS, the sections were stained with 3,3′-diaminobenzidine (DAB) for approximately 10–20 s, rewashed in PBS and counterstained with hematoxylin.
Statistical analysis
GraphPad Prism, version 7.0 and SPSS, version 22.0, Gene Cluster 3.0 and Gene Tree View software were used in this study. Univariate and multivariate Cox regression analyses were performed to assess the relative risk for potential factor. Kaplan–Meier curves were used to estimate the survival distributions. Then, the log-rank test was applied to value the statistical significance between different survival groups. Differential lncRNAs between low and high grades were filtrated using significance analysis of microarray (SAM) algorithm. DAVID (http://david.abcc.ncifcrf.gov/) was used for Gene Ontology analysis [22]. Gene set enrichment analysis (GSEA) was conducted using the GSEA software [23]. P < 0.05 was considered statistically significant.
Results
Aberrant expression and prognostic value of LINC00152 for patients with GBMs
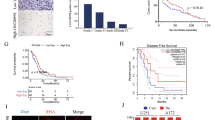
We combined differentially expressed lncRNAs in GBMs and low grade gliomas (LGGs) according to Reon et al. [24] and lncRNAs most up- or down-regulated in GBM according to Pastori et al. [25] to obtain 1296 annotated lncRNAs. SAM analysis was performed to compare the differential expressed lncRNAs between the low (WHO II/III) and high (WHO IV) grade glioma samples in CGGA and TCGA databases. Only those lncRNAs (SAM: Fold change > 1.5, Q value < 0.01) were considered as significant lncRNAs. Among those lncRNAs, a lncRNA LINC00152 towered above for its unknown role in glioma (Fig. 1a). We analyzed LINC00152 expression levels from whole mRNA profiling of the CGGA and TCGA cohorts. LINC00152 expression was significantly higher in WHO IV grade samples than WHO III and WHO II samples in CGGA database (P = 0.0428 and P < 0.0001, Fig. 1b). The same result revealed that LINC00152 expression was significantly higher in WHO IV glioma than that in other grades in TCGA database (P < 0.0001 and P < 0.0001, Fig. 1b). To further affirm the expression tendency of LINC00152 in glioma, we detected the level of LINC00152 in 35 samples of adult brain tissues by using qRT-PCR. As expected, LINC00152 had a higher level in WHO IV glioma samples (Fig. 1c). We employed the dichotomization to separate cases for depicting the survival curves according to the median value. Patients with higher LINC00152 expression had significantly shorter overall survival than those with lower LINC00152 expression, which highlighted the potential oncogenic character of LINC00152 for GBM patients (CGGA, P = 0.0022; TCGA, P = 0.0077, Fig. 1d). Next, we analyzed the clinicopathological features of patients through CGGA and TCGA databases, and found that the relative expression of LINC00152 was associated with TCGA subtype (P = 0.0003) and IDH status (P = 0.0019) in CGGA database and closely related to TCGA subtype (P = 0.0241) and chemotherapy (P = 0.0029) in TCGA database (Tables S1 and S2). Then, we conducted a univariate Cox regression analysis enrolling clinical and genetic variables of GBM patients from the CGGA and TCGA databases. We found that the expression of LINC00152, age, chemotherapy and radiotherapy were statistically associated with overall survival in CGGA database and the expression of LINC00152, age, IDH status and chemotherapy were statistically associated with overall survival in TCGA database. We evaluated the factors that contributed to overall survival using a multivariate Cox proportional hazards model. The result revealed that LINC00152 expression was independently correlated with overall survival (HR = 1.268, P = 0.005; HR = 1.536, P = 0.0005, respectively) after adjusting other factors in CGGA and TCGA databases (Table S3).
LncRNA profiling analysis in the CGGA and TCGA databases identifies a lncRNA LINC00152 in glioma. a Heatmaps were constructed using the differential lncRNAs between different grades of glioma identified by significance analysis of microarray algorithm in CGGA and TCGA databases. b The levels of LINC00152 were analyzed in CGGA and TCGA databases. c The level of LINC00152 was analyzed in 35 brain samples. d Kaplan–Meier curve was performed to evaluate overall survival time between LINC00152 high- and low-expression groups in CGGA and TCGA databases
Knockdown of LINC00152 suppresses GBM cell malignant behaviors
LINC00152 decreased in siRNA groups, particularly in the siRNA3 group (Fig. S1A). CCK-8 assay revealed that LINC00152 knockdown could inhibit the proliferation of LN229 and U87-MG cells (Fig. S1B). Transwell assay also showed that the invasive capacity was suppressed when LINC00152 decreased in LN229 and U87-MG cells (Fig. S1C). Colony formation assay illustrated that knockdown of LINC00152 could inhibit the colony formation of LN229 and U87-MG cells in vitro (Fig. S1D). Wound healing results demonstrated that inhibition of LINC00152 attenuated the LN229 and U87-MG cells migration (Fig. S1E).
Interference of LINC00152 inhibits NF-κB expression and nuclear transport
To clarify the cellular functions of LINC00152 in GBM, we clustered positively or negatively genes in the TCGA database according to LINC00152 expression level (Fig. 2a). GO analysis showed that LINC00152 positively-related genes were involved in the NF-κB signaling pathway (P < 0.01, Fig. 2b). In order to explore the effect of LINC00152 on the canonical NF-κB pathway, we detected the P65 and phosphorylated P65 (p-P65) protein levels and found that these two proteins decreased in LINC00152 down-regulated groups of LN229, U87-MG and N9 cells (Figs. 2c, S2A). In the meantime, the NF-κB transcriptional activity was suppressed when the LINC00152 decreased (Fig. 2d). P65 and p-P65 distribution in cytoplasm and nucleus decreased after suppressing LINC00152 in LN229, U87-MG and N9 cells (Figs. 2e, S2B). Furthermore, confocal microscopy analysis suggested that P65 decreased in both cytoplasmic and nuclear components of U87-MG and N9 cells (Figs. 2f, S2C).
Knockdown of LINC00152 inhibits NF-κB expression and nuclear transport. a. A heatmap of the LINC00152 positive-associated genes in GBM tissues sorted by the level of LINC00152 expression in TCGA database. b Gene ontology analysis of LINC00152 positively associated genes revealed several pathways. c The protein levels of P65 and phosphotylated-P65 were detected in siRNA and scramble group. d NF-κB luciferase reporter and Renilla plasmids were transfected to measure the influence of LINC00152 on NF-κB transcriptional activity. e The distributions of P65 and p-P65 in cytoplasm and nucleus were tested after suppressing LINC00152 in GBM cells. f P65 expression and the subcellular location were confirmed by confocal microscopy. Scale bars = 20 µm. *P < 0.05, **P < 0.01, ***P < 0.001
The functional role of LINC00152 involves in the proneural–mesenchymal transition
Previous research found that NF-κB pathway plays an important role in proneural–mesenchymal transition (PN–MES shift) [19], so it was necessary for us to seek the functional influence of LINC00152 during this transition. We found that the mesenchymal GBM had a higher LINC00152 expression, whereas the proneural GBM contained a lower LINC00152 expression (Fig. 3a) in CGGA database. In TCGA cohort, we found that LINC00152 expression was higher in the mesenchymal subtype than that in the proneural subtype (Fig. 3b). Next, we divided GBM patients from CGGA and TCGA cohorts into two groups on the basis of the expression of LINC00152. Similarly, GSEA between different groups reveals that patients with a high-level of LINC00152 were enriched in mesenchymal-associated gene set (Fig. 3c). It has been reported that in GBM miR-181d/MALT1 modulator axis attenuated mesenchymal phenotype characteristics [26] and miR-34a inhibition in proneural subtype increases the expression of its target PDGFRA and accelerates tumor growth [27], which indicates that ncRNAs are associated with GBM subtype profiles. Downregulation of LINC00152 induced both mRNA and protein expression of proneural markers (Olig2 and PDGFRA) while restrained the mesenchymal markers (CD44 and Vimentin) in LN229, U87-MG and N9 cells (Figs. 3d, e, S2 D, E).
Knockdown of LINC00152 attenuates proneural–mesenchymal transition. a, b The expression of LINC00152 was analyzed in subtypes of GBM in the CGGA and TCGA databases. c GSEA was conducted to analyze patients with different LINC00152 expression among GBM subtypes. d, e The mRNA and protein levels of PDGFRA, Olig2, Vimentin and CD44 were detected in siRNA and scramble groups. *P < 0.05, **P < 0.01, ***P < 0.001
Knockdown of LINC00152 could inhibit GBM progression in vivo
In consideration of the oncogenic effects of LINC00152 in vitro, we decided to extend our research to examine if down-regulated LINC00152 could suppress GBM progression in vivo. In accordance within vitro experiments, the intracranial tumor decreased (Fig. 4a, b) after inhibiting LINC00152. HE staining of xenograft tumors showed that tumor invasion decreased in the down-regulated LINC00152 group compared with control group (Fig. 4c). Down-regulated LINC00152 in U87-MG tumors was associated with prolonged survival (Fig. 4d). Immunohistochemistry assay showed that downregulation of LINC00152 could reduce the Ki-67, P65 and MMP9 protein expression (Fig. 4e).
Knockdown of LINC00152 inhibits GBM progression in vivo. a Luminescence imaging for lenti-NC versus lenti-LINC00152 RNAi group. b Tumor volume in the lenti-LINC00152 RNAi group was smaller than that in the lenti-NC group. c The tissue sections from representative tumors in each group were stained with HE; the black arrows indicated tumor invasion. Scale bars = 50 µm. d Kaplan–Meier curve was performed for evaluating overall survival time between lenti-NC and lenti-LINC00152 RNAi. e The levels of Ki67, P65 and MMP9 were analyzed through IHC. Scale bars = 100 µm. *P < 0.05, **P < 0.01, ***P < 0.001
LINC00152 functions as ceRNA and sponges miR-612 in GBM cells
In order to investigate how LINC00152 regulated NF-κB pathway and resulted in the proneural–mesenchymal transition. The RIP assay showed that LINC00152 and AKT2 could bind Ago2 in LN229 and U87-MG cells (Fig. 5a, b). We predicted that LINC00152 and AKT2 as potential targets of miR-612 (Fig. 5c). AKT2 was a potential target of miR-612 in previous research [28]. Then, we detected the level of miR-612 in LN229 and U87-MG cells treated by the scramble and siRNA3. MiR-612 increased in LN229 and U87-MG cells after LINC00152 was inhibited (Fig. 5d). Furthermore, the dual luciferase reporter assays revealed that miR-612 directly bound to the LINC00152 and AKT2 3′UTR regions (Fig. 5e, f). After overexpression of LINC00152, luciferase activity decreased in miR-612 WT groups, revealing that LINC00152 bound miR-612 (Fig. 5g).
LINC00152 functions as ceRNA and sponges miR-612 in GBM cells. a RIP–PCR assay of the enrichment of Ago2 on LINC00152 and AKT2 transcripts normalized to IgG in U87-MG and LN229 cells transfected with scramble or LINC00152. b RIP–PCR assay of the enrichment of Ago2 on LINC00152 and AKT2 transcripts normalized to IgG in U87-MG and LN229 cells transfected with scramble or siRNA3. c Predicted results showed the sequence of LINC00152 with highly conserved putative miR-612 binding sites and miR-612 with highly conserved putative AKT2 3′-UTR. d qRT-PCR analysis of miR-612 in GBM cells transfected with scramble or siRNA3. e, f Luciferase activity of LINC00152 and AKT2 3′-UTR in GBM cells upon transfection of miR-612 mimics. g Luciferase activity of miR-612 in GBM cells after overexpression of LINC00152. h Western blot analysis of indicated proteins in GBM cells after overexpression of LINC00152 upon transfection with miR-612 mimics. i Western blot analysis of indicated proteins in GBM cells after inhibiting of LINC00152 upon transfection with miR-612 mimics and inhibitor. j Western blot analysis of indicated proteins in GBM cells after overexpression of LINC00152 upon transfection with siAKT2. k Mechanism diagram of LINC00152 modulating GBM progression and proneural–mesenchymal transition through miR-612 dependent on AKT2/NF-κB pathway. *P < 0.05, **P < 0.01, ***P < 0.001
Moreover, the overexpression of miR-612 inhibited AKT2 and its downstream expression in LN229 and U87-MG cells. In contrast, miR-612 inhibitor could restore AKT2 and its downstream activity in LN229 and U87-MG knocking down LINC00152 (Fig. 5h, i). In the meantime, the knockdown of AKT2 could inhibit its downstream activity in LN229 and U87-MG overexpressing LINC00152 (Fig. 5j).
Discussion
Thousands of lncRNAs have been discovered, benefiting from large sequencing consortia such as ENCODE and FANTOM [29]. However, only a few of these lncRNAs have been successfully established. To manifest aberrant expression patterns of lncRNAs in glioma, we performed an analysis of lncRNAs in CGGA and TCGA mRNA databases. Subsequently, we identified a lncRNA, LINC00152 in glioma. The abnormal expression level of LINC00152 observed from the databases was further verified in glioma specimens, suggesting the oncogenic role of LINC00152. The high expression pattern of LINC00152 also predicted a poor outcome of patients. In other tumors, like non-small-cell lung cancer [30], tongue squamous cell carcinoma [31] and gastric cancer [32], LINC00152 could serve as a clinical prognostic predictor. Cluster analysis revealed gene profile differences between LINC00152 high versus low-expressing patients, which indicated that LINC00152 regulated genes involved in the NF-κB signaling pathway as determined via GO analysis. Our experimental results showed that LINC00152 affected a series of cellular biological functions via NF-κB transcriptional activity. Integrated genomic analysis has explicitly defined the classification of adult GBM into four subtypes [33]. Commonly, GBMs with mesenchymal subtype exhibited elevated levels of NF-kB pathway genes. Activation of NF-kB signaling has also been discovered to promote mesenchymal differentiation of GBM by regulating downstream transcriptional signaling [34]. Considering our experimental results, we suspected that LINC00152 had an association with GBM subtype and mesenchymal differentiation. Next, LINC00152 expression was analyzed in GBM subtype and revealed that it was low-expressed in PN compared with that in MES. MES differentiation induced resistance to chemotherapy is of great importance for GBM progression and recurrence [35]. A long non-coding RNA HIF1A-AS2 maintained mesenchymal features of GBM stem-like cells in the hypoxic microenvironment [36]. However, there is no available research until now that refers to the role of LINC00152 in the process of MES to PN transition. Our result showed that down-regulated lncRNA LINC00152 attenuates mesenchymal profile and induced proneural markers.
One of the main functions of lncRNAs is acting as endogenous miRNA sponges to bind to miRNAs and regulate their function of post-transcriptional control. Knockdown of X-inactive specific transcript (XIST) exhibits the tumor-suppressive effect in GBM stem cells via regulating miR-152 [37]. MALAT1 induces chemoresistance to temozolomide through restraining miR-203 and promoting synthesis of thymidylate synthase [38]. Taurine upregulated 1 enhances angiogenesis through blocking miR-299 in human GBM [39]. RP11-838N2.4 through blocking the functions of miR-10a induces the cytotoxic effects of temozolomide in GBM [40]. LINC00152 accelerates renal cell carcinoma progression through negatively regulating miR-205 [41] and facilitates gallbladder cancer metastasis by regulating HIF-1α via miR-138 [42]. LINC00152 also down-regulates miR-193a-3p to enhance MCL1 expression and promote gastric cancer progression [43]. In triple-negative breast cancer, LINC00152 accelerates tumor progression by regulating DNMTs [44].
The previous study reported that miR-612 negatively regulates colorectal cancer growth and metastasis by targeting AKT2 [28]. Besides, AKT/NF-κB axis widely exists in biological process and tumor progression. AKT/NF-κB signaling regulates expression of inflammatory genes [45, 46]. The metastatic ability of human breast cancer cells could be inhibited through the UPR inducer DPP23 by targeting the Akt/NF-κB/MMP-9 axis [47]. It has been proved that blocking AKT2 enhances sensitivity to chemotherapy through NF-κB (P65) in pancreatic carcinoma [48]. MK-2206 an AKT inhibitor suppresses the NF-κB activity in GBM [49]. In our work, LINC00152 acted as a miRNA sponge for miR-612 in GBM cells, negatively regulated miR-612 releases, which resulted in the elevated AKT2, activated NF-κB pathway to promote proneural–mesenchymal transition.
In summary, our research thoroughly showed that LINC00152 acted as a tumor oncogene with important prognostic value for patients with GBMs, and that LINC00152 regulated NF-κB/AKT2 pathway by capturing miR-612 to promote the proneural–mesenchymal transition (Fig. 5e). Our results demonstrate the regulatory mechanisms of LINC00152 and provide a novel strategy of lncRNA-based therapy in GBM.
References
Jiang T, Mao Y, Ma W, Mao Q, You Y, Yang X et al (2016) CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett 375(2):263–273. https://doi.org/10.1016/j.canlet.2016.01.024
Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ et al (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352(10):987–996. https://doi.org/10.1056/NEJMoa043330
Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C et al (2015) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro-oncology 17(Suppl 4):iv1–iv62. https://doi.org/10.1093/neuonc/nov189
Lien GS, Wu MS, Bien MY, Chen CH, Lin CH, Chen BC (2014) Epidermal growth factor stimulates nuclear factor-kappaB activation and heme oxygenase-1 expression via c-Src, NADPH oxidase, PI3K, and Akt in human colon cancer cells. PLoS ONE 9(8):e104891. https://doi.org/10.1371/journal.pone.0104891
Yang C, Li YS, Wang QX, Huang K, Wei JW, Wang YF et al (2017) EGFR/EGFRvIII remodels the cytoskeleton via epigenetic silencing of AJAP1 in glioma cells. Cancer Lett 403:119–127. https://doi.org/10.1016/j.canlet.2017.06.007
Holmes KM, Annala M, Chua CY, Dunlap SM, Liu Y, Hugen N et al (2012) Insulin-like growth factor-binding protein 2-driven glioma progression is prevented by blocking a clinically significant integrin, integrin-linked kinase, and NF-kappaB network. Proc Natl Acad Sci USA 109(9):3475–3480. https://doi.org/10.1073/pnas.1120375109
Zanotto-Filho A, Braganhol E, Schroder R, de Souza LH, Dalmolin RJ, Pasquali MA et al (2011) NFkappaB inhibitors induce cell death in glioblastomas. Biochem Pharmacol 81(3):412–424. https://doi.org/10.1016/j.bcp.2010.10.014
Consortium EP, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR et al (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447(7146):799–816. https://doi.org/10.1038/nature05874
Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N et al (2005) The transcriptional landscape of the mammalian genome. Science 309(5740):1559–1563. https://doi.org/10.1126/science.1112014
Spitale RC, Tsai MC, Chang HY (2011) RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics 6(5):539–543
Sahu A, Singhal U, Chinnaiyan AM (2015) Long noncoding RNAs in cancer: from function to translation. Trends Cancer 1(2):93–109. https://doi.org/10.1016/j.trecan.2015.08.010
Zhou X, Ren Y, Zhang J, Zhang C, Zhang K, Han L et al (2015) HOTAIR is a therapeutic target in glioblastoma. Oncotarget 6(10):8353–8365. https://doi.org/10.18632/oncotarget.3229
Jiang X, Yan Y, Hu M, Chen X, Wang Y, Dai Y et al (2016) Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J Neurosurg 124(1):129–136. https://doi.org/10.3171/2014.12.JNS1426
Wang Q, Zhang J, Liu Y, Zhang W, Zhou J, Duan R et al (2016) A novel cell cycle-associated lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA transcript and is a biomarker of progression in glioma. Cancer Lett 373(2):251–259. https://doi.org/10.1016/j.canlet.2016.01.039
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820. https://doi.org/10.1007/s00401-016-1545-1
Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD et al (2006) Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9(3):157–173. https://doi.org/10.1016/j.ccr.2006.02.019
Bhat KP, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F et al (2013) Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell 24(3):331–346. https://doi.org/10.1016/j.ccr.2013.08.001
Segerman A, Niklasson M, Haglund C, Bergstrom T, Jarvius M, Xie Y et al (2016) Clonal variation in drug and radiation response among glioma-initiating cells is linked to proneural-mesenchymal transition. Cell Rep 17(11):2994–3009. https://doi.org/10.1016/j.celrep.2016.11.056
Lau J, Ilkhanizadeh S, Wang S, Miroshnikova YA, Salvatierra NA, Wong RA et al (2015) STAT3 blockade inhibits radiation-induced malignant progression in glioma. Cancer Res 75(20):4302–4311. https://doi.org/10.1158/0008-5472.CAN-14-3331
Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang J et al (2018) Long noncoding RNA NEAT1, regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/beta-catenin pathway by scaffolding EZH2. Clin Cancer Res 24(3):684–695. https://doi.org/10.1158/1078-0432.CCR-17-0605
Liu X, Wang X, Du W, Chen L, Wang G, Cui Y et al (2014) Suppressor of fused (Sufu) represses Gli1 transcription and nuclear accumulation, inhibits glioma cell proliferation, invasion and vasculogenic mimicry, improving glioma chemo-sensitivity and prognosis. Oncotarget 5(22):11681–11694. https://doi.org/10.18632/oncotarget.2585
Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc 4(1):44–57. https://doi.org/10.1038/nprot.2008.211
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102(43):15545–15550. https://doi.org/10.1073/pnas.0506580102
Reon BJ, Anaya J, Zhang Y, Mandell J, Purow B, Abounader R et al (2016) Expression of lncRNAs in low-grade gliomas and glioblastoma multiforme: an in silico analysis. PLoS Med 13(12):e1002192. https://doi.org/10.1371/journal.pmed.1002192
Pastori C, Kapranov P, Penas C, Peschansky V, Volmar CH, Sarkaria JN et al (2015) The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci USA 112(27):8326–8331. https://doi.org/10.1073/pnas.1424220112
Yang F, Liu X, Liu Y, Liu Y, Zhang C, Wang Z et al (2017) miR-181d/MALT1 regulatory axis attenuates mesenchymal phenotype through NF-kappaB pathways in glioblastoma. Cancer Lett. https://doi.org/10.1016/j.canlet.2017.03.002
Silber J, Jacobsen A, Ozawa T, Harinath G, Pedraza A, Sander C et al (2012) miR-34a repression in proneural malignant gliomas upregulates expression of its target PDGFRA and promotes tumorigenesis. PLoS ONE 7(3):e33844. https://doi.org/10.1371/journal.pone.0033844
Sheng L, He P, Yang X, Zhou M, Feng Q (2015) miR-612 negatively regulates colorectal cancer growth and metastasis by targeting AKT2. Cell Death Dis 6:e1808. https://doi.org/10.1038/cddis.2015.184
Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M et al (2005) Antisense transcription in the mammalian transcriptome. Science 309(5740):1564–1566. https://doi.org/10.1126/science.1112009
Li N, Feng XB, Tan Q, Luo P, Jing W, Zhu M et al (2017) Identification of circulating long noncoding RNA Linc00152 as a novel biomarker for diagnosis and monitoring of non-small-cell lung cancer. Dis Mark. https://doi.org/10.1155/2017/7439698
Yu J, Liu Y, Guo C, Zhang S, Gong Z, Tang Y et al (2017) Upregulated long non-coding RNA LINC00152 expression is associated with progression and poor prognosis of tongue squamous cell carcinoma. J Cancer 8(4):523–530. https://doi.org/10.7150/jca.17510
Pang Q, Ge J, Shao Y, Sun W, Song H, Xia T et al (2014) Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biol 35(6):5441–5447. https://doi.org/10.1007/s13277-014-1709-3
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD et al (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17(1):98–110. https://doi.org/10.1016/j.ccr.2009.12.020
Kim SH, Ezhilarasan R, Phillips E, Gallego-Perez D, Sparks A, Taylor D et al (2016) Serine/threonine kinase MLK4 determines mesenchymal identity in glioma stem cells in an NF-kappaB-dependent manner. Cancer Cell 29(2):201–213. https://doi.org/10.1016/j.ccell.2016.01.005
Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z et al (2009) Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell 15(4):283–293. https://doi.org/10.1016/j.ccr.2009.02.015
Mineo M, Ricklefs F, Rooj AK, Lyons SM, Ivanov P, Ansari KI et al (2016) The long non-coding RNA HIF1A-AS2 facilitates the maintenance of mesenchymal glioblastoma stem-like cells in hypoxic niches. Cell Rep 15(11):2500–2509. https://doi.org/10.1016/j.celrep.2016.05.018
Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J et al (2015) Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett 359(1):75–86. https://doi.org/10.1016/j.canlet.2014.12.051
Chen W, Xu XK, Li JL, Kong KK, Li H, Chen C et al (2017) MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget 8(14):22783–22799. https://doi.org/10.18632/oncotarget.15199
Cai H, Liu X, Zheng J, Xue Y, Ma J, Li Z et al (2017) Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene 36(3):318–331. https://doi.org/10.1038/onc.2016.212
Liu Y, Xu N, Liu B, Huang Y, Zeng H, Yang Z et al (2016) Long noncoding RNA RP11-838N2.4 enhances the cytotoxic effects of temozolomide by inhibiting the functions of miR-10a in glioblastoma cell lines. Oncotarget 7(28):43835–43851. https://doi.org/10.18632/oncotarget.9699
Wang Y, Liu J, Bai H, Dang Y, Lv P, Wu S (2017) Long intergenic non-coding RNA 00152 promotes renal cell carcinoma progression by epigenetically suppressing P16 and negatively regulates miR-205. Am J Cancer Res 7(2):312–322
Cai Q, Wang Z, Wang S, Weng M, Zhou D, Li C et al (2017) Long non-coding RNA LINC00152 promotes gallbladder cancer metastasis and epithelial-mesenchymal transition by regulating HIF-1alpha via miR-138. Open Biol. https://doi.org/10.1098/rsob.160247
Huang Y, Luo H, Li F, Yang Y, Ou G, Ye X et al (2018) LINC00152 down-regulated miR-193a-3p to enhance MCL1 expression and promote gastric cancer cells proliferation. Biosci Rep. https://doi.org/10.1042/BSR20171607
Wu J, Shuang Z, Zhao J, Tang H, Liu P, Zhang L et al (2018) Linc00152 promotes tumorigenesis by regulating DNMTs in triple-negative breast cancer. Biomed Pharmacother 97:1275–1281. https://doi.org/10.1016/j.biopha.2017.11.055
Zhang G, Ghosh S (2001) Toll-like receptor-mediated NF-kappaB activation: a phylogenetically conserved paradigm in innate immunity. J Clin Investig 107(1):13–19. https://doi.org/10.1172/JCI11837
Kang J, Tae N, Min BS, Choe J, Lee JH (2012) Malabaricone C suppresses lipopolysaccharide-induced inflammatory responses via inhibiting ROS-mediated Akt/IKK/NF-kappaB signaling in murine macrophages. Int Immunopharmacol 14(3):302–310. https://doi.org/10.1016/j.intimp.2012.08.006
Shin SY, Kim CG, Jung YJ, Lim Y, Lee YH (2016) The UPR inducer DPP23 inhibits the metastatic potential of MDA-MB-231 human breast cancer cells by targeting the Akt-IKK-NF-kappaB-MMP-9 axis. Sci Rep 6:34134. https://doi.org/10.1038/srep34134
Chen D, Niu M, Jiao X, Zhang K, Liang J, Zhang D (2012) Inhibition of AKT2 enhances sensitivity to gemcitabine via regulating PUMA and NF-kappaB signaling pathway in human pancreatic ductal adenocarcinoma. Int J Mol Sci 13(1):1186–1208. https://doi.org/10.3390/ijms13011186
Huang K, Yang C, Wang QX, Li YS, Fang C, Tan YL et al (2017) The CRISPR/Cas9 system targeting EGFR exon 17 abrogates NF-kappaB activation via epigenetic modulation of UBXN1 in EGFRwt/vIII glioma cells. Cancer Lett 388:269–280. https://doi.org/10.1016/j.canlet.2016.12.011
Acknowledgements
This study was supported by (1) The Research Project of the Chinese Society of Neuro-oncology, CACA (CSNO-2016-MSD12); (2) The Research Project of the Health and Family Planning Commission of Heilongjiang Province (2017-201); and (3) The Harbin Medical University Scientific Research Innovation Fund (2017LCZX37).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
The study involving human participants and animals was approved by the ethics committees of participating hospitals, and all patients provided written informed consent.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cai, J., Zhang, J., Wu, P. et al. Blocking LINC00152 suppresses glioblastoma malignancy by impairing mesenchymal phenotype through the miR-612/AKT2/NF-κB pathway. J Neurooncol 140, 225–236 (2018). https://doi.org/10.1007/s11060-018-2951-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-018-2951-0