Abstract
Introduction
Temozolomide (TMZ) is the preferred chemotherapeutic drug approved for the Glioblastoma multiforme (GBM) treatment. However, resistance to TMZ is the most intractable challenge for treatment of GBM. Screening of miRNAs is becoming a novel strategy to reveal underlying mechanism of drug-resistance of human tumors.
Materials and methods
We conducted RNA sequencing (RNA-seq) for GBM cells treated continuously with TMZ 1 or 2 week or not. Bioinformatic analysis was used to predict targets of these altered miRNAs. Subsequently, we studied the potential role of miR-1268a in TMZ-resistance of GBM cells.
Results
Expression levels of 55 miRNAs were identified altering both after 1 and 2 weeks TMZ treatment. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were conducted to illuminate the biological implication and related pathways of predicted target genes. We showed that miR-1268a was downregulated after TMZ treatment and targeted ABCC1/MRP1, a membrane transporter contributing to drug resistance, using dual-luciferase assay. Furthermore, we confirmed overexpression of miR-1268a inhibited protein translation of ABCC1 and restored upregulated expression of ABCC1 due to TMZ. Inversely, knockdown of miR-1268a increased ABCC1 at protein level and enhanced upregulation of ABCC1 with TMZ treatment. In addition, our data indicated that miR-1268a enhanced TMZ sensitivity in GBM cells.
Conclusion
Through RNA-seq analysis, we discovered miR-1268a and elucidated its role in modulating TMZ-resistance of GBM cells by targeting ABCC1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma (GBM) is the most frequent and aggressive brain cancer with extremely poor prognostic outcome [1]. Current strategy for GBM treatment is composed of maximal surgical resection followed by radiotherapy with concomitant and adjuvant chemotherapy [2]. Temozolomide (TMZ), an oral alkylating agent, has been applied for newly diagnosed and recurrent malignant gliomas as the first-line chemotherapeutic reagent. However, the median survival period of GBM patients is only 14.6 months, due to the inherent or acquired resistance to chemo and radiotherapy [3, 4]. This study focus on the underlying mechanism of acquired chemotherapy resistance in GBM.
MicroRNAs (miRs) are short noncoding single-stranded RNAs. They post-transcriptionally affect gene expression by binding to regions in the 3′-UTR regions of mRNAs [5]. Concern about the role of miRNAs in drug-resistance of human tumors is increasing in recent years [6]. However, most of the studies targeted differences between sensitive and resistant cancer cells. Here, we performed whole transcriptome sequencing (RNA-seq) analysis of GBM cells after TMZ treatment continuously for 1 week or 2, to explore the mechanism for acquired chemotherapy resistance. With the same strategy, ABCC1 protein (also known as MRP1) was found differentially expressed after TMZ treatment by our previously proteomics analysis [7]. ABCC1 belongs to the C subfamily of ATP-binding cassette (ABC) family, is capable to extrude various anticancer drugs from the cell, contributing to cancer drug-resistance [8]. Different tumors, such as lung cancer, esophageal cancers, classical Hodgkin lymphoma, and colorectal cancer highly express ABCC1/ MRP1, resulting in resistance to a variety of anticancer drugs [8, 9]. Furthermore, previous studies showed the overexpression of ABCC1 in GBM cells enhance chemo-resistance by increasing drug efflux and reducing bioavailability of anticancer agents [10]. It has been identified that the expression of ABCC1 is negatively regulated by several miRNAs, such as miR-7 in small cell lung cancer, miR-326/miR-134 in breast cancer cell, miR-1291 in pancreatic carcinoma cell, embryonic kidney cell, etc [11,12,13,14]. But there is no report about miRNAs that regulate ABCC1 in TMZ resistance in GBM cells.
This study identified miR-1268a was significantly downregulated after TMZ treatment. MiR-1268a is one of the miRNAs related to embryogenesis and cell differentiation [15], and recent study suggested that miR-1268a is also involved with drug-resistance in diffusely infiltrating astrocytoma and hepatocellular carcinoma [16]. In addition, ABCC1 is one of the target of miR-1268a based on prediction of miRnada (v3.3a) and TargetScan(Version:7.0). In this study, we confirmed that miR-1268a is associated with drug-resistance of GBM cells by targeting ABCC1.
Materials and methods
Cell lines and culture
The U87 and LN229 GBM cells were purchased from ATCC (American Type Culture Collection, USA). Cells were maintained in Dulbecco’s Modifed Eagle’s Medium (DMEM; Gibco, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS; Gibco) at 37 °C in an incubator with 5% CO2. Cells were cultured in DMEM comprising TMZ (Sigma–Aldrich, St. Louis, MO, USA) (IC50 of U87, 200 µM; IC50 of LN229, 400 µM) or dimethyl sulfoxide (DMSO; Sigma–Aldrich) for 1 or 2 weeks, respectively, as described [7].
RNA isolation, reverse transcription and quantitative real-time PCR
Total RNA, including miRNAs, was extracted using Trizol reagents (Invitrogen, CA, USA) according to the manufacturer’s instructions. RNA samples were reversely transcribed into cDNA using PrimeScript RT reagent Kit (Takara, Dalian, China). cDNA samples were used as templates for qPCR amplifcations using the Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientifc, Pittsburgh, PA, USA).
Library construction and sequencing
18–30nt RNAs were enriched by polyacrylamide gel electrophoresis (PAGE) after being extracted. The 3′ adapters were added to enrich the 36–44nt RNAs. Then the 5′ adapters were ligated as well. The ligation products were reverse transcripted by PCR. After PCR amplification, products were enriched to construct a cDNA library and sequenced using Illumina HiSeqTM 2500 by Sagene Biotech Co.Ltd (Guangzhou, China).
Data analyses
Raw reads were filtered to get clean tags by removing dirty reads containing adapters or low quality bases. All of the clean tags were then searched against miRBase database (Release 21) (http://www.mirbase.org/) to identify miRNAs. The miRNA expression level was calculated and normalized to transcripts per million (TPM). The differentially expressed miRNAs were identified with a fold change ≥ 2 and P value < 0.05 in a comparison as significant. Miranda (v3.3a) and TargetScan (Version:7.0) were used to predict targets.
Functional annotation and pathway analysis
Predicted targets of differentially expressed miRNAs were mapped to GO terms in the Gene Ontology database (http://www.geneontology.org/) to illuminate the biological implication. The predicted targets of differentially expressed miRNAs were mapped to Kyoto encyclopedia of genes and genomes (KEGG) database. P-value was gone through FDR correction, taking FDR ≤ 0.05 as a threshold for determining significantly enriched GO terms and KEGG pathway.
GBM tissue samples
12 samples of primary GBM and 10 samples from recurrent GBM patients who were treated with TMZ were obtained from Nanfang hospital (Guangzhou, China). This study was approved by The Ethics Committee of Nanfang Hospital in accordance with the Helsinki Declaration, and written informed consent was obtained from all patients.
Immunohistochemistry (IHC)
Immunohistochemistry to detect ABCC1 in GBM specimens was performed as described previously [7]. Primary anti-ABCC1 antibody(#ab32574; Abcam; 1:200) was used in this assay.
R2: microarray analysis
GSE16011 database were obtained from R2: microarray analysis and visualization platform (http://hgserver1.amc.nl/cgibin/r2/main.cgi). Kaplan–Meier analysis and survival curves were carried out by using GraphPad Prism software program (version 5.0, GraphPad Software, San Diego, CA, USA). All cutoff values for separating high and low expression groups were determined by the online R2 database algorithm.
Cell transfection
U87 and LN229 cells were transiently transfected with 100 nmol/l of miR-1268a mimics (sense: 5′-CGGGCGUGGUGGUGGGGG-3′; antisense: 5′-CCCACCACCACGCCCGUU-3′), miR-1268a inhibitors (5′-CCCCCACCACCACGCCCG-3′) and miRNA negative control (miR-NC) (GenePharma, Shanghai, China) using Lipofectamine 2000 and OPTI-MEM I (Invitrogen USA, Carlsbad, California).
Dual-luciferase reporter assay
Luciferase reporter vectors were constructed containing wild-type or mutated 3′ UTR of ABCC1. U87 cells were co-transfected with synthetic miR-1268a mimics or inhibitor together with either vector. Cells were harvested after 48 h. Luciferase activities were tested based on manufacturer’s protocol (#E1910, Promega, Madison, WI, USA).
Protein extraction and western blot
Total protein were extracted with RIPA Buffer (#9806; Cell Signaling Technology, Danvers, MA, USA). For Western blot analysis, 30 µg of the whole lysates were separated on a 12% gel by electrophoresis and electrotransferred to 0.45µ m polyvinylidene fluoride membranes (Thermo Scientifc, Pittsburgh, PA, USA). Membranes were blocked in 5% milk in Tris-buffered saline (TBS, pH 7.6), then incubated with primary anti-ABCC1 antibody(#ab32574; Abcam; 1:500) overnight at 4 °C. Membranes were then washed three times with TBST for 5 min and incubated at 37 °C with an secondary HRP-conjugated anti-mouse IgG antibody (#7076; Cell Signaling Technology) at a dilution of 1:2000. Segments were photo-developed after further washes with TBST.
Cell proliferation assay
Cell Counting Kit-8 (CCK-8, Dojindo, Tokyo, Japan) was used to detect the GBM cells viability. 1 × 103 cells transfected with were planked in 96-well cell plates and subsequently treated with TMZ or DMSO. 100 µl medium containing 10% CCK-8 were substituted in each well. Cells were then incubated at 37 °C for another hour. Optical density (O.D.) was monitored at 450 nm. The detection last a week and measured every 24 h.
Nude mouse tumor cell xenograft assay
The protocol for the use of the mouse xenograft model was approved by the Experimental Animal Center of Southern Medical University (Guangzhou, China). The nude mice (female BALB/c-nu, 4 weeks old) were maintained in specific pathogen-free conditions. U87 cells stably expressing miR-1268a and NC were injected subcutaneously into right flank of each mouse (1 × 107 cells). 1 week later, the mice were subjected to daily intraperitoneal injection of TMZ (20 mg/kg for each mouse) for 2 weeks and equal amount of DMSO was used as a vehicle control. Mice were euthanized, and tumors were weighed and photographed.
Statistical analysis
All experiments were performed in triplicate. Data was presented as mean ± SD. Statistical analysis was performed by Student’s t-test using GraphPad Prism 6 software (San Diego, CA) and statistical significance is indicated (*p < 0.05).
Results
Differentially expressed miRNAs between control and TMZ treated GBM cells
By performing RNA-seq analysis, the expression levels of 55 miRNAs were found significantly changed (p-value < 0.05, fold change ≥ 2) after both 1 and 2 weeks TMZ treatment. The heatmap were given to display expression levels of the 55 miRNAs in different samples and to cluster miRNAs with similar expression pattern (Fig. 1a). Hierarchical cluster analysis indicated that the expression profile of these miRNAs could well distinct the TMZ treated samples from normal controls.
Differentially expressed miRNAs between control and TMZ treated GBM cells. a Heatmap show differential expression of miRNAs in groups of U87 cells under different treated conditions. b GO enrichment analysis of the top-ranking cellular component and molecular function according to − log10 (p-value), p-value < 0.05. c KEGG pathway enrichment analysis of the top 20. NC1 represents sample treated with DMSO for 1 week. NC2 represents sample treated with DMSO for 2 week. TMZ1 represents sample treated with TMZ for 1 week. NC2 represents sample treated with DMSO for 2 week
To investigate the correlated mechanisms of the acquired TMZ-resistance modulated by miRNAs, gene ontology (GO) analysis were conducted for the predicted targets of all differentially expressed miRNAs after TMZ treatment for 1 or 2 weeks, respectively. Enrichment analysis of cellular component (CC) and molecular function (MF) demonstrated that the predicted targets mostly related to membrane, transmembrane transporter activity and transporter activity (Fig. 1b). KEGG pathway analysis revealed that MAPK signaling pathway, Hedgehog signaling pathway and p53 signaling pathway were enriched (Fig. 1c).
Verification of the altered expression levels of miR-1268a and ABCC1 induced by TMZ
Combining the above analysis and our former proteomics research, we discovered ABCC1, a vital membrane transporter contributed to obtaining of drug-resistance, was elevated in TMZ treated U87 cells according to our former proteomics research. Conversely, miR-1268a, one of the differentially expressed miRNAs, downregulated after TMZ treatment in RNA-seq analysis, was predicted to target ABCC1 both in miRnada and TargetScan. To further explore the underlying mechanism of TMZ resistance, we detected 12 initial GBM samples and 10 recurrent cases of GBM samples from patients treated with TMZ. Our data showed that miR-1268a expression is lower in recurrent GBM specimens compared with initial specimens (Fig. 2a). Conversely, ABCC1 expression is significantly upregulated in recurrent samples (Fig. 2b, c). Survival analysis of ABCC1 expression in GSE16011 glioma database demonstrated that higher expression of ABCC1 was related to unfavorable prognosis (Fig. 2d). Subsequently, expression levels of miR-1268a and ABCC1 in U87 and LN229 were measured by using quantitative real-time PCR (qPCR). Significant miR-1268a downregulation and ABCC1 upregulation were observed in TMZ treated U87 and LN229 cells at RNA level (Fig. 3a, b). Furthermore, by Western blot, a marked upregulation of ABCC1 protein was found in TMZ treated sample (Fig. 3c).
MiR-1268a is downregulated and ABCC1 is upregulated in recurrent GBM tissues. a Relative miR-1268a expression levels were analyzed by qPCR in 12 initial GBM tissues and 10 recurrent GBM tissues and normalized to the U6 expression levels, **p < 0.01. b Relative ABCC1 expression levels in initial and recurrent GBM specimens, **p < 0.01. c Kaplan–Meier curves for overall survival in patients with high or low expression of ABCC1 in glioma patients using GSE16011 database
Validation of miR-1268a and ABCC1 altered expression in TMZ treated cells. a, b Relative expression levels of ABCC1 and miR-1268a mRNA in U87 and LN229 cells treated with TMZ for 1 or 2 weeks were detected by qPCR, DMSO was used as control. c Relative expression level of ABCC1 protein in U87 and LN229 cells treated with TMZ for 1 or 2 weeks were detected by western blot, β-actin expression served as an internal control. Student’s t tests and One-way ANOVA were performed. (*p < 0.05, **p < 0.01)
ABCC1 is a target of miR-1268a in GBM cells
Based on the bioinformatics analysis, ABCC1 may be a target of miR-1268a. To verified this, U87 cells were co-transfected with synthetic miR-1268a mimics together with luciferase reporter vectors with wild-type or mutated 3′ UTR of ABCC1 (Fig. 4a), exposed to TMZ or not. Marked declines of luciferase activity were observed after miR-1268a mimics were co-transfected with wild-type vector, but not with mutated-type, whether treated with TMZ or not (Fig. 4b), indicating a direct interaction of miR-1268a and 3′ UTR of ABCC1.
MiR-1268a directly targets ABCC1. a The putative binding sites between ABCC1 3′-UTR and miR-1268a sequences. The reporter products of wild type ABCC1 3′-UTR (ABCC1-WT) and mutated ABCC1 3′-UTR (ABCC1-MUT) are shown. b Dual-luciferase reporter assay was performed in U87 cells to monitor the relative luciferase activities of ABCC1-WT and ABCC1-MUT reporters. The firefly luciferase activity was normalized to renilla luciferase activity; *p < 0.05, **p < 0.01. c, d Expression of ABCC1 in U87 and LN229 cells transfected with miR-1268a mimics or inhibitor, plus TMZ or not. (c the mRNA levels of different samples, *p < 0.05. d the protein levels of different samples). β-actin expression served as an internal control. e Spearman’s correlation analysis of the correlation between the expression levels of ABCC1 and miR-1268a in GBM specimens
To further confirmed ABCC1 is a target of miR-1268a, we evaluated ABCC1 expression levels after transfecting miR-1268a mimics or inhibitor in U87 and LN229 cells. A marked decrease expression of ABCC1 was observed at protein, though not at mRNA level, treated with miR-1268a mimics. Moreover, miR-1268a mimics suppressed the upregulation of ABCC1 induced by TMZ, both at protein and mRNA levels (Fig. 4c, d). Increased ABCC1 protein expression, but not mRNA expression, was observed after transfecting miR-1268a inhibitor. Enhanced upregulation of ABCC1 induced by TMZ was also founded in U87 and LN229 cells treated with miR-1268a inhibitor (Fig. 4c, d). Furthermore, there was a significant inverse correlation between miR-1268a and ABCC1 in GBM specimens (Fig. 4e, r = − 0.4716). Combining these findings, we conclude that ABCC1 is a target of miR-1268a in GBM cells.
Overexpressing miR-1268a enhanced sensitivity of GBM cells to TMZ
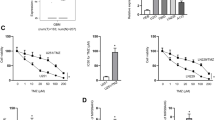
To further investigate the role of miR-1268a in TMZ therapy, we transfected miR-1268a mimics or inhibitor into U87 and LN229 cells and treated them with DMSO or TMZ. Cell viability was monitored using CCK-8 (Cell Count Kit-8) assay. The curves demonstrated that transfection of miR-1268a mimics did not affect the proliferative ability of cells alone, but markedly suppressed the cell viability after TMZ treatment, indicating that miR-1268a mimics enhanced sensitivity of U87 and LN229 cells to TMZ (Fig. 5a). Furthermore, it’s interesting that transfection of miR-1268a inhibitor induced ascent of cell viability, exposed to TMZ or not (Fig. 5a). Subsequently, a subcutaneous xenotransplanted GBM model was used to evaluate the function of miR-1268a in TMZ therapy in vivo. The results showed that after TMZ treatment, the volume and weight of xenografts group overexpressing miR-1268a significantly decreased compared with NC group. Thus, miR-1268a enhances the sensitivity of GBM cells to TMZ.
Mir-1268a modulates the chemo-sensitivity of GBM cells to TMZ. a CCK-8 assay was conducted to monitored cell viability in U87 and LN229 cells transfected with miR-1268a mimics or inhibitor, with or without TMZ treatment. b Tumor samples from subcutaneous xenograft model. c Weight of the excised tumors in different groups, **p < 0.01
Discussion
Drug-resistance of GBM to TMZ plays a crucial role in tumor recurrence and progression. Our study specially target on acquired TMZ resistance in GBM cells through transcriptome sequencing of miRNAs. U87, as a GBM cell line sensitive to TMZ but capable of obtaining TMZ-resistance, is suitable to be served as a cell model of studying the mechanism of acquired TMZ-resistance in GBM cells. Based on RNA-seq analysis, altered expression level of 55 miRNAs were identified in U87 cell line after TMZ treatment, containing some miRNAs previously shown to be involved in drug-resistance of cancer including glioma (miR-139-5p, miR-451a miRNA-1246, miR-140-5p, miR-34a-5p, miR-194-5p, miRNA-34c-5p, miRNA-1268a, miRNA-335-3p, miR-181b-5p) [16,17,18,19,20,21,22,23,24,25]. Among these, miR-1268a was one of the most significantly downregulated miRNA and was discovered to modified the effects of TMZ treatment on diffusely infiltrating astrocytoma (DIA) [26]. We then analyzed the miRNA expression data with miRNA target predictions by Miranda and TargetScan database. Functional annotation revealed that predicted targets of differentially expressed miRNAs significantly dysregulated in transmembrane transporter activity of GO terms, which indicate that altering expression of microRNAs play a role in acquired TMZ resistance by targeting membrane transporters. KEGG pathway analysis demonstrated MAPK signaling pathway, Hedgehog signaling pathway and p53 signaling pathway were significantly enriched, served as vital pathways in cancer drug-resistance [27,28,29,30].
Besides inherent drug-resistance, mechanisms of acquired TMZ resistance in GBM cells provide hints to overcome the refractoriness of GBM [31,32,33]. In our previous study, dynein, cytoplasmic 2, heavy chain 1 (DHC2) was shown to be elevated and involved in acquired resistance after TMZ treatment [7]. Similarly, in the same proteomics study, expression of ABCC1 was indicated to be upregulated after TMZ treatment, which is verified in this study subsequently. Consistently, in another research about acquired TMZ resistance in GBM cells, upregulation of ABCC1 was also identified in TMZ-resistant cells [31]. ABCC1/MRP1 plays a role in efflux of cytotoxic hydrophobic drugs in tumor cells, contributing to the drug-resistance of tumor [12, 34, 35]. Accumulating evidences have shown that ABCC1 enhanced the drug-resistance of primary and recurrent GBM cells [10, 36, 37], indicating elevated ABCC1 induced by TMZ contribute to the acquired resistance in GBM cells. Understanding of further mechanism about ABCC1 upregulation is crucial for reversing acquisition TMZ resistance. In the present study, we identified ABCC1 is a target of miR-1268a, using dual-luciferase assay.
Our data showed that miR-1268a was markedly downregulated in U87 and LN229 cells induced by TMZ treatment, resulting in the upregulation of ABCC1, thus contributing to the acquired TMZ resistance. Furthermore, overexpression of miR-1268a induced downregulation of ABCC1 at protein level and restore the upregulated expression of ABCC1 induced by TMZ. Conversely, knockdown of miR-1268a increased ABCC1 expression and furtherly augment the upregulation of ABCC1 with TMZ treatment. In addition, our data showed that overexpressing miR-1268a enhanced sensitivity of GBM cells to TMZ in vitro and in vivo. These findings supplies the proof that miR-1268a modulates drug-resistance of GBM cells to TMZ by targeting ABCC1.CCK-8 assays also revealed that inhibition of miR-1268a enhances GBM cells viability, plus TMZ or not. Since interference of ABCC1 did not affect GBM cells viability [10] and miR-1268a polymorphism modified DIA risk and prognosis [26], miR-1268a inhibitor may play a role in GBM cells proliferation because of its effect on altering different phenotypes of miR-1268a relative expression, but not targeting ABCC1.
In conclusion, combining data from RNA-seq analysis and proteomics analysis, we found miR-1268a and validated its role in mediating TMZ sensitivity in GBM cells by targeting ABCC1. Combination of miR-1268a and TMZ may be a promising treatment beneficial to GBM patients.
References
Cloughesy TF, Cavenee WK, Mischel PS (2014) Glioblastoma: from molecular pathology to targeted treatment. Annu Rev Pathol 9:1–25. https://doi.org/10.1146/annurev-pathol-011110-130324
Wen PY, Reardon DA (2016) Neuro-oncology in 2015: progress in glioma diagnosis, classification and treatment. Nat Rev Neurol 12(2):69–70. https://doi.org/10.1038/nrneurol.2015.242
Messaoudi K, Clavreul A, Lagarce F (2015) Toward an effective strategy in glioblastoma treatment. Part I: resistance mechanisms and strategies to overcome resistance of glioblastoma to temozolomide. Drug Discov Today 20(7):899–905. https://doi.org/10.1016/j.drudis.2015.02.011
Messaoudi K, Clavreul A, Lagarce F (2015) Toward an effective strategy in glioblastoma treatment. Part II: RNA interference as a promising way to sensitize glioblastomas to temozolomide. Drug Discov Today 20(6):772–779. https://doi.org/10.1016/j.drudis.2015.02.014
Hammond SM (2015) An overview of microRNAs. Adv Drug Deliv Rev 87:3–14. https://doi.org/10.1016/j.addr.2015.05.001
Ling H, Fabbri M, Calin GA (2013) MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov 12(11):847–865. https://doi.org/10.1038/nrd4140
Wang H, Feng W, Lu Y, Li H, Xiang W, Chen Z, He M, Zhao L, Sun X, Lei B, Qi S, Liu Y (2016) Expression of dynein, cytoplasmic 2, heavy chain 1 (DHC2) associated with glioblastoma cell resistance to temozolomide. Sci Rep 6:28948. https://doi.org/10.1038/srep28948
Cole SP (2014) Targeting multidrug resistance protein 1 (MRP1, ABCC1): past, present, and future. Annu Rev Pharmacol Toxicol 54:95–117. https://doi.org/10.1146/annurev-pharmtox-011613-135959
Greaves W, Xiao L, Sanchez-Espiridion B, Kunkalla K, Dave KS, Liang CS, Singh RR, Younes A, Medeiros LJ, Vega F (2012) Detection of ABCC1 expression in classical Hodgkin lymphoma is associated with increased risk of treatment failure using standard chemotherapy protocols. J Hematol Oncol 5:47. https://doi.org/10.1186/1756-8722-5-47
Tivnan A, Zakaria Z, O’Leary CN, Gel K, Pokorny D, Sarkaria JL, Prehn JN JHM (2015) Inhibition of multidrug resistance protein 1 (MRP1) improves chemotherapy drug response in primary and recurrent glioblastoma multiforme. Front Neurosci. https://doi.org/10.3389/fnins.2015.00218
Liu H, Wu X, Huang J, Peng J, Guo L (2015) miR-7 modulates chemoresistance of small cell lung cancer by repressing MRP1/ABCC1. Int J Exp Pathol 96(4):240–247. https://doi.org/10.1111/iep.12131
Liang Z, Wu H, Xia J, Li Y, Zhang Y, Huang K, Wagar N, Yoon Y, Cho HT, Scala S, Shim H (2010) Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem Pharmacol 79(6):817–824. https://doi.org/10.1016/j.bcp.2009.10.017
Lu L, Ju F, Zhao H, Ma X (2015) MicroRNA-134 modulates resistance to doxorubicin in human breast cancer cells by downregulating ABCC1. Biotechnol Lett 37(12):2387–2394. https://doi.org/10.1007/s10529-015-1941-y
Pan YZ, Zhou A, Hu Z, Yu AM (2013) Small nucleolar RNA-derived microRNA hsa-miR-1291 modulates cellular drug disposition through direct targeting of ABC transporter ABCC1. Drug Metab Dispos 41(10):1744–1751. https://doi.org/10.1124/dmd.113.052092
Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, Eaves CJ, Marra MA (2008) Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18(4):610–621. https://doi.org/10.1101/gr.7179508
Lu Y, Yao J, Huang X, Wang C, Wu X, Xia Q, Long X (2016) Prognostic significance of miR-1268a expression and its beneficial effects for post-operative adjuvant transarterial chemoembolization in hepatocellular carcinoma. Sci Rep. https://doi.org/10.1038/srep36104
Xu K, Shen K, Liang X, Li Y, Nagao N, Li J, Liu J, Yin P (2016) MiR-139-5p reverses CD44+/CD133+-associated multidrug resistance by downregulating NOTCH1 in colorectal carcinoma cells. Oncotarget 7(46):75118–75129. https://doi.org/10.18632/oncotarget.12611
Liu ZR, Song Y, Wan LH, Zhang YY, Zhou LM (2016) Over-expression of miR-451a can enhance the sensitivity of breast cancer cells to tamoxifen by regulating 14-3-3zeta, estrogen receptor alpha, and autophagy. Life Sci 149:104–113. https://doi.org/10.1016/j.lfs.2016.02.059
Hasegawa S, Eguchi H, Nagano H, Konno M, Tomimaru Y, Wada H, Hama N, Kawamoto K, Kobayashi S, Nishida N, Koseki J, Nishimura T, Gotoh N, Ohno S, Yabuta N, Nojima H, Mori M, Doki Y, Ishii H (2014) MicroRNA-1246 expression associated with CCNG2-mediated chemoresistance and stemness in pancreatic cancer. Br J Cancer 111(8):1572–1580. https://doi.org/10.1038/bjc.2014.454
Wei R, Cao G, Deng Z, Su J, Cai L (2016) miR-140-5p attenuates chemotherapeutic drug-induced cell death by regulating autophagy through inositol 1,4,5-trisphosphate kinase 2 (IP3k2) in human osteosarcoma cells. Biosci Rep. https://doi.org/10.1042/BSR20160238
Pu Y, Zhao F, Wang H, Cai W, Gao J, Li Y, Cai S (2016) MiR-34a-5p promotes the multi-drug resistance of osteosarcoma by targeting the CD117 gene. Oncotarget 7(19):28420–28434. https://doi.org/10.18632/oncotarget.8546
Dell’Aversana C, Giorgio C, D’Amato L, Lania G, Matarese F, Saeed S, Di Costanzo A, Belsito PV, Ingenito C, Martens JH, Pallavicini I, Minucci S, Carissimo A, Stunnenberg HG, Altucci L (2017) miR-194-5p/BCLAF1 deregulation in AML tumorigenesis. Leukemia. https://doi.org/10.1038/leu.2017.64
Tung SL, Huang WC, Hsu FC, Yang ZP, Jang TH, Chang JW, Chuang CM, Lai CR, Wang LH (2017) miRNA-34c-5p inhibits amphiregulin-induced ovarian cancer stemness and drug resistance via downregulation of the AREG-EGFR-ERK pathway. Oncogenesis 6(5):e326. https://doi.org/10.1038/oncsis.2017.25
Martin EC, Conger AK, Yan TJ, Hoang VT, Miller DF, Buechlein A, Rusch DB, Nephew KP, Collins-Burow BM, Burow ME (2017) MicroRNA-335-5p and -3p synergize to inhibit estrogen receptor alpha expression and promote tamoxifen resistance. FEBS Lett 591(2):382–392. https://doi.org/10.1002/1873-3468.12538
Chen Y, Li R, Pan M, Shi Z, Yan W, Liu N, You Y, Zhang J, Wang X (2017) MiR-181b modulates chemosensitivity of glioblastoma multiforme cells to temozolomide by targeting the epidermal growth factor receptor. J Neuro-Oncol 133(3):477–485. https://doi.org/10.1007/s11060-017-2463-3
Li X, Wang J, Xu A, Huang J, Meng L, Huang R, Wang J (2016) The microRNA-1268a rs28599926 polymorphism modified diffusely infiltrating astrocytoma risk and prognosis. International J Clin Exp Med 9(11):21615–21624. doi
Roesch A (2015) Tumor heterogeneity and plasticity as elusive drivers for resistance to MAPK pathway inhibition in melanoma. Oncogene 34(23):2951–2957. https://doi.org/10.1038/onc.2014.249
Zeng X, Zhao H, Li Y, Fan J, Sun Y, Wang S, Wang Z, Song P, Ju D (2015) Targeting Hedgehog signaling pathway and autophagy overcomes drug resistance of BCR-ABL-positive chronic myeloid leukemia. Autophagy 11(2):355–372. https://doi.org/10.4161/15548627.2014.994368
Ding J, Zhou XT, Zou HY, Wu J (2017) Hedgehog signaling pathway affects the sensitivity of hepatoma cells to drug therapy through the ABCC1 transporter. Lab Investig 97(7):819–832. https://doi.org/10.1038/labinvest.2017.34
Bertheau P, Lehmann-Che J, Varna M, Dumay A, Poirot B, Porcher R, Turpin E, Plassa LF, de Roquancourt A, Bourstyn E, de Cremoux P, Janin A, Giacchetti S, Espie M, de The H (2013) p53 in breast cancer subtypes and new insights into response to chemotherapy. Breast 22 (Suppl 2):S27-S29. https://doi.org/10.1016/j.breast.2013.07.005
Zeng H, Xu N, Liu Y, Liu B, Yang Z, Fu Z, Lian C, Guo H (2017) Genomic profiling of long non-coding RNA and mRNA expression associated with acquired temozolomide resistance in glioblastoma cells. Int J Oncol 51(2):445–455. https://doi.org/10.3892/ijo.2017.4033
Kitange GJ, Mladek AC, Carlson BL, Schroeder MA, Pokorny JL, Cen L, Decker PA, Wu W, Lomberk GA, Gupta SK, Urrutia RA, Sarkaria JN (2012) Inhibition of histone deacetylation potentiates the evolution of acquired temozolomide resistance linked to MGMT upregulation in glioblastoma xenografts. Clin Cancer Res 18(15):4070–4079. https://doi.org/10.1158/1078-0432.CCR-12-0560
Quail DF, Bowman RL, Akkari L, Quick ML, Schuhmacher AJ, Huse JT, Holland EC, Sutton JC, Joyce JA (2016) The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science 352(6288):d3018. https://doi.org/10.1126/science.aad3018
Ma J, Wang T, Guo R, Yang X, Yin J, Yu J, Xiang Q, Pan X, Tang H, Lei X (2015) Involvement of miR-133a and miR-326 in ADM resistance of HepG2 through modulating expression of ABCC1. J Drug Target 23(6):519–524. https://doi.org/10.3109/1061186X.2015.1015536
Benyahia B, Huguet S, Decleves X, Mokhtari K, Criniere E, Bernaudin JF, Scherrmann JM, Delattre JY (2004) Multidrug resistance-associated protein MRP1 expression in human gliomas: chemosensitization to vincristine and etoposide by indomethacin in human glioma cell lines overexpressing MRP1. J Neurooncol 66(1–2):65–70. doi
Cui Y, Lin J, Zuo J, Zhang L, Dong Y, Hu G, Luo C, Chen J, Lu Y (2015) AKT2-knockdown suppressed viability with enhanced apoptosis, and attenuated chemoresistance to temozolomide of human glioblastoma cells in vitro and in vivo. Onco Targets Ther 8:1681–1690. https://doi.org/10.2147/OTT.S83795
Lu XY, Cao K, Li QY, Yuan ZC, Lu PS (2012) The synergistic therapeutic effect of temozolomide and hyperbaric oxygen on glioma U251 cell lines is accompanied by alterations in vascular endothelial growth factor and multidrug resistance-associated protein-1 levels. J Int Med Res 40(3):995–1004. https://doi.org/10.1177/147323001204000318
Acknowledgements
This work was supported by National Natural Science Foundation of China (81372692, 81472315), National Key Technology Research and Development Program of the Ministry of Science and Technology of China (2014BAI04B01), Science and Technology Program of Guangdong (2016A020213006), Natural Science Foundation of Guangdong Province (2014A030313167).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study has been approved by the Ethics Committee of Southern Medical University and has been performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Li, Y., Liu, Y., Ren, J. et al. miR-1268a regulates ABCC1 expression to mediate temozolomide resistance in glioblastoma. J Neurooncol 138, 499–508 (2018). https://doi.org/10.1007/s11060-018-2835-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-018-2835-3








