Abstract
With advances in genomic profiling and sequencing technology, we are beginning to understand the landscape of the genetic events that accumulated during the neoplastic process. The insights gleamed from these genomic profiling studies with regards to glioblastoma etiology has been particularly satisfying because it cemented the clinical pertinence of major concepts in cancer biology—concepts developed over the past three decades. This article will review how the glioblastoma genomic data set serves as an illustrative platform for the concepts put forward by Hanahan and Weinberg on the cancer phenotype. The picture emerging suggests that most glioblastomas evolve along a multitude of pathways rather than a single defined pathway. In this context, the article will further provide a discussion of the subtypes of glioblastoma as they relate to key principles of developmental neurobiology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the landmark review by Hanahan and Weinberg [1], the authors distilled the essence of cancer into six distinct phenotypes, including evasion of apoptosis, self-sufficiency in growth signals, insensitivity to anti-growth signals, tissue invasion and metastasis, limitless replicative potentials, and sustained angiogenesis. The widely accepted paradigm suggests that cancer arises as a result of mutations or epigenetic events, which alter the functions of genes critical for attaining these phenotypes. These gene functions are intimately linked to the regulation of developmental processes [2]. As such, their aberrant functions inevitably lead to cell states that resemble stages during normal development. These cell states can be captured using genomic technologies to define distinct transcriptomal subtypes. With the advent of large scale genomic profiling, [3, 4], we now have a glimpse of the genetic events underlying glioblastoma pathogenesis as well as distinct transcriptomal signatures. In this review, the genomic profiles of glioblastoma will be reviewed in the context of the properties described by Hanahan and Weinberg. Transcriptomal subtypes of glioblastoma will be discussed in the context of developmental biology and the cell of origin.
Glioblastoma
Glioblastoma is the most common form of primary brain tumor, with dismal prognosis. The incidence of this tumor is fairly low, with 2–3 cases per 100,000 people in Europe and North America. Despite its rarity, overall mortality related to glioblastoma is comparable to the more prevalent tumors [5]. This is, in large part, due to the near uniform fatality of the afflicted patients. Indeed, glioblastoma is one of the most aggressive of the malignant tumors. Without treatment, the median survival is approximately 3 months [6]. The current standard of treatment involves maximal surgical resection followed by concurrent radiation therapy and chemotherapy with the DNA alkylating agent, temozolomide [7]. With this regimen, the median survival is approximately 14 months. For nearly all affected, this treatment combination remains palliative.
Studies carried out over the past three decades suggest that glioblastomas, like other cancers, arise secondary to the accumulation of genetic alterations. These alterations can take the form of epigenetic modifications, point mutations, translocations, amplifications, or deletions, and modify gene function in ways that deregulate cellular signaling pathways leading to the cancer phenotype [1]. The exact number and nature of genetic alterations and deregulated signaling pathways required for tumorigenesis remains an issue of debate [8], although it is now clear that CNS carcinogenesis requires multiple disruptions to the normal cellular circuitry [3, 4].
Cancer genomics
In the history of cancer research, the current decade will likely be known as the decade of cancer genomics. It is during our time that the marriage of technology and annotated specimen collection led to fruition to provide us a glimpse of the complex genomic landscape that underlies cancer pathogenesis. Impressively, these efforts demonstrated true collaborative spirits between clinicians and laboratory scientists with common goals of furthering translational science.
The Cancer Genome Atlas (TCGA) constitutes the largest of the genomic efforts. It is a comprehensive and coordinated effort to catalogue the genetic and epigenetic changes in the cancer genome, with goals of identifying those responsible for carcinogenesis. The project constitutes a joint effort of the National Human Genome Research Institute (NHGRI), National Cancer Institute (NCI), and the U.S. Department of Health and Human Services, and collects tumor specimen from major cancer centers spanning across the continental USA. The project aims to provide the genomic profile of 500 specimens of various cancer types using state-of-the-art platforms for sequencing, microRNA, mRNA, single-nucleotide polymorphisms, and methylation profiling. TCGA started as a pilot project in 2006 with focus on glioblastoma as the first cancer type for study. With the success of the pilot project, TCGA plans to expand its efforts to aggressively pursue 20 or more additional cancers.
While acknowledging the importance of the TCGA in cancer research, one cannot neglect the value of the pioneering genomic efforts that, in many ways, laid the groundwork for the TCGA [9]. This article will review the paradigms established by the various major genomic efforts, including the TCGA, in the context of the cancer phenotypes proposed by Hanahan and Weinberg [1] and fundamental tenets of developmental biology.
The cancer phenotype
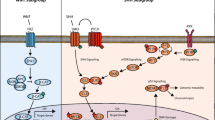
The aggregate of cancer research investigation spanning the past three decades suggest that cancer is a genetic disease characterized by mutations or epigenetic events that abrogate or compromise regulatory circuitry governing cell proliferation and homeostasis [8]. In the landmark review by Hanahan and Weinberg [1], the authors distilled the essence of these regulatory circuits into six distinct phenotypes, including evading apoptosis, self-sufficiency in growth signals, insensitivity to anti-growth signals, tissue invasion and metastasis, limitless replicative potentials, and sustained angiogenesis (Fig. 1). The following section will review the TCGA findings pertinent to these phenotypes.
Hallmarks of the cancer phenotype. TCGA revealed mutations that contributed to the cancer phenotype as proposed by Hanahan and Weinberg, including evading apoptosis, self-sufficiency in growth signals, insensitivity to anti-growth signals, tissue invasion and metastasis, limitless replicative potentials, and sustained angiogenesis. (Figure adapted from reference [1])
Self-sufficiency in growth signals—the receptor tyrosine kinase (RTK)/phosphoinosital 3 kinase (PI3K) signaling cascade
Active cellular proliferation in normal cells requires signals from its environment. These signals typically involve the binding of a transmembrane receptor to growth factors, extracellular matrix components, or cell surface components. This mitogenic signaling process is under stringent regulation in normal cells. Typically, multiple ligand-receptor interactions in a permissive cellular state are required before cellular proliferation can take place. This regulation minimizes the probability of dysregulated, autonomous cell growth [1, 10].
To abridge this stringent growth regulation, tumors often mutate the transmembrane receptors or their downstream effectors in ways that constitutively activate the pathway. The pathway most commonly mutated to achieve this end in glioblastoma involves the RTK–PI3K pathway [10, 11]. RTKs are cell surface receptors that are normally activated only in response to growth factor binding [10]. Results from the TCGA revealed that nearly all glioblastomas harbor activating mutations or amplifications in genes required for this signaling cascade [3, 4, 12, 13]. Epidermal growth factor receptor (EGFR) and platelet derived growth factor receptor (PDGFR) are two prototypical members of RTKs [3, 4, 13].
For EGFR and PDGFR, binding of the growth factor receptor to the ligand leads to homo- or hetero-dimerization of the receptor. This dimerization facilitates autophosphorylation of the cytoplasmic domains of the dimerized receptor at select tyrosine residues [10]. The phosphorylated tyrosine residue, in turn, recruits and binds to other signaling proteins on the cell membrane. In some cases, the phospho-tyrosine bound proteins serve as a platform for the recruitment of other effector proteins. In other cases, the bound protein undergoes a conformational change upon binding to the RTK and becomes activated in the process [10].
One of the critical cellular kinases that become activated upon binding to RTK is PI3K [14]. PI3Ks catalyze the phosphorylation of a critical component of the cell surface, phosphatidylinositol-4,5-isphosphate (PI(4,5)P2). This phosphorylation generates phosphatidylinositol-1,4,5-isphosphate (PI(1,4,5)P3), which in turn serves as a docking site for pro-proliferative down-stream effector proteins [11]. Thus, RTK activation transforms the cell membrane into a catalytic surface populated with a high density of pro-mitotic signaling molecules, ultimately leading to cell proliferation. Hydrolysis of (PI(1,4,5)P3) into (PI(4,5)P2) is catalyzed by a phosphatase termed phosphatase and tensin homology (PTEN), in turn, shuts off the RTK–PI3K pathway.
A number of investigations revealed activating mutations in the RTK–PI3K pathway [15–17], validating the importance of this pathway in glioblastoma pathogenesis. The large scale genomic efforts add to this literature in two ways. First, the effort revealed the wide prevalence of RTK–PI3K pathway activation in glioblastomas. In the TCGA report [3], activating mutations in the RTK–PI3K pathway are found in 88% of the 206 glioblastomas sequenced. Efforts by Parsons et. al. [4] revealed that activating mutations in the RTK–PI3K pathway are found in 50% of the 22 glioblastomas sequenced. Second, the TCGA effort revealed mutations in members of the RTK–PI3K pathway that were not previously implicated in glioblastoma pathogenesis. For instance, one of the effector proteins recruited to a phosphorylated RTK is Ras. Ras encodes a monomeric G-protein that cycles between an active form bound to GTP and an inactive form that binds to GDP [18]. It functions as a critical component of the pro-proliferative “catalytic surface”. Through a series of protein–protein interactions, RTK activation catalyzes the exchange of GDP for GTP in Ras, initiating signals required for cellular proliferation. The protein encoded by neurofibromatosis 1 (NF1) functions to catalyze the exchange of GTP for GDP in Ras, consequently preventing cell proliferation. While it is known that NF1 patients are predisposed to gliomagenesis [19], inactivating mutations in NF1 was not discovered in glioblastoma until recently [3, 4, 20, 21]. The TCGA results showed that approximately 20% of glioblastomas harbor loss of function mutations in NF1 [3, 4]. Importantly, mutations in NF1 appear to define a particular subtype of glioblastoma (see section on Transcriptomal subtypes). However, the oncogenic role of NF1 loss is complex and does not appear comparable to activation of Ras [3, 4, 13]. Overall, the available data suggest that NF1 is not an ubiquitously relevant Ras regulator.
Insensitivity to anti-growth signals—the RB axis
In addition to receiving pro-growth signals from their environment, cells also receive multiple anti-proliferative signals to prevent cell growth. These anti-growth signals, like their pro-mitotic counterparts, are sensed by the binding of transmembrane receptors to soluble factors, extracellular matrix components, or cell surface components.
Most of these anti-proliferative signals operate at the G1 phase of the cell cycle to trigger either 1) entry into a transient quiescent (G0) state or 2) entry into a post-mitotic, differentiated state. At the molecular level, nearly all of these signals converge at the retinoblastoma protein (RB) [1]. In quiescent cells, the RB protein is hyper-phosphorylated. This form of RB binds and sequesters the E2F family of transcription factors [22]. The genes transcribed by these transcription factors are essential for the G1–S transition of the cell cycle [23]. Phosphorylation of RB releases the sequestered E2F transcription factors and allows for cell growth. During normal cell cycle progression, induction of cyclin D1 and its associated cyclin-dependent kinases, CDK4 and CDK6, at the G1–S transition is responsible for the phosphorylation of RB. The kinase activity of the CDK4/6-cyclin D complex is under complex regulation, including the critical negative regulators CDKN2A (p16Ink4a), CDKN2B, and CDKN2C.
Pathway inactivating mutations in the RB pathway were described in glioblastomas prior to the large scale genomic efforts [24, 25]. Parsons et. al. [3, 4] and the TCGA validated these results and demonstrated that mutations and gene amplifications disrupting RB function are found in approximately 68–80% of glioblastomas, suggesting the critical importance of escaping anti-growth signals. Additionally, Genome-Wide Association Studies (GWAS) revealed that single nucleotide polymorphisms in the CDKN2A and CDKN2B have been identified as risk factors for glioma development [26, 27].
Evading apoptosis—the p53 axis
Apoptotic programs are inherent in all normal cells. These programs are activated by a number of physiologic signals during development and/or in response to cellular stress. Since the tumor state is associated with cellular stress capable of activating apoptosis (e.g., increased oxidative stress, increased DNA damage accumulation), inactivation of these programs constitute a critical step during carcinogenesis.
The regulation of apoptotic pathways is highly complex [28]. Broadly speaking, there are two pathways of apoptosis that converge on the activation of effector proteases (termed caspases), which ultimately trigger the pathognomonic DNA fragmentation, cell shrinkage, and membrane blebbing. The intrinsic cell death pathway (often termed the mitochondrial apoptotic pathway) involves the release of cytochrome c from the mitochondrial membrane space [29]. Binding of cytochrome c to a protein termed apoptosis protease-activating factor 1 (APAF-1), in turn, initiates the caspase cascade. In contrast, the extrinsic apoptotic pathway operates independently of mitochondria and is activated by direct signaling from cell surface receptors to the effector caspase [30].
Both intrinsic and extrinsic apoptotic programs are profoundly influenced by the p53 tumor suppressor protein [31]. TP53 encodes a transcription factor that regulates gene sets critical for cell cycle progression and apoptosis. Under normal conditions, p53 is a short-lived protein [32]. In response to cellular stress (for instance, DNA damage or oncogene expression), p53 undergoes post-translational modifications and protein–protein interactions that enhance its stability and transcriptional activity [32]. Key among the transcripts regulated by p53 are pro-apoptotic genes (including BAX and Puma) that facilitate both the intrinsic and extrinsic pathway [31]. Additionally, p53 interacts with a number of anti-apoptotic proteins to inhibit their function [31].
There are several lines of evidence that point to the importance of the p53 axis in glioblastoma pathogenesis. As with other critical pathways discussed, there was a body of literature implicating p53 pathway inactivation in glioblastomas [25, 33]. However, these studies implicate p53 pathway inactivation only in a subset of glioblastomas. The TCGA effort and the effort by Parsons et. al. [3, 4] added to the literature by demonstrating that the p53 axis is more broadly impaired in glioblastomas than previously thought. Mutations that inactivate this axis are found in greater than 70% of all glioblastoma specimens as reported by both studies. This understanding has led to more accurate murine modeling of glioblastoma by combined inactivation of p53 and PTEN [34].
Replicative potential
The definition of cancer as a continuous growing entity implies that normal cells exhibit a limited capacity for proliferation. Indeed, estimates based on tissue culture study suggest that most normal cells have the capacity for 50 doublings [35]. Studies over the past three decades suggest that the main reason for this limited life span involves progressive shortening of chromosomes due to loss of telomeres. Telomeres consist of thousands of six base pair sequence repeat elements that are located at the ends of every chromosome. Because of the inability of DNA polymerases to replicate the 3′ ends of chromosomal DNA, approximately 60 base pairs of the telomeric sequence is lost with each replicative cycle [36]. With progressive erosion of the telomeric sequence, the unprotected chromosomal ends participate in aberrant fusion events that inevitably result in cell death [37].
To overcome this inherent limitation, most cancer cells activate an enzyme called telomerase. Telomerase is a reverse transcriptase capable of elongating telomeres [38]. Various mechanisms are employed by tumors to activate telomerase in order to sustain continued cell growth. With regards to glioblastomas, single nucleotide polymorphisms in two genes encoding components of the telomerase (RTEL1 and TERT) have been identified as risk factors for glioma development in two independent GWAS studies [26, 27]. Additionally, elevated expression levels of TERT in glioblastoma are associated with decreased patient survival [39]. These studies suggest a critical importance of telomeric biology in glioblastoma growth and survival.
Angiogenesis
The intense proliferation of cancer cells require continued supply of oxygen and nutrients. Due to inherent limitations on the distance that oxygen and macromolecules can travel, virtually all cells in a tissue reside within 100 μm of a capillary. In xenograft model systems, solid tumors can only proliferate up to a size of 1–2 mm without the development of a new blood supply [40]. Thus, angiogenesis is a necessary pre-requisite of solid tumor growth.
One way that cancer cells signal angiogenesis is by secretion of soluble factors that bind to receptors present on the surface endothelial cells. A key soluble factor that functions in such capacity is the vascular endothelial growth factor (VEGF). VEGF binds to RTKs on the surface of endothelial cells to facilitate their proliferation—leading to angiogenesis [41]. Expectedly, clinical studies revealed that the expression level of VEGF correlated with both tumor aggressiveness and poor patient survival [42–44].
In normal cells, transcription of VEGF and other pro-angiogenic signaling factors are under strict regulation. The induction of Hypoxia inducible factor I (HIF1) is a pivotal element in this regulatory network [45]. HIF1 encodes a dimeric transcription factor consisting of two subunits: HIF1α and HIF1β. HIF1β is constitutively expressed irrespective of oxygen concentration, whereas HIF1α levels increase dramatically in response to hypoxia. The underlying mechanism for this regulation is that HIF1α is hydroxylated by HIF Prolyl-4-Hydroxylase (HPH) in the presence of di-oxygen (O2), iron, and α-ketoglutarate. The hydroxylated HIF1α is targeted for proteasome degradation. Without molecular oxygen, HIF1α is not hydroxylated and is free to dimerize with HIF1β to activate the transcription of downstream pro-angiogenetic factors.
Integrated analysis of genomic data in glioblastoma revealed recurrent mutations in the R132 residue of isocitrate dehydrogenase 1 (IDH1) [4], a gene largely responsible for the production of α-ketoglutarate. The TCGA data revealed that the IDH1 mutation is predominantly found in the proneural subtype of glioblastoma [13, 46] (see following section on Transcriptomal subtypes). The wildtype IDH1 normally functions as a homodimer that converts isocitrate to α-ketoglutarate [47]. Biochemical characterization of the R132 mutated IDH1 revealed that it functions to inhibit the process. Expectedly, glioblastoma harboring the R132 IDH1 mutation harbor decreased levels of α-ketoglutarate.
Given the importance of α-ketoglutarate in HIF1α degradation, some investigators have proposed that IDH1 mutations may predispose to increased HIF1α accumulation and increased VEGF secretion. These predictions were confirmed in a panel of primary glioblastoma specimens [48]. However, there are several issues in narrowly conceptualizing IDH1 mutations as a pro-angiogenic event. First, IDH1 mutations are frequently found in low grade gliomas where angiogenesis is essentially never observed [49]. Thus, while IDH1 mutations may contribute to angiogenesis, it is insufficient to initiate the process. Second, the R132 mutant IDH1 proteins exhibit a gain-of-function phenotype by generating R(-)-2-hydroxyglutarate, a carcinogenic metabolite [50]. In principle, this metabolite may impact a pleiotropy of cellular processes. Finally, any enzymes requiring α-ketoglutarate are, in principle, affected by the IDH1 mutations. Interestingly, studies in Acute Myeloid Leukemia (AML) revealed that IDH1 mutations are found in 15–30% of the afflicted patients [51, 52]. Mutational and epigenetic profiling of these patients revealed that IDH1 mutations closely associate with a specific hyper-methylation signature. Moreover, expression of IDH1 mutations induced global DNA hyper-methylation [53]. These results suggest that IDH1 mutations may lead to dysregulated epigenetic processes. Of note, many proteins required for epigenetic modulation, including the Jumonji-C domain histone demethylases, require α-ketoglutarate as a cofactor [54]. Ultimately, the mechanism of IDH1 pathogenesis remains unclear given current literature. A thorough discussion of this topic is beyond the scope of this review but can be found in the above referenced articles.
In glioblastomas without IDH1 mutation, alternate mechanisms are utilized to facilitate angiogenesis. It is somewhat intuitive that during normal development, periods of cellular proliferation must be coordinated with angiogenesis. Indeed, a large body of study suggests that gene functions that facilitate cell-autonomous growth or insensitivity to growth inhibition and apoptosis also tend to facilitate angiogenesis [55, 56]. It is likely that most glioblastoma cells attain angiogenesis by aberrant activation of such coordinated developmental programs. For instance, EGFR activation has been shown to up-regulate VEGF in both HIF dependent and independent manner [57]. Inactivation of Rb increases VEGF expression and angiogenesis in vivo [55]. Similarly, p53 normally up-regulates thrombospondin 1, an inhibitor of angiogenesis [58]; inactivation of p53 can facilitate angiogenesis by ablation of this up-regulation.
Invasion and metastasis
The ability to invade and metastasize constitutes the fundamental distinction between benign and malignant tumors. It is important to note that invasion refers not just to distortion of normal tissue secondary to tumor growth. Instead, it refers to a coordinated set of cellular activities to destroy and migrate into the surrounding normal tissue. Metastasis refers to the capacity to travel via circulation to a distant tissue site [40]. Glioblastoma is unique in that while it is one of the most invasive of cancers, it rarely metastasizes outside of the central nervous system.
It is a truism that cancer cells generally retain some general properties of the cell of origin. Since glioblastoma originates from astrocytes, which normally possess significant migratory capacity, the invasive nature of glioblastoma would be anticipated. During normal development, astrocytes migrate in a centripetal manner to establish a scaffold for neuro-blasts [59]. Additionally, in response to injury, astrocytes migrate to the affected region to form a gliotic scar [60]. This migratory capacity is the phenotypic expression of carefully orchestrated interactions between cellular cytoskeletal proteins, cell adhesion molecules, and extracellular matrix [40].
To date, the TCGA has not uncovered gain-of function mutations in these proteins. However, enhanced invasive properties have been associated with mutations establishing autonomous growth or suppressing apoptosis. For instance, aberrant EGFR activation results in increased expression and phosphorylation of cell adhesion molecules that ultimately lead to increased invasiveness [61]. Similarly, the p53 mutation drives cancer invasiveness by facilitating the recycling of integrin, a class of cell surface receptor that interacts with extracellular matrix during cell migration [62].
The aggregate of the data suggest that both angiogenesis and cell migratory properties are intimately integrated into a master circuitry controlled by critical proteins that dictate cellular response to growth or apoptotic signals. In this context, mutations facilitating self-autonomous growth or suppression of apoptosis also contribute to angiogenesis and cell invasion.
Cross-talk between canonical pathways
The conceptualization of distinct pathways contributing to the various critical phenotypes constitutes a simplification aimed to consolidate distinct biological concepts. The reality is that pathways mediating the cancer phenotype exhibit high degrees of cross-talk and functional redundancy. For instance, EGFR hyperactivation is associated with increased tumor growth (replicative potential), angiogenesis, and increased tumor motility [63]. Similarly, many genes mediating cell motility, telomere function, and angiogenesis are under transcriptional regulation by p53 and RB associated E2Fs [64].
Pathway of glioblastoma progression
It was previously thought that glioblastoma arises from the acquisition of a defined set of mutations that occur in a particular temporal order. This model is largely grounded on the framework established in colon cancer, where a series of genetic alterations characterizes different phases of neoplastic progression [65]. The framework is supported by the observation that Grade II astrocytomas typically harbor mutations in p53; Grade III astrocytomas harbor activating mutations/amplifications of CDKN2A (p16Ink4a); and Grade IV astrocytomas harbor mutations in PTEN and EGFR [66]. This data was interpreted to mean that glioblastoma results from sequential inactivation of the p53, RB, and RTK/PI3K axes.
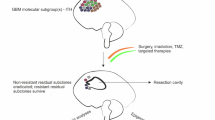
While such a paradigm may hold true for a subset of the secondary glioblastomas, the picture emerging from the genomic characterization of primary glioblastomas reveals a much more dynamic process [3, 4]. The profile of somatic mutations in different glioblastomas is highly variable. These results suggest that most glioblastomas, primary or secondary, evolve along a multitude of pathways in response to differing selective pressures to achieve the phenotypes described by Hanahan and Weinberg [67] (Fig. 2). This somewhat stochastic model of cancer progression further implies that mutations critical at one juncture in the neoplastic process may lose relevance as additional mutations are acquired. Thus, while a mutational profile constitutes an archeological profile of the history of the neoplasm, extrapolating therapeutic targets from such a profile may be challenging.
Pathway of glioblastoma pathogenesis. a Glioblastoma was previously thought to evolve along a linear progression of mutational accumulation. b The TCGA data suggests that most glioblastomas evolve along a multitude of pathways in response to differing selective pressures to achieve the cancer phenotypes
Transcriptomal subtypes
Though glioblastoma has been conceptualized as a single disease since Cushing and Bailey’s proposed classification scheme, it is widely appreciated that the term encapsulated significant histologic heterogeneity. This heterogeneity suggests distinct subtypes with differing physiologic states that are captured under the umbrella term “glioblastoma”. Following up on this hypothesis, there have been many efforts using distinct genomic profiling methods to tease out these subtypes. The number of subtypes varies depending on the study [9, 13, 68–70]. Interpretations of these results are difficult due to methodological differences in profiling platforms, bioinformatic extrapolation, and specimen collection. However, the aggregate of data suggests at least two distinct subtypes that reflect fundamental biologic behavior [9, 13, 70]. Importantly, these subtypes have been validated by independent studies. Further, the transcript signature parallels those obtained during distinct stages in neural development, suggesting the tumor may have arisen from different stages of neurogenesis [70].
The first subtype initially reported by Phillips et al. [70] and subsequently confirmed by the TCGA mRNA [13] and microRNA profiling [71]. The transcript signature resembles those of neuro-blasts and oligodendrocytes derived from fetal and adult brain [70]. The subtype harbors transcriptomal and clinical features that mirror those previously classified as secondary glioblastomas. Molecularly, proneural glioblastomas harbor mutations classically associated with the secondary glioblastomas [13]. Accordingly, Grade II and III gliomas harbor transcriptomal signatures most reminiscent of the proneural subtype [70]. Clinically, this subtype typically affects younger patients, is associated with improved overall survival [70], and responds poorly to concurrent radiation/temozolomide treatment upon disease progression [13].
Interestingly, IDH1mutations that are frequently found in secondary glioblastomas, are found up to 30% of primary proneural glioblastomas [13]. Importantly, methylation profiling, revealed that the nearly 80% of the IDH1 mutated glioblastoma, whether primary or secondary, harbor a particular global pattern of methylation, termed Glioma CpG Island Methylation Phenotype (G-CIMP) [72]. These studies suggest that the IDH1 mutated primary proneural glioblastomas may share cell states similar to those observed in secondary glioblastomas. The overall similarity in transcriptomal signature between the primary proneural glioblastomas and secondary glioblastoma appears independent of IDH1 status, suggesting molecular similarities between these two subclasses extend beyond IDH1 related physiology.
The second subtype that has emerged is characterized by a gene expression signature that mirror those observed in the neural stem cells of the forebrain [70], cultured astroglial cells [73], and tissue of mesenchymal origin [70]. The subtype is termed “mesenchymal” for the latter correlation. Like the proneural subtype, this second subtype was initially identified by Phillips et al. [70] and subsequently confirmed by the TCGA [13]. Both studies share the observation that patients afflicted with the mesenchymal subtype exhibit poorer clinical prognosis relative to the proneural subtype In support of these findings, the expression level of key genes in this signature, including VEGF and YKL40, were identified by several independent studies to associate poor prognosis in glioblastoma patients [42–44, 74]. Three other lines of evidence support the existence of the mesenchymal glioblastoma subtype. First, this subtype is highly enriched for mutations inactivating NF1, suggesting a common genetic etiology [13]. Second, the mesenchymal signature appear driven a common transcriptional network. Expression of two key critical factors in this network (STAT3 and CEBPβ) can establish the mesenchymal signature and enhance tumor aggressiveness in murine models [75]. Finally, microRNA profiling revealed that mesenchymal subtype glioblastoma share common signatures, though the data suggested that further subtypes exist within the mesenchymal class [71].
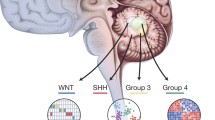
Other glioblastoma subtypes proposed by the TCGA and Phillips et al. [13] are as reviewed in Fig. 3. However, most (if not all) of these subtypes have not been as rigorously validated as the proneural and the mesenchymal. The emerging literature suggests that the proneural and mesenchymal subtypes define the two poles in the spectrum of molecular glioblastoma physiology [9, 13, 70]. It remains unclear whether the other proposed subtypes constitute a “forced fit” of a set of truly heterogeneous biology, a gradation of phenotypes between the two extreme poles, or a genuine subtype whose biologic basis remains to be understood.
There is significant debate with regards to the origin of the distinct transcriptomal subtypes. On one extreme is the thought that the subtypes originate from the same cell type with differences driven by distinct signaling pathways. The other extreme suggests that subtypes are determined by the same signaling pathways activated in a different cell of origin. The observation that the same canonical pathways are altered irrespective of subtype would tend to support the latter hypothesis. However, it is conceivable that different genes thought to participate in the same canonical pathway may modulate processes distinct of that pathway. Such functions may contribute to the distinct transcriptomal subtypes. Still, it is conceivable that differences in signaling and cell of origin both contribute to subtype formation. A hybrid stand posits that certain mutations arrest the developmental program at stages that are particularly to oncogenesis by specific molecular events. This critical debate awaits experimental resolution.
Limitation of genomic profiling and clinical impact
There are many critical aspects of cancer biology that fundamentally cannot be addressed by profiling an excised tumor mass. The critical interactions between glioblastoma and its microenvironment [76] and the cross-talk between subclonal heterogeneity inherent within every tumor [77] are but a few examples of the limitations. However, in broad strokes, the genomic profiling efforts have validated and consolidated the clinical pertinence of the fundamental biologic paradigms. Further, these efforts refined the umbrella term of glioblastoma into subtypes of distinct physiology. Unfortunately, clinical grade methodologies for routine transcriptomal subtyping are not yet available. Reports of the association between IDH1 mutations and favorable prognosis hold promise for biomarker development [4, 13, 46, 72]. These correlations await validation in prospective clinical trials. Similarly, the reported sensitivity of IDH1 mutation expressing glioblastomas to pharmacologic inhibitors of glutaminase [78] awaits further investigation. Future development of tools for subtyping, biomarker development, and therapeutic strategies grounded in the genomic landscape of the particular glioblastoma will facilitate clinical trial designs. Ultimately, meaningful therapeutic gain can be achieved only when agents are directed toward the Achille’s heel inherent within the distinct physiologies of different glioblastomas.
Summary
The past three decades of study in cancer research has generated a sophisticated conceptual framework for the process of neoplastic transformation. The framework suggests that genetic and epigenetic events inactivating critical pathways that regulate several key aspects of cellular function are an etiology. These cellular functions can be categorized as self-sufficiency in growth signaling, evasion of apoptosis, insensitivity to anti-growth signals, tissue invasion, and limitless replicative potential and angiogenesis. This framework has largely been validated by a large scale, high-throughput characterization of the genomic and epigenomic landscape in glioblastomas. The picture emerging from these analyses suggests that most glioblastomas evolve along a multitude of pathways in response to differing selective pressures to achieve the cancer phenotypes. Transcript based analysis revealed distinct subtypes with potential implications with regards to the cell of origin and developmental neuro-biology. The dynamic interplay of growth dysregulation and the cell of origin during the neoplastic transformation process harbors vital implications with regards to therapeutic development. Our understanding of this interplay will be advance by refined resolution of the genomic landscape (using second and third generation technologies) and interpreting these landscapes in the context of fundamental biologic tenets. As such, the escalating cost of genomic technology development/application must be balanced with funding for advancing fundamental biologic understanding. In the context of the economic limitations increasingly imposed on funding for scientific investigations, visionary leadership will be needed in resource allocation when balancing these critical needs.
References
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100:57–70
Zelenka PS (1990) Proto-oncogenes in cell differentiation. Bioessays 12:22–26
GA TC (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455:1061–1068
Parsons DW, Jones S, Zhang X, Lin JC-H, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu I-M, Gallia GL, Olivi A, Mclendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Hartigan J, Smith DR, Strausberg RL, Marie SKN, Shinjo SMO, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321:1807–1812
Wen PY, Kesari S (2008) Malignant gliomas in adults. N Engl J Med 359:492–507
Walker MD, Alexander E Jr, Hunt WE, MacCarty CS, Mahaley MS Jr, Mealey J Jr, Norrell HA, Owens G, Ransohoff J, Wilson CB, Gehan EA, Strike TA (1978) Evaluation of BCNU and/or radiotherapy in the treatment of anaplastic gliomas. A cooperative clinical trial. J Neurosurg 49:333–343
Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996
Stratton MR, Campbell PJ, Futreal PA (2009) The cancer genome. Nature 458:719–724
Huse JT, Phillips HS, Brennan CW (2011) Molecular subclassification of diffuse gliomas: seeing order in the chaos. Glia 59:1190–1199
Schlessinger J (2000) Cell signaling by receptor tyrosine kinases. Cell 103:211–225
Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB (2005) Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov 4:988–1004
Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C, Chin L, DePinho RA (2007) Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science 318:287–290
Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN, Network CGAR (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17:98–110
Cantley LC (2002) The phosphoinositide 3-kinase pathway. Science 296:1655–1657
Clarke ID, Dirks PB (2003) A human brain tumor-derived PDGFR-alpha deletion mutant is transforming. Oncogene 22:722–733
Soroceanu L, Kharbanda S, Chen R, Soriano RH, Aldape K, Misra A, Zha J, Forrest WF, Nigro JM, Modrusan Z, Feuerstein BG, Phillips HS (2007) Identification of IGF2 signaling through phosphoinositide-3-kinase regulatory subunit 3 as a growth-promoting axis in glioblastoma. Proc Natl Acad Sci USA 104:3466–3471
Sugawa N, Ekstrand AJ, James CD, Collins VP (1990) Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci USA 87:8602–8606
Downward J (2003) Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 3:11–22
Walker L, Thompson D, Easton D, Ponder B, Ponder M, Frayling I, Baralle D (2006) A prospective study of neurofibromatosis type 1 cancer incidence in the UK. Br J Cancer 95:233–238
McGillicuddy LT, Fromm JA, Hollstein PE, Kubek S, Beroukhim R, De Raedt T, Johnson BW, Williams SM, Nghiemphu P, Liau LM, Cloughesy TF, Mischel PS, Parret A, Seiler J, Moldenhauer G, Scheffzek K, Stemmer-Rachamimov AO, Sawyers CL, Brennan C, Messiaen L, Mellinghoff IK, Cichowski K (2009) Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell 16:44–54
Zhu Y, Harada T, Liu L, Lush ME, Guignard F, Harada C, Burns DK, Bajenaru ML, Gutmann DH, Parada LF (2005) Inactivation of NF1 in CNS causes increased glial progenitor proliferation and optic glioma formation. Development 132:5577–5588
Knudsen ES, Wang JY (2010) Targeting the RB-pathway in cancer therapy. Clin Cancer Res 16:1094–1099
Buchkovich K, Duffy LA, Harlow E (1989) The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell 58:1097–1105
Costello JF, Plass C, Arap W, Chapman VM, Held WA, Berger MS, Su Huang HJ, Cavenee WK (1997) Cyclin-dependent kinase 6 (CDK6) amplification in human gliomas identified using two-dimensional separation of genomic DNA. Cancer Res 57:1250–1254
Henson JW, Schnitker BL, Correa KM, von Deimling A, Fassbender F, Xu HJ, Benedict WF, Yandell DW, Louis DN (1994) The retinoblastoma gene is involved in malignant progression of astrocytomas. Ann Neurol 36:714–721
Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, Hoang-Xuan K, El Hallani S, Idbaih A, Zelenika D, Andersson U, Henriksson R, Bergenheim AT, Feychting M, Lonn S, Ahlbom A, Schramm J, Linnebank M, Hemminki K, Kumar R, Hepworth SJ, Price A, Armstrong G, Liu Y, Gu X, Yu R, Lau C, Schoemaker M, Muir K, Swerdlow A, Lathrop M, Bondy M, Houlston RS (2009) Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet 41:899–904
Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S, Giannini C, Halder C, Kollmeyer TM, Kosel ML, LaChance DH, McCoy L, O’Neill BP, Patoka J, Pico AR, Prados M, Quesenberry C, Rice T, Rynearson AL, Smirnov I, Tihan T, Wiemels J, Yang P, Wiencke JK (2009) Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet 41:905–908
Fan TJ, Han LH, Cong RS, Liang J (2005) Caspase family proteases and apoptosis. Acta Biochim Biophys Sin (Shanghai) 37:719–727
Lowe SW, Cepero E, Evan G (2004) Intrinsic tumour suppression. Nature 432:307–315
Ashkenazi A, Dixit VM (1999) Apoptosis control by death and decoy receptors. Curr Opin Cell Biol 11:255–260
Haupt S, Berger M, Goldberg Z, Haupt Y (2003) Apoptosis—the p53 network. J Cell Sci 116:4077–4085
Harris CC (1996) p53 tumor suppressor gene: from the basic research laboratory to the clinic—an abridged historical perspective. Carcinogenesis 17:1187–1198
Watanabe T, Yokoo H, Yokoo M, Yonekawa Y, Kleihues P, Ohgaki H (2001) Concurrent inactivation of RB1 and TP53 pathways in anaplastic oligodendrogliomas. J Neuropathol Exp Neurol 60:1181–1189
Holland EC (2001) Gliomagenesis: genetic alterations and mouse models. Nat Rev Genet 2:120–129
Hayflick L (1965) The limited in vitro lifetime of human diploid cell strains. Exp Cell Res 37:614–636
Zhang X, Mar V, Zhou W, Harrington L, Robinson MO (1999) Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev 13:2388–2399
Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA (2000) Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406:641–645
Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR (2007) Protein composition of catalytically active human telomerase from immortal cells. Science 315:1850–1853
Alonso MM, Fueyo J, Shay JW, Aldape KD, Jiang H, Lee OH, Johnson DG, Xu J, Kondo Y, Kanzawa T, Kyo S, Bekele BN, Zhou X, Nigro J, McDonald JM, Yung WK, Gomez-Manzano C (2005) Expression of transcription factor E2F1 and telomerase in glioblastomas: mechanistic linkage and prognostic significance. J Natl Cancer Inst 97:1589–1600
Leber MF, Efferth T (2009) Molecular principles of cancer invasion and metastasis (review). Int J Oncol 34:881–895
Veikkola T, Alitalo K (1999) VEGFs, receptors and angiogenesis. Semin Cancer Biol 9:211–220
Fuller GN, Hess KR, Rhee CH, Yung WK, Sawaya RA, Bruner JM, Zhang W (2002) Molecular classification of human diffuse gliomas by multidimensional scaling analysis of gene expression profiles parallels morphology-based classification, correlates with survival, and reveals clinically-relevant novel glioma subsets. Brain Pathol 12:108–116
Godard S, Getz G, Delorenzi M, Farmer P, Kobayashi H, Desbaillets I, Nozaki M, Diserens AC, Hamou MF, Dietrich PY, Regli L, Janzer RC, Bucher P, Stupp R, de Tribolet N, Domany E, Hegi ME (2003) Classification of human astrocytic gliomas on the basis of gene expression: a correlated group of genes with angiogenic activity emerges as a strong predictor of subtypes. Cancer Res 63:6613–6625
Sallinen SL, Sallinen PK, Haapasalo HK, Helin HJ, Helen PT, Schraml P, Kallioniemi OP, Kononen J (2000) Identification of differentially expressed genes in human gliomas by DNA microarray and tissue chip techniques. Cancer Res 60:6617–6622
Sharp FR, Bernaudin M (2004) HIF1 and oxygen sensing in the brain. Nat Rev Neurosci 5:437–448
Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773
Yan H, Bigner DD, Velculescu V, Parsons DW (2009) Mutant metabolic enzymes are at the origin of gliomas. Cancer Res 69:9157–9159
Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, Ding J, Lei Q, Guan KL, Xiong Y (2009) Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science 324:261–265
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114:97–109
Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462:739–744
Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D, Holland KB, Whitman SP, Becker H, Schwind S, Metzeler KH, Powell BL, Carter TH, Kolitz JE, Wetzler M, Carroll AJ, Baer MR, Caligiuri MA, Larson RA, Bloomfield CD (2010) IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J Clin Oncol 28:2348–2355
Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, Fulton LA, Locke DP, Magrini VJ, Abbott RM, Vickery TL, Reed JS, Robinson JS, Wylie T, Smith SM, Carmichael L, Eldred JM, Harris CC, Walker J, Peck JB, Du F, Dukes AF, Sanderson GE, Brummett AM, Clark E, McMichael JF, Meyer RJ, Schindler JK, Pohl CS, Wallis JW, Shi X, Lin L, Schmidt H, Tang Y, Haipek C, Wiechert ME, Ivy JV, Kalicki J, Elliott G, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson MA, Baty J, Heath S, Shannon WD, Nagarajan R, Link DC, Walter MJ, Graubert TA, DiPersio JF, Wilson RK, Ley TJ (2009) Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361:1058–1066
Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun Z, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Lowenberg B, Licht JD, Godley LA, Delwel R, Valk PJ, Thompson CB, Levine RL, Melnick A (2010) Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18:553–567
Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439:811–816
Gabellini C, Del Bufalo D, Zupi G (2006) Involvement of RB gene family in tumor angiogenesis. Oncogene 25:5326–5332
Hoeben A, Landuyt B, Highley MS, Wildiers H, Van Oosterom AT, De Bruijn EA (2004) Vascular endothelial growth factor and angiogenesis. Pharmacol Rev 56:549–580
Haley JD, Gullick WJ (eds) (2008) EGFR signaling networks in cancer therapy. Humana Press, New York
Rak J, Mitsuhashi Y, Sheehan C, Tamir A, Viloria-Petit A, Filmus J, Mansour SJ, Ahn NG, Kerbel RS (2000) Oncogenes and tumor angiogenesis: differential modes of vascular endothelial growth factor up-regulation in ras-transformed epithelial cells and fibroblasts. Cancer Res 60:490–498
Jacobsen CT, Miller RH (2003) Control of astrocyte migration in the developing cerebral cortex. Dev Neurosci 25:207–216
Ridet JL, Malhotra SK, Privat A, Gage FH (1997) Reactive astrocytes: cellular and molecular cues to biological function. Trends Neurosci 20:570–577
Micallef J, Taccone M, Mukherjee J, Croul S, Busby J, Moran MF, Guha A (2009) Epidermal growth factor receptor variant III-induced glioma invasion is mediated through myristoylated alanine-rich protein kinase C substrate overexpression. Cancer Res 69:7548–7556
Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, Lukashchuk N, Gillespie DA, Ludwig RL, Gosselin P, Cromer A, Brugge JS, Sansom OJ, Norman JC, Vousden KH (2009) Mutant p53 drives invasion by promoting integrin recycling. Cell 139:1327–1341
Nakamura JL (2007) The epidermal growth factor receptor in malignant gliomas: pathogenesis and therapeutic implications. Expert Opin Ther Targets 11:463–472
Sherr CJ, McCormick F (2002) The RB and p53 pathways in cancer. Cancer Cell 2:103–112
Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL (1988) Genetic alterations during colorectal-tumor development. N Engl J Med 319:525–532
Gladson CL, Prayson RA, Liu WM (2010) The pathobiology of glioma tumors. Annu Rev Pathol 5:33–50
Salk JJ, Fox EJ, Loeb LA (2010) Mutational heterogeneity in human cancers: origin and consequences. Annu Rev Pathol 5:51–75
Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, Lamborn KR, Berger MS, Botstein D, Brown PO, Israel MA (2005) Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci USA 102:5814–5819
Nutt CL, Mani DR, Betensky RA, Tamayo P, Cairncross JG, Ladd C, Pohl U, Hartmann C, McLaughlin ME, Batchelor TT, Black PM, von Deimling A, Pomeroy SL, Golub TR, Louis DN (2003) Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res 63:1602–1607
Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K (2006) Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9:157–173
Kim TM, Huang W, Park R, Park PJ, Johnson MD (2011) A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res 71:3387–3399
Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K (2010) Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17:510–522
Gunther HS, Schmidt NO, Phillips HS, Kemming D, Kharbanda S, Soriano R, Modrusan Z, Meissner H, Westphal M, Lamszus K (2008) Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene 27:2897–2909
Tanwar MK, Gilbert MR, Holland EC (2002) Gene expression microarray analysis reveals YKL-40 to be a potential serum marker for malignant character in human glioma. Cancer Res 62:4364–4368
Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, Lasorella A, Aldape K, Califano A, Iavarone A (2010) The transcriptional network for mesenchymal transformation of brain tumours. Nature 463:318–325
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674
Fox EJ, Salk JJ, Loeb LA (2009) Cancer genome sequencing—an interim analysis. Cancer Res 69:4948–4950
Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, Tsukamoto T, Rojas CJ, Slusher BS, Rabinowitz JD, Dang CV, Riggins GJ (2010) Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res 70:8981–8987
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ng, K., Kim, R., Kesari, S. et al. Genomic profiling of glioblastoma: convergence of fundamental biologic tenets and novel insights. J Neurooncol 107, 1–12 (2012). https://doi.org/10.1007/s11060-011-0714-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-011-0714-2