Abstract
Dermatophytes are a group of closely related fungi that nourish on keratinized materials for their survival. They infect stratum corneum, nails, and hair of human and animals, accounting the largest portion of fungi causing superficial mycoses. Huge populations are suffering from dermatophytoses, though the biology of these fungi is largely unknown yet. Reasons are partially attributed to the poor amenability of dermatophytes to genetic manipulation. However, advancements in this field over the last decade made it possible to conduct genetic studies to satisfying extents. These included genetic transformation methods, indispensable molecular tools, i.e., dominant selectable markers, inducible promoter, and marker recycling system, along with improving homologous recombination frequency and gene silencing. Furthermore, annotated genome sequences of several dermatophytic species have recently been available, ensuring an optimal recruitment of the molecular tools to expand our knowledge on these fungi. In conclusion, the establishment of basic molecular tools and the availability of genomic data will open a new era that might change our understanding on the biology and pathogenicity of this fungal group.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dermatophytes are a group of highly specialized keratinophilic fungi [1]. Species of this closely related group invade keratinized tissues like skin, hair, and nails of human and animals. Huge populations are infected with these organisms worldwide; nonetheless, key factors of mechanism they employ to cause infections are largely obscure. One approach to elaborate biology and pathogenicity-related elements of fungi is reverse genetics [2]. However, studies of such kind have drawn less attention, partially due to difficulties associated with genetic manipulation of these fungi.
Although it is on small scale yet, studies exploring the biology and potential pathogenetic factors of some dermatophytic species have being inflowing over the last decade [3–7], thanks to efforts aimed at developing indispensable tools for their genetic manipulation. Of those species that received particular attention were the anthropophilic dermatophyte Trichophyton rubrum [4] and the zoophilic dermatophytes Arthroderma vanbreuseghemii (teleomorph of Trichophyton mentagrophytes) [8] and Arthroderma benhamiae (teleomorph of T. mentagrophytes) [7], considering their medical significance and amenability to genetic manipulation. Recent advancements toward dermatophytes included transformation methods, dominant selectable markers, marker recycling system, down-regulation systems, and improving gene-targeting efficiency [8–13]. Along with these molecular tools, efforts have been intensified to expand the genome sequences library available for dermatophytes [14, 15], in addition to T. rubrum expression database [4, 5].
The recent advancements in dermatophytes, represented by establishing molecular tools and creating databases, ensure the optimal conditions to investigate the biological significance of these fungi. Herein, we present a comprehensive review of the up-to-date advancements pertaining in genetic manipulations for dermatophytes.
Transformation Method
Dermatophytes have recently witnessed major advancements with the release of genomic data of some of its species, along with the accumulation of information on their expressional profiling such as expressed sequence tag (EST) sequencing and cDNA-based microarrays [4–7, 14, 15]. These improvements make it much easier to conduct functional analyses of specific endogenous genes by genetic manipulation, undoubtedly boosting our understanding of the pathogenicity of dermatophytes and their host invading mechanisms. Gene transfer by transformation (genetic transformation) is the cornerstone for such functional genomics of fungal pathogens. It has provided powerful tools for their genetic manipulation, including heterologous gene expression, targeted gene inactivation and insertional mutagenesis. Following early reports of successful genetic transformation in model filamentous fungi such as Aspergillus nidulans and Neurospora crassa, polyethylene glycol (PEG)-mediated transformation of protoplasts (or spheroplasts), which are produced through cell wall degradation by lytic enzymes, has become the most common procedure for the introduction of exogenous genes into filamentous fungi [16, 17]. Transformation of protoplasts is carried out in an osmotically stabilized solution containing a high concentration of PEG and 10–50 mM of CaCl2 to activate the uptake of foreign DNAs to the fungal cells. Subsequently, protoplasts are regenerated on an osmotically stabilized selection medium containing appropriate antibiotics or lacking essential nutrients, depending on the selectable marker to be used. Since the first successful transformation of the dermatophyte T. mentagrophytes [18], random (ectopic) or homologous integration of exogenous genes by such protoplasts/PEG-mediated procedures has also been utilized for transformation of a variety of dermatophytic species [9, 19–23]. However, several reports have pointed out the low efficiency of protoplasts/PEG-mediated transformation in these species [18, 19, 23]. Only few comparative data on transformation efficiency are available, but interspecies variations are expected. The reasons for this are likely multifactorial, but it has been already assumed that the quality of protoplasts (e.g., their origins and stability) should be closely related to the transformation efficiency. Therefore, preparation of protoplasts requires special attention. Moreover, digestibility of fungal cell walls by certain lytic enzyme batches is sometimes variable. Thus, each individual step of protoplast preparation procedures, especially cell wall digesting reaction by lytic enzymes must be monitored, and the optimal reaction conditions have to be determined. In contrast to A. nidulans and N. crassa, standard procedures to produce high-quality protoplasts that are readily applicable to many species have not been well established for dermatophytes. Our group has practically experienced that transformation procedures, including protoplast preparation, established for a selected species are not necessarily applicable to other species and require species-specific modifications. Therefore, to some extent, the use of protoplasts for genetic transformation of dermatophytes is laborious and time-consuming.
To eliminate problems caused by the use of protoplasts, alternative procedures have been explored. Dobrowolska and Stączek [24] used electroporation to transform T. rubrum, allowing random integration and successful expression of exogenous genes, i.e., hygromycin B phosphotransferase gene (hph) and enhanced green fluorescent protein gene (eGFP). This rapid procedure had been previously described for a variety of filamentous fungi including Aspergillus species and N. crassa [25, 26]. In general, electroporation is used to transform protoplasts, but germinating conidia of filamentous fungi are also used. During electroporation, protoplasts or conidia are exposed briefly to a high amplitude electric field, which permeabilizes the membrane to allow the uptake of foreign DNAs. In case of germinating conidia, pretreatment of conidial preparations with cell wall digesting enzymes is more effective to increase the transformation efficiencies. However, electroporation requires special equipment to treat fungal cells electrically and is likely to cause a high frequency of multiple integrations of foreign DNA into the host genome. This may be disadvantageous for manipulations of specific endogenous genes such as targeted gene disruption and insertional mutagenesis. Restriction enzyme-mediated integration (REMI) was developed to overcome such disadvantage [27]. REMI involves transforming protoplasts or conidia with a linearized plasmid vector and the restriction enzyme used to linearize this vector. The restriction enzyme gains access to the host nucleus and stimulate creation of double-strand breaks in the genome. The restriction enzyme-produced DNA ends react with those of the linearized plasmid vector, leading to the integration of the vector into the genome at the restriction enzyme recognition site. REMI was also reported to improve the transformation efficiency in some filamentous fungi [28, 29]. Kaufman et al. [23] and Dobrowolska and Stączek [24] used a combination of REMI and protoplasts/PEG-mediated procedure or electroporation to transform T. mentagrophytes and T. rubrum, respectively, but the frequency of the single-copy integration of the foreign DNA and the transformation efficiency are both made unclear. Almost simultaneously, another approach was introduced into dermatophytes that is Agrobacterium tumefaciens-mediated transformation (ATMT) system. Agrobacterium tumefaciens is a gram-negative plant pathogenic bacterium, which has the ability to create crown gall tumors in plants. This bacterium can transfer the T-DNA (transfer DNA) region from the tumor-inducing plasmid (Ti plasmid) to the host plant nuclear genome. ATMT system has long been used for genetic manipulation of higher plants. Agrobacterium tumefaciens is also able to transfer the T-DNA to a very wide variety of fungi and fungal tissues under appropriate conditions [30–32]. The first successful transformation of dermatophytes by ATMT was in A. vanbreuseghemii [8]. For this species, the transformation efficiency via ATMT system was shown to be significantly higher, compared with the traditional protoplasts/PEG-mediated procedure. As in other fungal species, the majority of the transformants obtained by ATMT showed random integration of a single copy of the T-DNA into the host genome. Targeted gene disruption was also achieved by ATMT. Afterward, the ATMT system was applied to the model dermatophyte A. benhamiae [33], implying that ATMT system is an efficient and simple tool for transformation by random integration of T-DNA and for manipulation of specific exogenous genes, e.g., targeted gene disruption by homologous integration. However, as in the protoplasts/PEG-mediated procedure, we have already experienced that our routine ATMT procedure established for A. vanbreuseghemii was not fully functional in the transformation of Microsporum canis. Therefore, even in ATMT, there are several variables that should be optimized for individual species, and species-specific modifications are required.
Improving Gene-Targeting Efficiency
Reverse genetics is an approach that has been used widely to study the biology of prokaryotes and eukaryotes [2]. Similar to other filamentous fungi, though, its implementations in dermatophytes are restricted, partially due to the low efficiency of site-specific mutagenesis [34]. Despite decades since the first successful transformation system in dermatophytes [18], the number of cases where site-directed mutagenesis was conducted in this fungal group has not been ample. Among many other factors, the issue is ascribed to the tendency of dermatophytes to integrate exogenous DNA ectopically. During DNA-mediated transformations, cells recognize the introduced DNA constructs as DNA double-strand breaks (DSBs). In living cells, DNA damage provokes repairing process through either homologous recombination (HR) or nonhomologous end-joining (NHEJ) pathways [35]. While Saccharomyces cerevisiae preferentially uses HR to repair its DSBs (a process involving homologous sequences) [36], NHEJ-mediated repairing is dominant in human, insects, plants and most of fungi, including dermatophytes, leading to an ectopic integration of the exogenous DNA, no matter how much homology it shares with the host cell’s chromosomal DNAs. Hence, achievement of site-specific mutations to obtain desired genotypes would require screening a large proportion of generated putative transformants.
To improve the amenability of filamentous fungi to gene-targeting studies, two strategies might be hypothesized, enhancing the mechanism behind HR events or impairing NHEJ pathway. Considering the complexity involved in enhancing HR repair pathway, however, disrupting proteins recruited in NHEJ pathway has been routinely applied for many ticklish fungal species [37, 38]. Although conserved along eukaryotes, NHEJ pathway seems to show species-specific variations [35]. In general, NHEJ-mediated DNA repair process is initiated after the KU70–KU80 heterodimer recognizes and binds to broken DNA ends. Ultimately, the process is catalyzed by DNA ligase IV (Lig4), which forms a complex with Xrcc4 by joining at BRAC1 domains [39, 40].
Notably, a previous study reported that disruption of mus-51 (Ku70) and mus-52 (Ku80) genes in N. crassa resulted in 100 % gene-targeting frequency, compared with 10–30 % of their wild-type strain [41]. In dermatophytes, Yamada et al. [42] were the first to report enhanced gene replacement events in A. vanbreuseghemii deficient in its Ku80 (TmKu80). It was demonstrated that homologous recombination frequency during disruption of the areA/nit-2-like nitrogen regulatory gene (tnr) improved 33-fold in the ΔTmKu80 strain, compared with its wild type. The study also showed a high frequency of gene replacement events of 67 % in disruption of Tri m4 gene, encoding a putative serine protease [42]. Thereafter, more comprehensive data on improving gene-targeting frequency were reported by Alshahni et al. [11]. They demonstrated that disruption of TmLIG4 (Lig4) in A. vanbreuseghemii could enhance gene replacement frequency up to 93 %. Utilizing ΔTmLIG4 as a host strain, homologous recombination frequencies in four loci (tnr, TmSSU1, TmFKBP12 and TmKu80) were investigated in a comparative study involving the wild-type and ΔTmKu80 strains as well [11]. Homologous recombination occurred at frequencies of 53–93 %, 35–73 % and 0–40 % in ΔTmLIG4, ΔTmKu80, and their wild-type strain, respectively. In a separate study, targeted gene deletion frequency of ACUE (AbenACUE) locus in A. benhamiae was 98 % in ΔKu70, compared with 22 % in its wild-type strain, leading to a significant increase in HR frequency [22]. Table 1 compares the level of enhancements in HR frequency in dermatophytes.
Targeting NHEJ pathway has undoubtedly led to a significant advancement in genetic studies in dermatophytes both quantitatively and qualitatively [3, 10, 11]. Even with host strains deficient in their NHEJ pathway, though, improving HR frequency is a multifactor process. Site-specific integration also relies on such factors as transformation methods, selectable markers, loci, and length of homologous sequence stretches; the longer homologous sequences, the higher HR frequency. On the other hand, disruption of NHEJ pathway increases gene-targeting efficiency, but it also lowers transformation frequency. It was two fold less in ΔTmLIG4, compared with its wild-type strain [11], although it was still enough to obtain the desired genotypes.
Dermatophytic host strains deficient in their NHEJ pathway, i.e., ΔKu70, ΔTmKu80 and ΔTmLIG4, displayed no discernible phenotypic differences from their parent strains, in terms of growth ability, sporulation rate, and sensitivity to DNA damaging agents [11, 22, 42]. Moreover, ΔKu70 did not develop adverse effects on its virulence in guinea pig infection model [22]. Accordingly, improving site-directed integration in dermatophytes by disruption of NHEJ pathway is a powerful tool to conduct genetic studies; however, unanticipated genetic variations cannot be excluded too, especially with a report showing higher genomic instability in mouse cells deficient in its KU [43].
Marker and Reporter Genes
Selectable markers are indispensable tools for any genetic transformation, allowing detection and selection for desired genotypes and, thus, downstream analyses. The first successful genetic transformation in fungi has documented the use of inositol auxotrophy as a selectable marker in N. crassa [16]. Later studies reported the selection of genetic transformants based on resistance against antibiotics [44, 45]. Hence, selection of genetically modified cells can be obtained either through auxotrophic markers that require strains unable to synthesize organic compounds necessary for their survival, or dominant selectable markers conferring resistance against antibiotics or other toxins [16, 44, 45]. A very common example of auxotrophic marker is URA3 gene, which encodes an essential enzyme in pyrimidine biosynthesis in S. cerevisiae, orotidine-5′-phosphate decarboxylase [46]; however, these gene markers are restricted to auxotrophic host strains and spontaneous complementation might occur, limiting their applications. On the contrary, dominant selectable markers do not have prerequisites for host strains, as long as they are susceptible to the chemicals in question.
In the first successful report of genetic transformation in dermatophytes, T. mentagrophytes was genetically transformed using resistance against hygromycin B [18]. Transformation cassette included hph gene from Escherichia coli under the control of Cochliobolus heterostrophus promoter 1 (P ch ) [47, 48]. The hph gene codes for hygromycin B phosphotransferase, which inactivates hygromycin B through phosphorylation [49, 50]. Hygromycin B is an aminocyclitol antibiotic that inhibits protein synthesis in both prokaryotes and eukaryotes through interfering translocation and causing misreading [51, 52]. Moreover, Gonzalez et al. [18] demonstrated that transformants with hph cassette were mitotically stable, marking the first dominant selectable marker in dermatophytes. However, transformation frequency was low (0.004–6 per µg of vector and per 1 × 106 viable protoplasts), a result attributed to the poor functionality of P ch in T. mentagrophytes [18]. Thereafter, Yamada et al. [19] successfully recruited again the selectable marker gene hph in A. vanbreuseghemii and M. canis. Although yielded similar low transformation frequency, they proved that P ch was functional in A. vanbreuseghemii, a fact evidenced by its ability to drive the expression of enhanced green fluorescence protein (eGFP) gene. Selection of genetic transformations based on hygromycin B resistance was further demonstrated in other dermatophytes, e.g., T. rubrum [20, 21] and A. benhamiae [22].
Although the hygromycin B-mediated marker was proven to provide tight selection, genetic transformations in dermatophytes were limited to a single event, inasmuch as no other markers documented in these fungi. Therefore, exploring functions of members in gene families that work in high consistency, based on reverse genetics, is almost impossible with one selectable marker. A good example is genes coding for extracellular proteases, factors widely believed to participate in infections, which work in splendid orchestration to compensate deficiency in secretion of one another. Accordingly, the need for further selectable markers has emerged as an inevitable way to construct multiple mutations. Yamada et al. [19] reported the second dominant selectable marker applicable for dermatophytes in A. vanbreuseghemii. The maker based on E. coli neomycin phosphotransferase gene (nptII) [53], whose product inactivates geneticin (G418), an aminoglycoside antibiotic that specifically inhibits protein synthesis mainly in eukaryotes. The marker cassette constituted of nptII gene under the control of P ch promoter and the terminator sequence of A. nidulans tryptophan C gene (T trpC ) [54]. Moreover, this marker was proved to be dominant, given the ectopic integration of the nptII cassette into the chromosomes of A. vanbreuseghemii. The applicability of this marker in A. benhamiae was demonstrated too [22].
The establishment of the third dominant selectable marker came about in 2010 by Alshahni et al. [55] based on resistance against the aminoglycoside antibiotic nourseothricin. The resistance against the antibiotic is conferred by nourseothricin acetyltransferase gene (nat1) derived from Streptomyces noursei [56]. The marker cassette harbored nat1 gene under the control of the P act Cryptococcus neoformans actin promoter [57] and the T trp1 C. neoformans Trp1 terminator [58]. This selectable marker was also proven dominant.
Double sequential transformations based on resistance against hygromycin B and geneticin have been documented for several dermatophytes [11, 22]. The addition of nourseothricin selectable marker made it possible to conduct triple sequential mutations in A. vanbreuseghemii (unpublished data). Nevertheless, three selectable markers are still insufficient to study gene families like subtilisin where multiple mutations are required due to the compensatory nature of secretion process in dermatophytes. Hence, a marker recycling system was established in A. vanbreuseghemii, precluding the need to develop further selection tools (refer to the section of “Selectable Marker Recycling” below). Table 2 lists the selection cassettes based on their chronological order.
While selectable markers allow screening for genetically transformed cells, reporter genes enables studying function of genes of interest. Two reporter genes have been tested in dermatophytes GFP [19, 23] and lacZ [12]. Kaufman et al. [23] used sgfp gene to track development of human explants infection by T. mentagrophytes. The reporter gene was under the control of A. nidulans trpC promoter and terminator. Although functioning in the dermatophyte, level of fluorescence was variable among transformants [23]. Moreover, eGFP gene was successfully expressed in A. vanbreuseghemii under the control of P ch , a fact evidenced by illuminating the fungal hyphae under fluorescence microscopy [19]. However, no fluorescence was observed with transformation construct lacking any promoter. On the other hand, the E. coli lacZ reporter was utilized in A. vanbreuseghemii to test two regulatable copper-responsive promoters (refer to the section of “Down-Regulation of Target Genes” below) [12]. The product of this gene is β-galactosidase that hydrolyzes the colorimetric substrate o-nitrophenyl-d-galactopyranoside into galactose and o-nitrophenol, whose color is yellow. Under the inducible promoters, enzyme activity was observed; however, without a promoter, no such activities were detected, implying that A. vanbreuseghemii does not have intrinsic β-galactosidase activity [12].
Down-Regulation of Target genes
In spite of the developments observed in reverse genetics, generating knockout cells have not always been straightforward. It is often encountered by topology-related difficulties of targeted sequences or lethality issues pertaining essential functions of targeted loci; therefore, alternative approaches are needed. Originally described in the nematode Caenorhabditis elegans [59], RNA interference (RNAi) has been established as another choice to silence a given gene or gene family through post-transcriptional inhibition pathway. In filamentous fungi, a commonly used approach is gene silencing through expression of a long hairpin RNA [60]. It is based on the expression of inverted repeats (up to 1 kb long) under the control of proper promoter. In dermatophytes, the first report of gene down-regulation based on RNAi was by Vermout et al. [13] in M. canis. They utilized integrative plasmid constructs to down-regulate SUB3 and DPPIV genes, coding for endopeptidase three of the subtilisin family and dipeptidyl peptidase IV, respectively. Using this approach, it was possible to obtain desired transformants; however, variations were observed between phenotypic effects and mRNA depletion among individual RNA-silenced transformants [13], a phenomenon that is frequently encountered in other fungi [61]. Moreover, it bears noting that this sort of gene silencing can be further improved by using regulatable promoters, allowing suppression of gene expression in dosage-dependent manner [62, 63].
Another option to down-regulate genes is using inducible promoters, DNA sequences that allow switching gene expression on/off conditionally. The first report in dermatophytes came from Iwata et al. [12] who introduced conditional knockdown systems in A. vanbreuseghemii based on two copper-responsive promoters derived from T. rubrum. These were the promoter of copper transporter gene CTR4 (P CTR4 ) and that of copper efflux pump gene CRP1 (P CRP1 ), which can turn the expression of gene they control off or on, respectively, with addition of copper. It was demonstrated that gene regulation by P CTR4 is tighter than by P CRP1 in A. vanbreuseghemii, a fact evidenced by their variable abilities to regulate the lacZ reporter gene [12]. Notably, time course of transcriptional repression of lacZ gene under the control of P CTR4 showed a significant decrease in the mRNA level within only 30 min, whereas other nutrients-based knockdown systems require several hours to achieve targeted repression levels [64]. Furthermore, P CTR4 -based down-regulation system was employed to study two endogenous essential genes in A. vanbreuseghemii, ERG1 that encodes squalene epoxidase involved in the ergosterol biosynthesis pathway and TRP5 that encodes tryptophan synthase in the tryptophan biosynthesis pathway. The down-regulation of the two genes resulted in stunted growth ability [12], implying that this copper-responsive promoter is functional in A. vanbreuseghemii. Thereafter, P CTR4 was utilized to study functions of calcineurin A subunit-coding gene (TmcanA) in A. vanbreuseghemii, where null mutants of this gene were not possible to produce [3]; the repression of TmcanA was dose-dependent. Moreover, another useful exploitation of P CTR4 was in flippase (FLP) recombinase-mediated marker recycling system (refer to the section of “Selectable Marker Recycling” below).
Selectable Marker Recycling
Despite the poor amenability of dermatophytes to genetic manipulation, compared with many other fungi, improvements in basic molecular tools established for them have taken their research up to the next level [8, 11, 12, 22, 42]. However, further technical advancements in genetic manipulation tools and methodologies are required to expand our understanding on infections by these pathogens. In general, various molecules are involved in the infection of pathogenic microbes, a fact evidenced by differential gene expression profiles at various stages during the infection process. Therefore, it is difficult to study the functions and roles of given factors in developing infections by manipulating only one of their relevant genes, besides it is unlikely to reveal the whole mechanism. Similarly, dermatophytes selectively activate appropriate gene sets in response to various growth conditions during their infection. A representative example is the secretion of a variety of endoproteases, which are involved in dermatophytoses. Dermatophytes have unique compensatory protease secretion machinery, whose synergetic effects cause an infection. Apart from the secreted endoprotease gene families, genomes of dermatophytes are enriched in particular families of genes encoding fungal specific kinases (including pseudokinases) and proteins containing the LysM domain, which binds chitin and potentially related carbohydrates. Therefore, multiple genetic manipulations are required for functional analyses of members of such gene families in order to reveal the relationships among individual factors of complex networks. Considering that each single genetic transformation event requires unique selectable marker, numerous markers are required to accomplish this. However, only three dominant selectable markers are currently available for genetic manipulations of dermatophytes: hygromycin B, geneticin (G418) and nourseothricin (refer to the section of “Marker and Reporter Genes” above). Employing these three markers, it is possible to generate up to triple mutations in a single host strain by homologous recombination. Accordingly, studying large gene families, such as the secreted endoprotease families, subtilisins, deuterolysins and fungalysins, will be impossible.
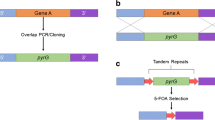
Selectable marker recycling system has emerged as an indispensable approach to generate multiple mutations in host strains that are limited in their selection tools. Just a few years ago, our group succeeded in recycling the geneticin-resistant gene (nptII) cassette mediated by the FLP recombinase-recombination target (FRT) site-specific recombination system in A. vanbreuseghemii [10]. This FLP/FRT recombination system, which is derived from S. cerevisiae [65], is the eukaryotic equivalent of the Cre/loxP-mediated site-specific recombination system that was originally described in bacteriophage P1 [66]. Naturally encoded within the S. cerevisiae 2 micron plasmid [67], FLP does not require cofactors and uses a phosphotyrosine intermediate for energy, like Cre recombinase. The FLP actively catalyzes specific recombination between two 34-bp FLP recognition targets (FRTs). A total of four FLP molecules and two FRT sequences are utilized for every FLP-mediated recombination event. The asymmetric region dictates whether excision (FRT sequences in the same orientation) or inversion (FRT sequences in inverted orientation) of the intervening DNA sequence occurs after recombination. Excision of the selectable markers mediated by the FLP/FRT recombination system has already been successfully applied in a variety of organisms including two filamentous fungi, Penicillium chrysogenum and Sordaria macrospora [68, 69]. Developing a functional and reliable selectable marker recycling system mediated by the FLP recombinase in dermatophytes was encountered by two major problems. The first was the lack of conditional promoters to timely control the FLP recombinase-mediated excision of the selectable marker. The marker cassette has to be excised after transformation for optimal target gene disruption. Trichophyton rubrum CTR4 promoter (P CTR4 ), a copper-repressible promoter [12], was fully functional as it controlled on/off switch of flp gene expression, and prevented spontaneous loss of the selectable marker cassette during the selection process of transformants. The other problem was the instability in production of the FLP recombinase. The previous study in P. chrysogenum strongly suggested that optimization of the codon usage for each amino acid in the flp gene to the pattern of the host species should be a prerequisite for functional FLP/FRT-mediated site-specific recombination in filamentous fungi. In practice, our group confirmed that the pathogenic yeast Candida albicans-adapted flp gene, which has almost identical nucleotide sequence with the native flp gene, failed to function in A. vanbreuseghemii. In contrast, a synthetic flp (Avflp) gene using codons optimized based on the codon usage bias of the closely related zoophilic dermatophyte A. benhamiae was fully functional in this dermatophyte. This has led to a successful excision of the selectable marker cassette via FLP-mediated site-specific recombination between the FRT sequences, producing mutant strains free of any selectable markers or lacking multiple endoprotease genes (see Fig. 1). However, little is currently known on the exact mechanism through which codon optimization improves the FLP-mediated site-specific recombination efficiency in filamentous fungi.
Recycling of the selectable marker (nptII) mediated by the FLP/FRT site-specific recombination system. a Schematic representation of the targeted gene disruption vector. The vector harbors the FLP/FRT module including the nptII and Avflp cassettes. The Avflp cassette consists of T. rubrum copper-repressible promoter (P CTR4 ), a synthetic codon-optimized flp (avflp) gene and Cryptococcus neoformans trp1 terminator (T trp1 ). b Schematic representation of the target locus before and after excision of the FLP/FRT module. Site-specific recombination between the flanking FRT sequences was carried out by conditional expression of Avflp gene after transformation for targeted gene disruption. FRT FLP recognition target
We have already confirmed that the developed FLP/FRT recombination system is applicable to A. benhamiae (unpublished data). In the recent years, A. benhamiae has been selected as a model dermatophyte at molecular level for several reasons including a relatively fast growth, availability of genome sequence and annotation, and existence of an animal model of infection. Thus, recycling of selectable markers via FLP-mediated site-specific recombination will provide new perspectives for in vitro and in vivo studies targeting complex networks of genes encoding redundant products such as secreted endoproteases in dermatophytes, which require multiple gene deletions for functional dissection. On the other hand, it is noteworthy that FLP/FRT recombination system mediated introduction of consecutive mutations into the same host may lead to undesirable secondary effects, such as inversions or deletions between these sites, due to the presence of multiple chromosomal FRT sequences. To reduce these risks, therefore, a combination of the FLP/FRT system and other available site-specific recombination systems, such as Cre/loxP and phiC31 integrase/att [70], may have to be considered.
Genomes and Transcriptomes
Attempts to establish molecular tools for dermatophytes have coincided with parallel efforts to establish collections of transcriptome and genome databases. The first reports came by Wang et al. [71] who lunched comprehensive T. rubrum Expression Database (TrED) covering topics such as pathogenic mechanisms, metabolism, and signal transduction [4, 5]. These significant contributions paved the way for constructing cDNA microarray from T. rubrum to study its transcriptional profile during growth on protein sources [6] and differential gene expression of A. benhamiae in vivo during infection of guinea pig and in vitro growth on keratin [7]. Burmester et al. [14] published the first genome sequences of the two dermatophytes A. benhamiae and Trichophyton verrucosum that were sequenced by Hans Knöll Institute. They also conducted a global transcriptome profiling analysis of A. benhamiae during infection of human keratinocytes. Very soon afterward, a comparative genomic study was conducted on five dermatophytes, T. rubrum, T. tonsurans, T. equinum, M. canis, and M. gypseum that were sequenced by Broad Institute [15]. Currently, 19 annotated dermatophyte genomes are available in the Dermatophyte Comparative Database by Broad Institute [72]. The availability of these data with the establishment of molecular tools will undoubtedly contribute for greater revelation of mysteries behind this unique fungal group.
References
Ajello L. Natural history of the dermatophytes and related fungi. Mycopathol Mycol Appl. 1974;53:93–110.
Bhadauria V, Banniza S, Wei Y, Peng YL. Reverse genetics for functional genomics of phytopathogenic fungi and oomycetes. Comp Funct Genomics. 2009. doi:10.1155/2009/380719.
Alshahni MM, Shimizu K, Yoshimoto M, et al. Genetic and phenotypic analyses of calcineurin A subunit in Arthroderma vanbreuseghemii. Med Mycol. 2016;54:207–18.
Wang L, Ma L, Leng W, et al. Analysis of part of the Trichophyton rubrum ESTs. Sci China Ser C. 2004;47:389–95.
Wang L, Ma L, Leng W, et al. Analysis of the dermatophyte Trichophyton rubrum expressed sequence tags. BMC Genomics. 2006;7:255.
Zaugg C, Monod M, Weber J, et al. Gene expression profiling in the human pathogenic dermatophyte Trichophyton rubrum during growth on proteins. Eukaryot Cell. 2009;8:241–50.
Staib P, Zaugg C, Mignon B, et al. Differential gene expression in the pathogenic dermatophyte Arthroderma benhamiae in vitro versus during infection. Microbiology. 2010;156:884–95.
Yamada T, Makimura K, Satoh K, Umeda Y, Ishihara Y, Abe S. Agrobacterium tumefaciens-mediated transformation of the dermatophyte, Trichophyton mentagrophytes: an efficient tool for gene transfer. Med Mycol. 2009;47:485–94.
Yamada T, Makimura K, Hisajima T, Ito M, Umeda Y, Abe S. Genetic transformation of the dermatophyte, Trichophyton mentagrophytes, based on the use of G418 resistance as a dominant selectable marker. J Dermatol Sci. 2008;49:53–61.
Yamada Y, Maeda M, Alshahni MM, Monod M, Staib P, Yamada T. Flippase (FLP) recombinase-mediated marker recycling in the dermatophyte Arthroderma vanbreuseghemii. Microbiology. 2014;160:2122–35.
Alshahni MM, Yamada T, Takatori K, Sawada T, Makimura K. Insights into a nonhomologous integration pathway in the dermatophyte Trichophyton mentagrophytes: efficient targeted gene disruption by use of mutants lacking ligase IV. Microbiol Immunol. 2011;55:34–43.
Iwata A, Alshahni MM, Nishiyama Y, Makimura K, Abe S, Yamada T. Development of a tightly regulatable copper-mediated gene switch system in dermatophytes. Appl Environ Microbiol. 2012;78:5204–11.
Vermout S, Tabart J, Baldo A, Monod M, Losson B, Mignon B. RNA silencing in the dermatophyte Microsporum canis. FEMS Microbiol Lett. 2007;275:38–45.
Burmester A, Shelest E, Glöckner G, et al. Comparative and functional genomics provide insights into the pathogenicity of dermatophytic fungi. Genome Biol. 2011;12:R7.
Martinez DA, Oliver BG, Gräser Y, et al. Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. mBio. 2012;3:e00259-12.
Mishra NC. DNA-mediated genetic changes in Neurospora crassa. J Gen Microbiol. 1979;113:255–9.
Tilburn J, Scazzocchio C, Taylor GG, Zabicky-Zissman JH, Lockington RA, Davies RW. Transformation by integration in Aspergillus nidulans. Gene. 1983;26:205–21.
Gonzalez R, Ferrer S, Buesa J, Ramon D. Transformation of the dermatophyte Trichophyton mentagrophytes to hygromycin B resistance. Infect Immun. 1989;57:2923–5.
Yamada T, Makimura K, Uchida K, Yamaguchi H. Reproducible genetic transformation system for two dermatophytes, Microsporum canis and Trichophyton mentagrophytes. Med Mycol. 2005;43:533–44.
Ferreira-Nozawa MS, Silveira HC, Ono CJ, Fachin AL, Rossi A, Martinez-Rossi NM. The pH signaling transcription factor PacC mediates the growth of Trichophyton rubrum on human nail in vitro. Med Mycol. 2006;44:641–5.
Fachin AL, Ferreira-Nozawa MS, Maccheroni W Jr, Martinez-Rossi NM. Role of the ABC transporter TruMDR2 in terbinafine, 4-nitroquinoline N-oxide and ethidium bromide susceptibility in Trichophyton rubrum. J Med Microbiol. 2006;55:1093–9.
Grumbt M, Defaweux V, Mignon B, et al. Targeted gene deletion and in vivo analysis of putative virulence gene function in the pathogenic dermatophyte Arthroderma benhamiae. Eukaryot Cell. 2011;10:842–53.
Kaufman G, Horwitz BA, Hadar R, Ullmann Y, Berdicevsky I. Green fluorescent protein (GFP) as a vital marker for pathogenic development of the dermatophyte Trichophyton mentagrophytes. Microbiology. 2004;150:2785–90.
Dobrowolska A, Stączek P. Development of transformation system for Trichophyton rubrum by electroporation of germinated conidia. Curr Genet. 2009;55:537–42.
Chakraborty BN, Patterson NA, Kapoor M. An electroporation-based system for high efficiency transformation of germinated conidia of filamentous fungi. Can J Microbiol. 1991;37:858–63.
Sánchez O, Aguirre J. Efficient transformation of Aspergillus nidulans by electroporation of germinated conidia. Fungal Genet Newsl. 1996;43:48–51.
Schiestl RH, Petes TD. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:7585–9.
Sánchez O, Navarro RE, Aguirre J. Increased transformation frequency and tagging of developmental genes in Aspergillus nidulans by restriction enzyme-mediated integration (REMI). Mol Gen Genet. 1998;258:89–94.
Thon MR, Nuckles EM, Vaillancourt LJ. Restriction enzyme-mediated integration used to produce pathogenicity mutants of Colletotrichum graminicola. Mol Plant Microbe Interact. 2000;13:1356–65.
de Groot MJ, Bundock P, Hooykaas PJ, Beijersbergen AG. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol. 1998;16:839–42.
Gouka RJ, Gerk C, Hooykaas PJ, et al. Transformation of Aspergillus awamori by Agrobacterium tumefaciens-mediated homologous recombination. Nat Biotechnol. 1999;17:598–601.
Sugui JA, Chang YC, Kwong-Chung KJ. Agrobacterium tumefaciens-mediated transformation of Aspergillus fumigatus: an efficient tool for insertional mutagenesis and targeted gene disruption. Appl Environ Microbiol. 2005;71:1798–802.
Zhang X, Wang Y, Chi W, Shi Y, Chen S, Lin D, Jin Y. Metalloprotease genes of Trichophyton mentagrophytes are important for pathogenicity. Med Mycol. 2014;52:36–45.
Fincham JRS. Transformation in fungi. Microbiol Rev. 1989;53:148–70.
Clancy S. DNA damage & repair: mechanisms for maintaining DNA integrity. Nat Educ. 2008;1:103.
Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–808.
Takahashi T, Masuda T, Koyama Y. Enhanced gene targeting frequency in ku70 and ku80 disruption mutants of Aspergillus sojae and Aspergillus oryzae. Mol Genet Genomics. 2006;275:460–70.
Choquer M, Robin G, Le Pêcheur P, Giraud C, Levis C, Viaud M. Ku70 or Ku80 deficiencies in the fungus Botrytis cinerea facilitate targeting of genes that are hard to knock out in a wild-type context. FEMS Microbiol Lett. 2008;289:225–32.
Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–14.
Critchlow SE, Jackson SP. DNA end-joining: from yeast to man. Trends Biochem Sci. 1998;23:394–8.
Ninomiya Y, Suzuki K, Ishii C, Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci USA. 2004;101:12248–53.
Yamada T, Makimura K, Hisajima T, Ishihara Y, Umeda Y, Abe S. Enhanced gene replacements in Ku80 disruption mutants of the dermatophyte Trichophyton mentagrophytes. FEMS Microbiol Lett. 2009;298:208–17.
Hande PM. Orchestration of telomeres and DNA repair factors in mammalian cells: implications for cancer and ageing. In: Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000. http://www.ncbi.nlm.nih.gov/books/NBK6524/.
Kaster KR, Burgett SG, Ingolia TD. Hygromycin B resistance as dominant selectable marker in yeast. Curr Genet. 1984;8:353–8.
Dickman MB. Whole cell transformation of the alfalfa pathogen Colletotrichunm trifolii. Curr Genet. 1988;14:241–6.
Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast; 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–6.
Gritz L, Davies J. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene. 1983;25:179–88.
Turgeon BG, Garber RC, Yoder OC. Development of a fungal transformation system based on selection of sequences with promoter activity. Mol Cell Biol. 1987;7:3297–305.
Leboul J, Davies J. Enzymatic modification of hygromycin B in Streptomyces hygroscopicus. J Antibiot. 1982;35:527–8.
Pettinger RC, Wolfe RN, Hoehn MM, Marks PN, Dailey WA, McGuire JM. Hygromycin I. Preliminary studies on the production and biological activity of a new antibiotic. Antibiot Chemother. 1953;3:1268–78.
Cabañas MJ, Vázquez D, Modolell J. Dual interference of hygromycin B with ribosomal translocation and with aminoacyl-tRNA recognition. Eur J Biochem. 1978;87:21–7.
Singh A, Ursic D, Davies J. Phenotypic suppression and misreading Saccharomyces cerevisiae. Nature. 1979;277:146–8.
Beck E, Ludwig G, Auerswald EA, Reiss B, Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–36.
Mullaney EJ, Hamer JE, Roberti KA, Yelton MM, Timberlake WE. Primary structure of the trpC gene from Aspergillus nidulans. Mol Gen Genet. 1985;199:37–45.
Alshahni MM, Makimura K, Yamada T, Takatori K, Sawada T. Nourseothricin acetyltransferase: a new dominant selectable marker for the dermatophyte Trichophyton mentagrophytes. Med Mycol. 2010;48:665–8.
Krügel H, Fiedler G, Smith C, Baumberg S. Sequence and transcriptional analysis of the nourseothricin acetyltransferase-encoding gene nat1 from Streptomyces noursei. Gene. 1993;127:128–31.
Cox GM, Rude TH, Dykstra CC, Perfect JR. The actin gene from Cryptococcus neoformans: structure and phylogenetic analysis. J Med Vet Mycol. 1995;33:261–6.
Perfect JR, Rude TH, Penning LM, Johnston SA. Cryptococcus neoformans TRP1 gene by complementation in Saccharomyces cerevisiae. Gene. 1992;122:213–7.
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE. Potent and specific genetic interference by double stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11.
Li L, Chang SS, Liu Y. RNA interference pathways in filamentous fungi. Cell Mol Life Sci. 2010;67:3849–63.
Kadotani N, Nakayashiki H, Tosa Y, Mayama S. RNA silencing in the phytopathogenic fungus Magnaporthe oryzae. Mol Plant Microbe Interact. 2003;16:769–76.
Barton LM, Prade RA. Inducible RNA interference of brlAbeta in Aspergillus nidulans. Eukaryot Cell. 2008;7:2004–7.
Goldoni M, Azzalin G, Macino G, Cogoni C. Efficient gene silencing by expression of double stranded RNA in Neurospora crassa. Fungal Genet Biol. 2004;41:1016–24.
Monteiro MC, De Lucas JR. Study of the essentiality of the Aspergillus fumigatus triA gene, encoding RNA triphosphatase, using the heterokaryon rescue technique and the conditional gene expression driven by the alcA and niiA promoters. Fungal Genet Biol. 2010;47:66–79.
Sadowski PD. The Flp recombinase of the 2-microns plasmid of Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1995;51:53–91.
Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J Mol Biol. 1981;150:467–86.
Broach JR, Guarascio VR, Jayaram M. Recombination within the yeast plasmid 2 μm circle is site-specific. Cell. 1982;29:227–34.
Kopke K, Hoff B, Kück U. Application of the Saccharomyces cerevisiae FLP/FRT recombination system in filamentous fungi for marker recycling and construction of knockout strains devoid of heterologous genes. Appl Environ Microbiol. 2010;76:4664–74.
Kopke K, Hoff B, Bloemendal S, Katschorowski A, Kamerewerd J, Kück U. Members of the Penicillium chrysogenum velvet complex play functionally opposing roles in the regulation of penicillin biosynthesis and conidiation. Eukaryot Cell. 2013;12:299–310.
Belteki G, Gertsenstein M, Ow DW, Nagy A. Site-specific cassette exchange and germline transmission with mouse ES cells expressing phiC31 integrase. Nat Biotechnol. 2003;21:321–4.
T. rubrum Expression Database. http://www.mgc.ac.cn/TrED. Retrieved 4 Feb 2016.
Dermatophyte Comparative Database. http://www.broadinstitute.org/annotation/genome/dermatophyte_comparative. Retrieved 4 Feb 2016.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Alshahni, M.M., Yamada, T. Genetic Manipulations in Dermatophytes. Mycopathologia 182, 33–43 (2017). https://doi.org/10.1007/s11046-016-0039-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-016-0039-y