Abstract
Segmentation of pathological lung regions from the body regions is the most challenging part in any Computer Aided Diagnosis (CAD) system due to the presence of Pathological Bearing Regions (PBR) which lies on the lung’s periphery. Here, a unique pathological lung segmentation method called reference-model based segmentation that uses shape property of human lung is proposed. This method trounces the difficulties in segmentation from traditional approaches by examining the shape knowledge of lung. The proposed segmentation approach constructs a reference lung model from input slices using a novel Sampling Lines Algorithm (SLA) and extracts the shape features. The segmentation work is validated using dataset consisting of Digital Imaging and Communications in Medicines (DICOM) standard chest CT images of seven patients from cancer institute Chennai, nine patients from Gemini Scans, Chennai and fourteen patients from Lung Image Data-base Consortium image collection (LIDC-IDRI). The segmentation method’s performance is analyzed against widely used segmentation methods namely Graph Cuts (GC), Region Growing (RG), Active Contour and Flood Fill in terms of accuracy, specificity, sensitivity, overlap score, Jaccard index, and also Dices similarity coefficients (DSC). The numerical outcomes specify that the proposed work attains an improved result against widely used segmentation techniques.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Pulmonary diseases are the primary reasons for hospitalization and deaths around the world [29]. The disorders in the human lung are owing to the diseases affecting various lung components such as airways, air sacs, interstitium, blood vessels, lung pleura, and chest walls. The human death rate due to lung disorders is elevated than other disorders in the human body [6]. Lately, the Electrocardiogram (ECG) is employed to disease analysis which was introduced in [25, 35]. Computed Tomography (CT) images are widely utilized to identify the abnormal patterns present in the human lungs [24, 26]. Segmentation algorithm inclines to highlight and segment the irregular patterns. These irregular patterns in the input slice exhibit structures like a signet ring, a tree in bud and ground glass patterns are commonly referred to as Region of Interest (ROI). The ROIs in the CT slice can well be on account of the existence of pathological regions affected because of various lung diseases, blockage in nerves, concussions, and internal fractures and possibly a tumor [15]. Segmenting the diseased lung tissues is complicated owing to the following reasons. First, the challenge in segmenting a diseased region from a CT slice is the timely examination of pathologies in the human lung [28]. Second, segmentation of lung tissues is complicated owing to the large size and anatomy of human lungs, the position of Pathological Bearing Regions (PBR) in addition to minor differences betwixt normal and diseased tissues [21]. Third, segmentation of pathological bearing regions becomes difficult due to its uneven size, jagged shape and possesses an unpredictable growth over an extent depending on the lung disease. Finally, Additional challenge in segmenting the lung regions manually is time-consuming and the results may vary depending on the initialization of the seed points in segmentation algorithm [3, 23].
Segmentation methods utilized in CAD, for instance, thresholding, shape, region and also edge-based methods lack accuracy as they segment the pathological bearing regions having high density and laying on the lung’s periphery [20]. In such scenarios, the information regarding the shape features of the lung can well be used [10]. Thresholding centered segmentation methods don’t consider the spatial traits of lungs. Thresholding based methods are impacted by abnormalities in lung images. The region- centred segmentation techniques are used in the segmentation of normal lung with less pathologic conditions, but their accuracy is low in cases where the location dense pathologies and normal tissues are extremely close. In all the region- centred segmentation methods like region-growing, graph-cut and watershed method, an assortment of a seed point is a chief concern as its accuracy depends on its manual initialization of the algorithm. Region-based methods also experience more false negative values within the lung region. In regions that are debarred from vascular and pleural effusion, the region-centred methods fail [1]. Edge-centred segmentation algorithms have expensive computation time and they perform inefficiently when seed point initialization is not close to the boundary. Further, it achieves less accuracy when lung tissues are divided using smooth boundaries [37]. The deep convolutionals neural networks is used for end-to-end training and saliency detection in [13].
A reference-based segmentation using sampling lines method is proposed to overcome the difficulties faced by traditional approaches. The proposed segmentation work includes validation on the healthiness of the segmented lung. The segmentation technique proposed here consists of pre-processing phase, Image Analysis phase, Reference Model (RM) phase, and Validation Phase. The segmentation method functions by reading healthy CT slices of the patients belonging to a specific age group as input. In the preprocessing subsystem, image noises, patient information, and other irrelevant background are removed. Image Analysis sub-system extracts the geometric features of the lung in the input slice and stores in a database. Shape model subsystem then generates edges of RM as of the input dataset using the sampling line algorithm. The sampling line algorithm studies the shape knowledge about similar dataset by extracting the edge coordinates of the lungs in the input CT image. Finally, the validation subsystem combines the shape information and generated feature vector (FV) of the reference lung models to the input slice and validates both the shape property and healthiness of the segmented input slice using a distance metric.
This paper is written as follows, Section 2 contains the literature survey, Section 3 contains the formation of the proposed work, Section 4 consist results as well as discussion, and section 5 consists of the conclusion and section 6 contains references.
2 Related researches: Review
A segmentation model for detecting the lung shape from pulmonary CT image dataset is presented by Hu et al. [17]. The presented segmentation model comprised three stages explicitly, optimal thresholding, identifying and separation of the left as well as right lungs followed by a succession of morphological operations. The lung tissues were extracted as of the input slices by carrying out optimal thresholding method. Dynamic programming technique was adapted to find the anterior in addition to posterior junction lines. Authors used morphological erosion to separate both lungs and then dilatation was implemented to approximate the lung shape devoid of reconnecting the 2 lungs. To lessen the computational time, authors applied their lung separation technique only to the slices where the left along with right lung needed to be separated for processing. The authors tested their work on datasets consisting of eight normal patients scanned at regular intervals with 90% fundamental capacity. The dataset was acquired as of Imatron Corporation, California. Authors inferred that the segmented boundaries of the obtainable model to the borders traced by manual experts are similar. Authors included an optional boundary smoothing step to reduce the deviation between boundaries extracted manually and by the presented method. It was observed from the dataset that, authors tested their automated model with lungs vital capacity. This segmentation model formed the base for numerous pieces of literature in the segmentation field.
A shape-based segmentation algorithm is presented by Dawoud et al. [8] to extract lung shape by adapting iterative thresholding. The presented segmentation model involved 3 phases. In the initial phase, the author developed a statistical model to extort the lung shape from healthy input images. Preliminary extraction of the lung shape from the healthy images was done by radiologists. In the second phase, the healthy lung shape obtained from the first phase was used as a reference image to segment the lung shape from the test CT images [31]. The geometric deviation between the segmented input lung and the reference lung was computed using Mahalanobis distance metric. In the event of relatively small deviation, the lung shape was adjusted utilizing Active Shape Modeling (ASM) technique to enhance the accuracy. The dataset was obtained from the Japanese Society of Radiological Technology (JSRT) database was used to evaluate the presented segmentation model. JSRT data-base consists of 247 chest radiographs acquired as of 13 institutions from Japan and also one institution from the USA. The JSRT data-base also included manual segmentation results carried out by experienced radiologists. Overlap percentage and contour distance were used as performance measures and the author claimed that the presented model was effective compared to widely used shape models, for instance, Actives Appearance Model (AAM) as well as Actives Shape Model (ASM). The presented system achieved better overlap percentage of 0.94 compared to 0.90 of ASM and 0.85 of AAM methods. The primary concern in the presented work regarded the initial extraction of lung shape done by the radiologist. The extraction of lung shape slightly vary based on the experience and knowledge of the radiologist and mightn’t be accurate at all times.
Adaptive Borders Marching (ABM) segmentation is presented by Pu et al. [27] consists of a 3-step pre-processing subsystem which used Gaussian technique to eradicate the noise. After noise removal, optimal thresholding was carried out to identify lung regions in addition to flood filling technique to separate the non-lung regions from lung regions. After pre-processing, lung borders were traced by border tracing algorithm which identified the border region as a sequence of pixels on a particular direction. To include the juxtapleurals nodules on the lung borders, the authors presented a new technique called ABM Algorithm. Dataset used to evaluate the presented technique consists of 20 subjects that included 16 male and 4 female subjects. The dataset was acquired from Siemens Medical Systems, Germany. Authors compared their presented method against widely used rolling ball operator. Centred on the experimental results on twenty datasets, the presented method was able to include all the 67 juxtapleurals nodules identified the radiologists were included and added with an elevated rate of per nodule sensitivity. Nevertheless, in a few cases, the presented ABM technique included minor non-lung tissue instead of juxtapleural nodules in the segmented lungs parenchyma.
A completely automated method for parenchyma segmentation and parenchyma reconstruction by including juxtapleural nodules was presented by Wei et al. [34] involves three phases. In the 1st stage, optimal iterative thresholding was done for binarization to identify lung shape. In the 2nd stage, if both lungs were connected, the authors discussed the separation of left along with the right lungs using centre location method. Centre location method calculated the centre of the lungs parenchyma region of left and also right lungs to carry out the separation of the lungs. In the last phase, the extracted lung parenchyma was repaired to comprise the juxtapleurals nodules. Juxtapleural inclusion was done by widely used chain codes algorithm and also the Bresenham lines algorithm. Chain codes algorithm focused on joining the linked components in an image. Bresenham line algorithm determined a set of straight points that ought to be chosen to form a straight line to join two points [2]. The dataset used for evaluating the presented method was gotten from Sheng Jing Hospital, China. Based on experimental results, this method attained as lower computational expenditure, good overlap score of 95.24%, better inclusion accuracy of 98.6% and needed no user interaction. However, authors evaluated their work to other recent parenchyma extraction methods apart from rolling ball operator.
A completely automated segmentation technique to extract the lung shape as of chest CT images utilizing Insight Tool kit (ITK) was presented by Heuberger et al. [16]. ITK stands as a widely utilized open source tool used for registration and segmentation of medical images. The segmentation technique had five steps. First, optimal thresholding was carried out for the binarization of the input image. In the 2nd step, non-lung tissues were identified and removed. In the third step, Gaussian smoothing was done to eliminate the noises [22]. Airway removal was done by eliminating the regions where the pixel intensity falls behind the certain threshold limit. The non-lung regions with a threshold level lesser than 20 pixels were eradicated. In the fourth step, widely used rolling ball operator was implemented to lung edges for parenchyma extraction [9]. In the end step, lung separation was done using the centroid method if both lungs were linked. The dataset used by the offered method consists of 153 lung CT images from the CasIMAGE lung database. The results showed that the segmentation approach works well when the Pathological Bearing Regions (PBRs) lied internally in the lung [5]. The algorithm failed to segment the lung region if the PBR was peripherally placed and the radius of the rolling ball was less than peripheral PBR. However, for some input slices, the segmentation algorithm using ITK eliminated lung lobes together with the background elimination. Authors stated that the offered segmentation technique was simple and effective and had added performance scrutiny of the presented algorithm against widely used segmentation algorithms.
A Cybernetics Microbial Detection System (CMDS) was suggested by Dinesh Jackson Samuel R and Rajesh Kanna B [11]. The CMDS was further split into 2 sub-systems, namely data acquisition (DA) system in addition to Microbial recognition system. In the DA system, scanning movements for smear examination are automated by a programmable micro-scopic stage. Different sorts of scanning movements for specimen assessment were done using the programmable microscopic stage. Subsequent to the acquisition, the image or video was given to software for pre-processing and image recognition. In the pre-processing phase, the acquired image was re-scaled to fit into the image recognition system. In image recognition, the deep neural network layers and classifiers were added. For classification, the kernel-based Support Vector Machine (SVC) was used. Experiments with tuberculosis (TB) dataset obtain better results in case detection using VGG19 + SVM (86.6% of accuracy) compared with either Visual Geometry Group (VGG16) or VGG19.
The microscopic TB detection system assists human in diagnosing the disease rapidly was presented by Dinesh Jackson Samuel R and Rajesh Kanna B [12]. This methodology consists of three chief stages, explicitly DA system, recognition system, and deep transfer learning. The DA system captured or recorded all field of views while scanning the specimen. This process was automated by a programmable microscopic stage which scanned the specimen in defined scanning patterns. Subsequent to the acquisition, data were given to the recognition system for categorization of infected along with the non-infected field of views. This system used deep learning nets to classify infected as well as the non-infected field of view images. This system used transfer learning along with fine-tuning in DeepNet models. A complete search of images with a large blend of parameters was performed to discover the best parameter settings. This process was called cross-validation. This can well be applied only once to the dataset beforehand to discover the source domain. On the DeepNet model’s development, reduction of layers was considered for reduced computational complexity during screening.
A lung segmentation technique centred on an enhanced GC algorithm as of the energy function was proposed by Shuangfeng et al. [7]. Initially, the lung CT images were modelled with Gaussians mixture models (GMMs); in addition, the optimized distribution parameters can well be attained with expectation maximization (EM). Secondly, regarding the image edge information, the Sobels operator was taken to detect as well as extort the lung image edges, in addition, the lung image edges information was employed to ameliorate the boundary penalty item of the GC energy function. At last, the enhanced energy function of the GC algorithm is attained, then the corresponding graph was generated, and lung was segmented with the smallest cut theory.
It can be inferred that diseases causing pathologies can lie on lung parenchyma and periphery of the lung. Thresholding centred methods, RG methods, Actives Contour Methods (ACM), Rolling Ball operators are widely used for segmenting the images. In recognizing the lung shape, iterative thresholding is widely used. Edge detecting operators are applied to extort the lung parenchyma. The prime concern with thresholding methods are thresholding can only be used if the pathological bearing region lies on lung parenchyma and cannot be applied when the PBR’s lie in the periphery of the lung. Active contour methods usually contain self-adjustable splines that detect the lung shapes and extract the lung parenchyma. ACM methods don’t work on all datasets automatically because they need user level interaction for initializing the snake and captures only shape information and not information about pathologies. Rolling Ball operator widely used operator for extracting the lung shape. Rolling ball operator tends to include the PBRs on the parenchyma and juxtapleural nodules whilst extracting the lung shape. However, the concern with this method is a selection of ball radius, the selection of Rolling ball operator radius must be done with precise often results in over segmentation or under segmentation.
3 System framework
The system framework of the proposed segmentation method is shown in Fig. 1. The proposed framework encompasses 4 phases explicitly preprocessing phase, image analysis phase [4, 30]. RM generation phase, and validation phase. A healthy lung slice identified by the radiologist is inputted to the pre-processing phase. Pre-processing subsystem is done to make the slice apt for other processing. The preprocessing phase consists of noise removal, optimal thresholding, and background removal, and airway removal, identification of lung components and separation of lung components. After implementing the image preprocessing, the slice is inputted to RM generation phase and also image analysis phase. Noise removal is executed to remove various noises like random noise, Salt and pepper, Gaussian noise, and other noises produced due to the scanning mechanisms by different CT scanners. In the proposed method, the Weiner filter mainly eliminates the additive noise, in addition, reverses the blurring concurrently, and the advantage of this filter is that it solves the random noise problem.
Widely adapted optimal thresholding is done for image binarization to recognize the lung components. Background exclusion removes the non-body pixels surrounding the lung. The non-body pixels in the thresholded image consist of air adjoining the body, the lung tissues, and Patient artifacts. The pixels (non-body) that are connected outside to the lung component are considered as background pixels and removed by setting it to 0. Edges detection is executed to identify the lung components. Edges on an image are a collection of points upon which pixel intensity changes suddenly. Discrete Laplacian operator is utilized to extort the edges of an image. The Laplacian technique searches for the zero crossings on the 2nd-derivative of the image of find edges. Finally, in the preprocessing phase, separation of lung components is done only if both the lung components left lung and also the right lung appear to be connected.
The Edge detected slice is given as input to the image analysis subsystem. The image analysis subsystem carries out the extraction of geometrical features of the human lung and preparation of FV. RM phase carries out sampling line construction algorithm and extraction of seed point coordinates. Seed point coordinates give the location where the sampling line intersects the lung border. RM phase is accountable for the generation of reference lung model. In the validation phase, a test slice is given as input. Pre-processing phase, Image Analysis phase, and Shape extraction phase are done on the input slice. The input slice is segmented, and the FV generated. This FV of the input slice compared with the FV of the RM to ascertain the healthiness of the segmented input slice.
3.1 Preprocessing phase
3.1.1 Noise removal
-
Input: Healthy chest CT slices.
-
Step1: Eliminate the noises existing in the input images using the Wiener filter stated below.
Where H(x, y) is degradation function, K is the constant used to approximate the noise, R(x, y) represents the image after applying wiener function and G(x, y) is degraded input image.
Output: Denoised CT slice.
3.1.2 Optimal thresholding
-
Input: Denoised CT Slice.
-
Step 1: A global threshold level T for the inputted image is determined by Otsu’s Method [36].
-
Step 2: The image divided by T is categorized into 2 groups, i.e., G1 and G2. G1 consists of the complete pixel values that are lesser than the thresholdT, in addition, G2 consists of the complete pixel values which are bigger than T. The threshold level is ascertained by the Otsu’s threshold method.
-
Step 3: The average intensity values α1, α2are computed for G1 and also G2.
-
Step 4: The optimum threshold is obtained using
Where, Ti + 1 Is the threshold (i + 1)th iteration.
Step 5: go over steps 1–4 until there is no change in ThresholdT = Ti + 1for the image.
Step 6: whenT = Ti + 1, the image is split into 2 classes with pixel intensities greater than Tand lesser thanT. Each pixel with pixels intensities greater than T is fixed to 0 to represent body pixels, and other pixels are fixed to 1to represents non-body pixels.
Output: Thresholded slice.
3.1.3 Background removal
-
Input: Thresholded Slice.
-
Step1: Generate a binary mask.
-
Step2: Perform AND operation over Thresholded image with a binary mask as given below.
Where, Dt(x, y) is the Thresholded image, BM(x, y) is the Binary mask and O(x, y) is Intermediate output image.
Step3: Complement the image as
Where, O′(x, y)denotes the complement of the intermediate output image.
Step 4: Perform AND operation over the complemented image with binary mask defined,
Where, BE = Background eliminated image.
Output: Background eliminated slice.
3.1.4 Airway removal
-
Input: Background eliminated slice.
-
Step 1: Set the pixels which are not connected to lung component as 0 to remove the airway from the lung.
-
Output:Thresholded slice with airways removed.
3.1.5 Identification of lung components
-
Input: Thresholded slice with airways removed.
-
Step 1: Compute the Laplacian L(x, y) operator for the image
Where, I is the input image.
Step 2: The discrete approximations to the Laplacian operator [33] is employed to extract the edges of the lung region is given below.
-
Output: Edge detected slice.
3.1.6 Lung separation
-
Input: Edge detected slice.
-
Step 1: The left, as well as, right lungs are separated utilizing the morphological process discussed in [19]. The morphological process is implemented for separating the right as well as left lungs. Let SP signify the set of lung pixels in a single slice. For separating the left, as well as right lungs here, compute a new set Sα using an n-fold operation
Where, ⊕ denotes a binary morphological operation and (BS)4 denotes a four-connected binary structuring element.
Steps from 3.1.1 to 3.1.6 are repeated for all input slices on the image directory.
Output:
- (i)
Edge detected left lung.
- (ii)
Edge detected right lung.
3.2 Image analysis phase
3.2.1 Extraction of geometric features and database storage
The shape features represent the geometric structure of the edge detected lung. Features are extorted for each input slice, stored in a data-base.
- a)
Area: this is basically a value which gives the number of pixels in lung region. Area function returns a scalar value. Area feature represents the shape property of the ROI. The lung region’s area is stated as,
-
b)
Bounding Box: Bounding box is the area defined within two latitudes and two longitudes. This is constructed around the left and also right lungs to recognize the maximum extent coordinates of major along with minor axis respectively. The Bounding box attribute is represented as,
-
c)
Convex Area: Convex area gives the number of pixels within the polygon. The convex area is computed as,
Where (x1, y1), (x2, y\2) and (xn, +yn) are the coordinates of the convex polygon.
- d)
Eccentricity: Eccentricity which encompasses the same 2nd-moments like the region, returned in the form of a scalar. This stands as the ratio of the distance betwixt the foci of the ellipse and its major axis length and is the measure of the deviation of PBR from the ellipse Eccentricity value usually ranges as of ‘0’ to ‘1’. Eccentricity is formulated as,
-
e)
Equiv-diameter: this is the diameter of a circle that is similar to the area of interest. This feature gives the circle diameter that covers the lung regions. It returns a scalar value in pixels. Equiv-Diameter is computed using,
-
f)
Extent: Extent gives the ratio of white pixels on the region to the white pixels on the bounding box. The values are returned in the form of a scalar. Extent is defined as,
-
g)
Filled Area: Filled area computes the number of white pixels on a filled image within the bounding box. It returns a scalar value. The filled area is represented as,
-
h)
Major Axis: Major axis length gives the number of white pixels present in the major axis. This is the segment of the line that links the vertical extreme coordinates of the left along with the right lung. The major axis is returned in the form of scalar value. Major Axis is represented as,
-
i)
Minor Axis: Minor axis length gives the number of white pixels present in the minor axis. This is the segment of the line that links the horizontal extreme coordinates of the left and also right lung. The minor axis is returned in the form of scalar value. It is stated as.
-
j)
Orientation: Orientation stands as the angle betwixt the x-axis and the major axis of the ellipse which encompass the same 2nd-moments as the region, returned in the form of a scalar. The value is in the form of degrees, ranging as of ‘-90’ to ‘90’ degrees. Orientation gives the angular deviation betwixt the x-axis and the major axis and orientation is returned in the form of a scalar value.
-
k)
Perimeter: Distance about the region’s boundary. It is returned in the form of a scalar. Perimeter is computed utilizing the distance betwixt every couple of coordinates in the lungs border. The perimeter can well be computed using,
-
l)
Solidity: Solidity is estimated as the ratio between area and the convex area. Solidity measures the concavity and also convexity of a particular shape. Compute solidity using,
Feature extraction steps
Input: Edge detected image
Step 1: The shape features are extracted stored in a database relation named data and data1.
Step 2: Repeat Step 1 for all the slices in the input directory.
The relations of data and data1 are listed as,
Data (Slice id, Area, Bbox, C_Area, Ecc, ED, Extent, fill_area, Mj_axis, Mn_axis, Orientation, Perimeter, Solidity).
Data1 (Slice id, Area, Bbox, C_Area, Ecc, ED, Extent, fill_area, Mj_axis, Mn_axis, Orientation, Perimeter, Solidity)
Where, data and data1 refer to the shape features of the left along with right lung respectively.
Output: Extracted shape features of the left lung (data) and also right lung (data1).
3.2.2 Feature vector preparation
-
Input: Data and Data1
-
Step 1: Compute the average of each feature for the left lung.
-
Step 2: Store the values attained as of step 1 as an feature vector as
-
Step 3: Repeat Step 1 along with Step 2 for right lung.
-
Output: FVs for data and data1.
3.3 Shape model subsystem
3.3.1 Sampling line construction algorithm
-
Input: Edge detected left lung along with right lung from section 3.1.6.
-
Step 1: Compute the Major axis length.
-
Step 2: Set the number of sampling lines to be constructed on the major axis.
-
Step 3: Ascertain the distance intervals between each sampling lines to be constructed.
-
Step 4: For the number of sampling lines,
- a)
For each row, scan the image from left to right, identify the left most sampling pointci(x, y). ci(x, y) gives the coordinate location of the directional scan meets the left side lung edge.
- b)
On finding ci(x, y), on the same row continue the scan identify the rightmost sampling point cj(x, y).
- c)
If ci(x, y) = cj(x, y),
- i.
Store the sampling points in a 2-D Array.
Else,
- d)
If ci(x, y) ≠ cj(x, y),
- i.
Store the sampling points in a 2-D Array.
- ii.
Join ci(x, y) and cj(x, y) to constructs a sampling line.
- a)
-
Step 5: Repeat the steps 1 to 4 for all the edge detected images and store the coordinate values in a 2-Dimensional array. The array is defined as Data_sp [Slice_id, line_id, ci(x, y), cj(x, y)]for left lung and Data1_sp [Slice_id, line_id,ci(x, y), cj(x, y)] for right lung. Where, Data_sp and Data1_sp represent the data seed points. Pseudo code of the sampling line construction algorithm is shown in below Fig. 2.
Output: 2-dimensional arrays consisting of seed points of all images.
3.3.2 Seed point database storage
-
Input: 2-dimensional array consisting of seed points of all images.
-
Step 1: Write the 2-dimensional array to the database.
-
Step 2: For every sampling line in every input slice,
-
i.
calculate the average location of every seed point using,
Where ci(x, y), and cj(x, y) gives the coordinate location where the sampling linesl1 intersects the lung border and K implies the number of images.
Output: Database storage containing the average seed point location of all the seed points.
3.3.3 Reference model generation
It is formed from healthy lung slices of patients which give a deep knowledge about lung shapes and how they differ based on age.
Input: Database storage encompassing the average seed point location of all the seed points.
Step 1: Read the average seed point location from the database.
Step 2: Connect the coordinates to make the reference lung model.
Step 3: Write the generated reference lung model to an image.
Step 4: Repeat the steps 1–3 for multiple reference models.
Output: Multiple reference models
3.3.4 Edge enhancement
Post processing is done to enhance the edge of the reference model generated via the system.
Input: 1) Reference shape model from 3.3.3.
Step1: Initialize the active contour model [38] to enhance the edges of the reference lung. The ACM takes both local intensity mean and variance information to typify the local intensity distribution.
Output: Edge enhanced reference model.
3.3.5 Feature vector preparation
-
Input: Edge enhanced reference model.
-
Step 1: Compute the average of each feature for the left lung. In feature vector preparation, considered only the left lung.
-
Step 2: Store the values attained from step 1 as an feature vector as
-
Step 3: Repeat Step 1 and Step 2 for right lung.
-
Output:
-
i.
feature vector for the reference model.
-
ii.
Edge-enhanced reference model.
3.4 Validation phase
3.4.1 Healthiness of Slice
-
Input:
-
i.
feature vector of Reference model.
-
ii.
feature vector of input slice.
-
iii.
Generated reference model.
-
iv.
Segmented input slice.
Determining the healthiness of the input slice is done by comparing the FV of the RM and FV of the input slice. If the deviation between vectors of the input slice and RM is significant, then the input slice is unhealthy. The deviation is computed utilizing distance metrics. The results are labelled manually as 0 if the FV of the test slice is healthy and has a lesser deviation from the FV of the actual RM. The results are labeled as 1 if there is a significant deviation between the FV computed from the input slice to the FV computed from the RM, which means the results between 0 and 1 signifying the test slice of the image belonging to positive and negative.
Output:
- i.
Generated reference model.
- ii.
Segmented input slice.
- iii.
0 - Healthy input slice.
- iv.
1- Unhealthy input slice.
4 Results and discussion
The proposed method is implemented in MATLAB. For the proposed segmentation method, the input dataset consists of more than 2000 Healthy CT Images (512 × 512) of 10 patients between the age group 40–60 years collected from Cancer Institute-Chennai, Government Hospital-Chennai, Lung Image Data-base Consortium image collection (LIDC-IDRI) and other scan centres across Chennai, India. The healthy CT images were identified by the expert radiologists and were given as input to generate the reference shape model.
4.1 Experimentation results
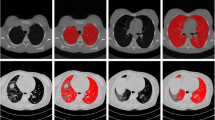
The experimentation snapshots of each step in the proposed approach are presented here. Figure 3 displays the Input CT slice of the image to be segmented. This is a DICOM standard [512 × 512] CT slice. The output of the pre-processing phase after carrying out background removal and airway removal as discussed in section 3.1.3 and 3.1.4 is shown in Fig. 4. The edge detected image using Laplacian operator in section 3.1.5 is exhibited in Fig. 5. The major lung components are identified, labelled and separated. The separated lung components are exhibited in Figs. 6 and 7. The proposed method of generating RM is done by constructing sampling lines greater than line count 100 on the major lung components using Sampling Line Algorithm conferred in section 3.3.1 is exhibited in Figs. 8 and 9. The process is repeated for all the healthy slices and RM is generated for both the lung components based on sections 3.3.2 and 3.3.3 as exhibited in Figs. 10 and 11. Finally, the segmented image based the RM of left as well as right lung components are displayed in Figs. 12 and 13.
4.2 Performance metrics
4.2.1 Ground truths
It is basically the true and accurate segmentation that is labelled by human experts. Ground truth (GT) is generated from the same data-set that is used in the segmentation method.
4.2.2 Jaccard index (J)
It is also called a Jaccard similarity coefficient is a metric used to assess the similarity betwixt the GT and segmented Image. Jaccard index outcomes vary from 0 to 1. In Jaccard index, the GT, as well as segmented image, is compared using the formula stated below,
Where A alludes to the GT labelled by experts and B alludes to the segmented image resulting as of the proposed technique.
4.2.3 Dice similarity coefficient
It is used for comparing the similarity betwixt the GT and segmented image. The value of Dice coefficient ranges as of ‘0’ to ‘1’. This is computed using,
Where A implies the GT labelled by experts and B implies the segmented image resulting as of the proposed work.
4.2.4 Accuracy
It is utilized as a numerical measure of a result classification test. Higher Accuracy indicates a better system performance. Accuracy is stated as,
4.2.5 Specificity
It is stated as similarity of the background pixels recognized by the GT along with the presented technique. Specificity is also labelled as True Negative Rate (TNR) Specificity is computed using,
4.2.6 Sensitivity
It is stated as lung region correctly recognized by the radiologist and by the proposed method. It is also called as True Positive Rate (TPR). The sensitivity of the presented approach can be computed using,
Where TP implies the true positive is the similarity of the foreground pixels identified by GT computation against the proposed work. TN implies true negative is the foreground pixels incorrectly segmented by proposed truth against the GT. FP stands for false positive, which is the similarity of back-ground pixels recognized by GT computation against the proposed technique. FN stands for false negative is the background pixels wrongly segmented by proposed method against the GT.
4.2.7 Overlap score
It represents the similarity betwixt the segmentation outcomes. Overlap score is given by,
Where,
- A, B:
represents the segmentation outcomes by means of the proposed work and GT method performed manually.
4.3 Result analysis
Performance analysis of the proposed work is evaluated by comparing the GT against Jaccard index as well as DSC of the widely used segmentation approaches such as RG, GC, and also Flood fill as displayed in Table 1. Evaluation parameters for instance Accuracy, Specificity and Sensitivity are calculated for the proposed method against widely used segmentation techniques is displayed in Table 2 for the dataset considered.
For the considered input dataset, proposed segmentation method gives improved results over RG, GC and Flood fill Algorithms chiefly due to following limitations, RG segmentation methods are initialised with seed points as well as analyses the neighborhood pixels. This algorithm suffers in segmentation when the difference betwixt intense pathological bearing regions and other non-lung tissues are very minimal. In this scenario, the RG algorithm suffers from accuracy and may require additional post-processing steps. Moreover, selection of initial seed points to begin the segmentation requires high manual intervention and analysis may vary depending on various radiologists.
Edge-based (Active Contour) fails to segment the lung region when initial seed points are not near the right boundary. This segmentation achieves a lower accuracy due to convergence criteria which may segment incorrect lung boundaries when pathological bearing regions lie in the lung periphery. In such cases, it also suffers from differentiating minor non-lung tissues from lung tissues and involves long computational times. The Flood fill algorithm suffers in performance due to the closeness of pathological bearing regions and difficulty in selecting a start point which requires manual intervention which can impact the segmentation method’s performance. Graphical depiction of the comparison of the segmentation techniques of (a) right lung, as well as (b) left lung, as shown in Fig. 14.
GC algorithm suffers from the inability to identify the lung regions from the body regions due to the property of minimum cut and the gradient information that is used to find the optimal boundaries in the lung regions. Identification of optimal boundaries is a concern if the input data-set has more pathological bearing regions than usual. The algorithm tends to include minor and thin non-lung tissues as lung regions which tend to affect the accuracy. Graphical depiction of the above Table 2 value is shown in Fig. 15.
5 Conclusion
A Segmentation method based on sampling lines approach to segment lung shape from the input slice is proposed. The proposed work not only segments the lung from the input slices but also validates the healthiness of the segmented lung by comparing it with the RM. The RM is generated as of healthy lung slices of patients which gives a deep knowledge about lung shapes and how they differ based on age. The RM helps to identify the lung border regions of patients more accurately than other methods. To recognize the lung shape with more accuracy, further enhancements in sampling lines algorithm are studied and carried out to generate more precise RMs. The lung shape might vary based on the height of the patient for the same age group. There are chances that the RM may not fit with the lung of that patient produces some marginal error. The problem is currently under research by constructing an intelligent system to generate the shape model for specific from measuring the lung height and age of patients. By incorporating future works, this method can further reduce the dependency of experts and helps to monitor the growth of lung shape and pathologies over time. This reference-based model segmentation can well be implemented to other body organs.
References
Ahmed Memon N, Mirza A, Gilani A (2006) Segmentation of lungs from CT scan images for early diagnosis of lung cancer
An H, Wang D, Pan Z, Chen M, Wang X (2018) Text segmentation of health examination item based on character statistics and information measurement. CAAI Transactions on Intelligence Technology 3(1):28–32
Arun V (2017) Seed point selection of segmentation of lung in HRCT images. Int J Adv Res Comput Sci 8:868–872. https://doi.org/10.26483/ijarcs.v8i9.5205
BalaAnand M, Karthikeyan N, Karthik S (2018) Designing a framework for communal software: based on the assessment using relation modelling. Int J Parallel Prog. https://doi.org/10.1007/s10766-018-0598-2
BalaAnand M, Sivaparthipan CB, Karthikeyan N, Karthik S Early Detection and Prediction Of Amblyopia By Predictive Analytics Using Apache Spark. International Journal of Pure and Applied Mathematics (IJPAM) - Scopus - ISSN: 1314-3395 (on-line version) 119(15):3159–3171
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424
Dai S, Lu K, Dong J (2015) Lung segmentation with improved graph cuts on chest CT images. In 2015 3rd IAPR Asian Conference on Pattern Recognition (ACPR), pp. 241–245, IEEE
Dawoud A (2011) Lung segmentation in chest radiographs by fusing shape information in iterative thresholding. IET Comput Vis 5(3):185–190. https://doi.org/10.1049/iet-cvi.2009.0141
Deng Q, Wu S, Wen J, Xu Y (2018) Multi-level image representation for large-scale image-based instance retrieval. CAAI Transactions on Intelligence Technology 3(1):33–39
Dhara AK, Mukhopadhyay S, Dutta A, Garg M, Khandelwal N (2016) A combination of shape and texture features for classification of pulmonary nodules in lung CT images. J Digit Imaging 29. https://doi.org/10.1007/s10278-015-9857-6
Dinesh Jackson Samuel R, Kanna R (2018) Cybernetic microbial detection system using transfer learning. Multimed Tools Appl
Dinesh Jackson Samuel R, Rajesh Kanna B (2018) Tuberculosis (TB) detection system using deep neural networks. Neural Comput & Applic:1–13
Dong S, Gao Z, Pirbhulal S, Bian G-B, Zhang H, Wu W, Li S (2019) IoT-based 3D convolution for video salient object detection. Neural Comput & Applic:1–12
Dong J, Lu K, Dai S, Xue J, Zhai R (2018) Auto-segmentation of pathological lung parenchyma based on region growing method. In: Huet B, Nie L, Hong R (eds) Internet multimedia computing and service. ICIMCS 2017. Communications in Computer and Information Science, vol 819. Springer, Singapore
Gaidel A (2017) Method of automatic ROI selection on lung CT images. Procedia Engineering 201:258–264. https://doi.org/10.1016/j.proeng.2017.09.612
Heuberger J, Geissbühler A, Müller H (2005) Lung CT segmentation for image retrieval using the insight toolkit (ITK). Medical Imaging and Telemedicine
Hu S, Hoffman EA, Reinhardt JM (2001) Automatic lung segmentation for accurate quantitation of volumetric X-ray CT images. IEEE Trans Med Imaging 20(6):490–498
Ju W, Xiang D, Zhang B, Wang L, Kopriva I, Chen X (2015) Random walk and graph cut for co-segmentation of lung tumor on PET-CT images. IEEE Trans Image Process 24(12):5854–5867
Li W, Nie SD, Cheng JJ (2007) A fast automatic method of lung segmentation in CT images using mathematical morphology. In: Magjarevic R, Nagel JH (eds) World congress on medical physics and biomedical engineering 2006. IFMBE proceedings, vol 14. Springer, Berlin, Heidelberg
Mansoor A, Bagci U, Foster B, Xu Z, Papadakis GZ, Folio LR, Udupa JK, Mollura DJ (2015) Segmentation and image analysis of abnormal lungs at CT: current approaches, challenges, and future trends. Radiographics: A Review Publication of the Radiological Society of North America, Inc 35(4):1056–1076
Mansoor A, Bagci U, Xu Z, Foster B, Olivier KN, Elinoff JM, Suffredini AF, Udupa JK, Mollura DJ (2014) A generic approach to pathological lung segmentation. IEEE Trans Med Imaging 33(12):2293–2310
Maram B, Gnanasekar JM, Manogaran G et al (2018) SOCA. https://doi.org/10.1007/s11761-018-0249-x
Mesanovic N, Grgic M, Huseinagic H, Males M, Skejić E, Muamer S (2019) Automatic CT image segmentation of the lungs with region growing algorithm
Mets OM, Vliegenthart R, Gondrie MJ, Viergever MA, Oudkerk M, Koning HJ, Mali WP, Prokop M, Klaveren RJ, Graaf YV, Buckens CF, Zanen P, Lammers JJ, Groen HJ, Isgum I, Jong PA (2013) Lung Cancer screening CT-based prediction of cardiovascular events. JACC Cardiovasc Imaging 6(8):899–907
Pirbhulal S, Zhang H, Wu W, Mukhopadhyay SC, Zhang Y-T (2018) Heartbeats based biometric random binary sequences generation to secure wireless body sensor networks. IEEE Trans Biomed Eng 65(12):2751–2759
Pu J, Gu S, Liu S, Zhu S, Wilson D, Siegfried JM, Gur D (2012) CT based computerized identification and analysis of human airways: a review. Med Phys 39(5):2603–2616
Pu J, Roos J, Yi CA, Napel S, Rubin GD, Paik DS (2008) Adaptive border marching algorithm: automatic lung segmentation on chest CT images. Comput Med Imaging Graph 32(6)
Shoaib M, Naseem R, Dar A (2013) Automated segmentation of lungs in computed tomographic images. Eur J Sci Res 98:45–54
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics, 2018. CA Cancer J Clin 68(1):7–30. https://doi.org/10.3322/caac.21442
Sivaparthipan CB, Karthikeyan N, Karthik S (2018) Designing statistical assessment healthcare information system for diabetics analysis using big data. Multimed Tools Appl
Tan Z, Zhang S, Wang R (2018) Stable stitching method for stereoscopic panoramic video. CAAI Transactions on Intelligence Technology 3(1):1–7
Vučković V, Arizanović B, Le Blond S (2017) Generalized N-way iterative scanline fill algorithm for real-time applications. J Real-Time Image Proc. https://doi.org/10.1007/s11554-017-0732-1
Wang X (2007) Laplacian operator-based edge detectors. IEEE Trans Pattern Anal Mach Intell 29:886–890. https://doi.org/10.1109/TPAMI.2007.1027
Wei Y et al (2012) A fully automatic method for lung parenchyma segmentation and repairing. J Digit Imaging 26:483–495
Wu W, Pirbhulal S, Sangaiah AK, Mukhopadhyay SC, Li G (2018) Optimization of signal quality over comfortability of textile electrodes for ECG monitoring in fog computing based medical applications. Futur Gener Comput Syst 86:515–526
Zhang Y, Yan H, Zou X, Tao F, Zhang L (2016) Image threshold processing based on simulated annealing and OTSU method. In: Jia Y, Du J, Li H, Zhang W (eds) Proceedings of the 2015 Chinese intelligent systems conference, Lecture notes in electrical engineering. Springer, Berlin, Heidelberg
Zheng L, Lei Y (2018) A review of image segmentation methods for lung nodule detection based on computed tomography images. MATEC Web of Conferences 232:02001. https://doi.org/10.1051/matecconf/201823202001
Zhou S, Wang J, Zhang S, Liang Y, Gong Y (2016) Active contour model based on local and global intensity information for medical image segmentation. Neurocomputing 186:107–118
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Dharmalingham, V., Kumar, D. A model based segmentation approach for lung segmentation from chest computer tomography images. Multimed Tools Appl 79, 10003–10028 (2020). https://doi.org/10.1007/s11042-019-07854-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-019-07854-0