Abstract
The recent trends in medical image segmentation and analysis are often used in many real world applications for analyzing different objects of interest. Analyzing the brain Magnetic Resonance (MR) images is found to be difficult task because of the existence of intensity non-uniformity. Although numerous models have been proposed to handle brain MR image segmentation, it is still a challenge to effectively approximate the Intensity Non-Uniformity (INU) and improve greater segmentation accuracy. Hence, an integrated energy minimization approach, namely adaptive weighted fuzzy region based optimization algorithm is developed for brain MR image segmentation. These adaptive fuzzy regions are iteratively weighted to estimate their membership values assigned to each pixel with respect to energy. Also, the optimal weighting parameter, membership values to each region, and bias fields are iteratively estimated and updated. Further, this algorithm is compared with the recent energy minimization approaches in simulated brain MR image dataset. The results of the quantitative evaluations demonstrate that the proposed algorithm gives more reliable segmentation and better accuracy in spite of initialization, noise, and intensity non-uniformity.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Segmentation of image is a basic process in image analysis and computer vision. It is used in different modalities of medical image analysis. During the MR image acquisition process the various intensity levels for different tissues are obtained by changing the acquisition parameters. The MR images give a better contrast compared with those of Computerised Tomography (CT). Therefore, MR images are widely used for medical image segmentation. The recognition of brain shape and structure in MR image is most essential in neuroscience functional activation mapping, brain growth learning and neuro anatomical variability investigation. The brain Magnetic Resonance (MR) image segmentation involves active research areas such as volumetric analysis of different cells, tumour detection, tissue classification, etc. In particular, it is very hard to partition the images into disjoint regions due to considerable spatial variations in pixel intensities caused by physical constraints in acquisition sensitivities [10].
Conventional segmentation algorithms are unable to segment the MR images due to the existence of INU. The bias estimation and correction are the pre-processing steps to rectify the intensity non-uniformity. Bias field approximation is a key task in medical image processing. The existing bias correction algorithms are generally classified into two types namely, prospective and retrospective. Prospective techniques deal with specific hardware and sequences during the process of acquisition, whereas the retrospective techniques work mainly based on spatial information provided in terms of intensity in the image [4].
In the early days, the bias field was corrected using the filtering methods. Later, this was well approximated by linear and spline approximation algorithms [15]. The N3 algorithm is belongs to retrospective methods used for bias field estimation and correction, which maps intensity histograms into probability distributions of the given image. It is enhanced by applying fast and robust B-spline approximation technique named N4 algorithm [16, 17]. Likar (2001) et al. proposed the information theoretic approach for correcting intensity non-uniformity by deriving parametric polynomials as a set of basis functions known as entropy minimization [12].
Chunming Li (2014) et al. proposed energy minimization technique for combined bias correction and segmentation. It represents the given image as two multiplicative elements. The energy minimization process is optimized to estimate these multiplicative elements [11]. But this method is not significant for higher level noisy images. Kaihua Zhang (2010) et al. proposed a new variational scheme to estimate the bias and segmentation simultaneously. The INU is modelled as the means and variances of Gaussian distribution. Each region is described with maximum likelihood (ML) energy function and it is applied to the entire image using Bayesian learning approach [23, 25].
Huibin Chang (2016) et al. proposed high-order and regularized modified technique for bias field calculation and correction [1]. This technique is designed using piecewise constant regularization and data fitting term. The smooth regularization is achieved by multi-resolution algorithm. But, it is difficult to process T1 weighted low contrast images using this model.
Fuzzy C-Means (FCM) clustering method is broadly applied to segment different regions as well [21]. Medical image segmentation is one of the important domains for identifying different regions of interest. Handling INU in brain MR image involves a tedious process. So, the Bias Corrected Fuzzy C-Means (BCFCM) has been developed to remove the bias field and segment the image simultaneously. Fuzzy local information based FCM algorithm enhances the clustering performance [7]. Based on FCM, many variations have been introduced to effectively segment the brain MR image for last 3 yrs. These variations include Modified FCM [14], Multi-scale, Multi-block (MsbFCM) [22], Rough Set FCM, Generalized Rough Set FCM, Adapted Non Local FCM [2], Bias Corrected FCM (BCFCM), Global Bias Corrected FCM (GBCFCM) [5], Kernel Generalized FCM [26] and the hybrid techniques [8] are introduced with the combination of watershed and FCM [9]. All the FCM based methods are sensitive to initializations.
Level set formulation with energy minimization method has been proposed to segment different kinds of tissues in the brain MR image. The intensity non-homogeneity is a slowly varying mixture with the original image is embedded in brain MR images [2, 10]. The bias calculation and segmentation are performed simultaneously to give accurate segmentation results. Different approaches have been designed for tissue segmentation using level set method. These approaches include variational multiphase level set [6, 23], statistical and variational multiphase level set [25], longitudinally guided level set [19] and unified approach using Expectation Maximization algorithm with level set [13]. Active Contour method (ACM) has been built using the variational level set method [27]. The energy is defined using local intensity fitting measure that attracts the edge, but it ends at the object boundaries [18].
Zexuan Ji (2012) et al. developed generalized rough FCM algorithm for tissue segmentation. In this approach, each region is represented using fuzzy regions, which is controlled using weighting parameter. The bias field and weighting parameter are estimated iteratively [5]. The thresholds are estimated for rough region based on random initialization of mean for each region, but this approach may not be efficacious to rectify the bias which in turn will result in lesser segmentation accuracy.
Ying wang (2013) et al. proposed a method for image segmentation using new local region based level set. The weighted least squares are used to categorize each and every region. The parameters of locally linear classifier are iteratively updated and the global segmentation is obtained by integrating the energy functions to entire image. The segmentation result is very sensitive to the statistical measure of the Gaussian kernel. The result of the final segmentation is affected by the non-convexity of the energy function [20].
Haili Zhang (2013) et al. presented variational approach to simultaneous multiphase bias calculation and segmentation. To deal with the problem of non-convex and non-smooth minimization, the maximum a-posterior theory of pixel density functions is used. This model relaxes the constraint on the characteristic functions [24].
Chunming Li (2014) et al. developed new energy minimization technique to estimate the intensity non-uniformity for correction and segmentation jointly using Multiplicative Intrinsic Component Optimization (MICO). The robustness and segmentation accuracy of the model depend on the choice of weighting parameter [11].
Kaihua Zhang (2015) et al. developed level set with energy minimization technique for MR image segmentation. The inhomogeneous objects and original signals are modelled and calculated adaptively using the means and variances of Gaussian distributions. The ML approach used for calculating energy function is described on the entire image domain that simultaneously performs bias estimation and energy minimization for segmentation. The different initializations of the images yield similar segmentation results which point to the robustness of this approach. This locally statistical level set method approximates the bias field by considering forward difference in temporal derivatives [25]. As there are no partial derivatives, it is difficult to accurately estimate bias field. But, to some extent, the segmentation accuracy depends on region scale parameter and satisfies the condition of partial volume effect.
In this article, an integrated energy minimization technique is proposed to approximate the bias and tissue segmentation. The proposed adaptive weighted fuzzy region based bias calculation and tissue segmentation is performed in succession to describe spatial properties of the MR image into different fuzzy regions. Each fuzzy region is handled in different adaptive weighting parameter to characterize them into intensity non-uniformity and original image. Thus the objective function is solved using modified energy minimization formulation iteratively. The algorithm proposed in this article mainly deals with robust to initialization sensitivity, INU and accurate segmentation.
2 Background
The given input brain MR image I is represented in the following form.
where I(x) is referred as the gray value of the input image at pixel x, b(x) is the spatial variations in the observed image referred to as bias field, F(x) is an original image, and n(x) is noise modelled as Gaussian. The bias can be approximated based on their property that it is varying slowly to whole image. The bias field b(x) is estimated as the linear combination of optimal coefficients w1, w2, …, wM as a column vector w = (w1, w2, …, wM)Tand basis functions g1(x), g2(x), …, gM(x) as a column vector G(x) = (g1(x), g2(x), …, gM(x))T. The calculation of bias field achieves subjective accuracy with adequately huge number of linear functions. The bias field is represented in vector form and it is calculated as follows.
where R = (P + 1)(P + 1)/2 is the polynomial count gk(x) and \( {w}_k\mathfrak{\in}\mathfrak{R},k=1,\dots, R \) are the real valued coefficients and P is the degree of those polynomials.
The original image F(x) can be approximated to piecewise constant based on their property that acquires N different constant numbers u1, u2, …, uN for each disjoint region Ω1, Ω2, …, ΩN respectively. The brain tissue segmentation based on pixel intensity is the tedious task due to intensity non-uniformity. Therefore, each region Ω1and Ω2 is characterized by using membership function defined as mi(x) = 1 for all x is belongs to I(y)Ωi and mi(x) = 0 for x is not belongs to Ωi for the case of two regions. For the N regions case, the different fuzzy values are assigned to\( {m}_i^q(x) \)between 0 and 1 that should assure\( \sum \limits_{i=1}^N{m}_i^q(x)=1 \), where q is the fuzzifier. The original image F(x) is approximated based on the membership functions mi(x) and constants ui as follows
The bias corrected image is generated by using calculated bias field and original image as follows I(x)/b(x).
For each region, three different adaptive fuzzy regions are estimated to apply the thresholds that are automatically approximated with respect to distance from every pixel I(k) with different intensity values hj of the image. The distance is estimated as given below.
where ηx is circular neighbourhood of pixel x defined with a radius r and centred at each point. |ηx| is the cardinality, v_min and v_ max are the minimum and maximum intensity levels, and V is the total intensity levels. Each pixel x is defined with distance vector dx = {dx(h1), dx(h2), …, dx(hV)}. The thresholds can be derived using the distance vector dx as follows.
where dx_minand dx_max are the minimum and maximum of distance vector dx respectively. To estimate the fuzzy regions for each region, the distance from every pixel x and mean value of all pixels in that region is calculated using the distance equation in Eq. (4). After that, the True Positive region TP(fu), True Negative region TN(fu) and Neutral region NL(fu) can be estimated as follows.
3 Adaptive weighted fuzzy region based optimization
It is an energy formulation for adaptive weighted fuzzy region based optimization to calculate the bias and brain image tissue segmentation modelled in Eq. 1. The problem of calculating bias b(x) and original image F(x) of the observed brain MR image I(x) is the two-stage iterative process. The formulation of energy is given below.
3.1 Energy formulation
The characteristics of the unknown bias field and original image are exploited for finding their values concerned. The property of bias field is a slowly varying mixture approximated with a set of linear combination of functions. These functions are orthogonal polynomials as shown in Eq. 2 and the property of true image takes N different values approximated as piecewise constant shown in Eq. 3. The representation of the energy minimization can be expressed in terms of\( m={\left({m}_1^q,{m}_2^q,\dots, {m}_N^q\right)}^T \), u=(u1, u2, …, uN)Tand w = (w1, w2, …, wM)T.
The summation and integration order is changed to solve the above equation and it takes the following form.
where λi is the adaptive weighting parameter for the ith tissue. Introduction of fuzzy region to the above function changes the objective function into the following form.
These three fuzzy regions represent the lower and the higher approximations of each region. Therefore, all the pixels in this region are considered to be true positive region TP(fu) and their membership value is assigned to be one. All the pixels in the True Negative region TN(fu) are excluded from that region, so that the membership value is assigned to zero. The exclusion of the True Negative region TN(fu) from true positive region that changes the objective function into the following form.
3.2 Adaptive weighting parameter calculation
The adaptive weighting parameter λ is calculated to adjust the pixels in each region. Normally, the parameter takes larger value for the true positive region and smaller value for the true negative region to approximate the pixel belongs to that region. The true positive region basically covers maximum pixels of the region. On the other hand, pixels belonging to true negative region must be lesser. The adaptive weighting parameter is estimated based on percentage of pixels covered by the true positive region within the region that will adaptively adjust the possibility of the pixel belong to that region and it is updated iteratively as follows:
where px is the region label assigned for each pixel x is estimated based on the maximum membership value as follows.
3.3 Energy minimization
The energy minimization process can used to estimate and update E(m, u, w) iteratively with the use of the variables m, c and w, specified the other two revised in earlier iteration.
3.3.1 Optimization of region descriptor c
The given updated values of m and w, the energy E(m, u, w) is optimized with the use of region descriptor \( \widehat{u}={\left({\widehat{u}}_1,\dots, {\widehat{u}}_N\right)}^T \) is given by
3.3.2 Optimization of optimal coefficient and bias field
The given updated values of m and u, the energy E(m, u, w) is optimized by using optimal coefficient w is realized by solving the equation \( \frac{\partial E}{\partial w}=0. \)
where A and B are R – dimensional column vector and R × R matrix respectively as follows
By solving the equation \( \frac{\partial E}{\partial w}=0 \)takes the linear form Bw = A. From this solution of the equation is obtained by \( \widehat{w}={B}^{-1}A \). The optimal coefficient is expressed in terms of m and u as follows
Based on the optimal coefficient column vector w, the bias field is calculated as follows.
3.3.3 Optimization of membership function
For given updated values of w and c, the energy E(m, u, w) is optimized based on the membership function \( m=\widehat{m}={\left({\widehat{m}}_1,\dots, {\widehat{m}}_N\right)}^T \) for the case q > 1 is given by
For the case q = 1 is given by
The optimal coefficient w is approximated with respect to spatial information and the derived set of linear orthogonal polynomials that result in effective approximation of the bias field. The adaptive weighting parameter λ is updated adaptively to cover the pixels of the respective region for obtaining robust and accurate segmentation. The proposed Adaptive Weighted Fuzzy Region based energy Minimization (AWFRM) algorithm is implemented to segment the brain image. The tissues in brain image dataset are White Matter (WM), Gray Matter (GM) and Cerebrospinal fluid (CSF). An iterative scheme for minimization of the energy formulation is summarized as below.
3.4 AWFRM algorithm
-
Step 1:
Initialize the region descriptors and membership values for all regions {u1, u2, …, uN} and {m1, m2, …, mN} randomly.
-
Step 2:
Calculate the three adaptive fuzzy regions for every region using Eq. (5).
-
Step 3:
Update the optimal coefficient w as \( \widehat{w} \) in Eq. (9) and update the bias field b as \( \widehat{b} \) in Eq. (10).
-
Step 4:
Calculate and update the membership value m as \( \widehat{m} \) in Eq. (11) for the case q > 1 or the Eq. (12) for the case q = 1.
-
Step 5:
Assign each pixel x with a region label px according to the Eq. (7).
-
Step 6:
Estimate the adaptive weighting parameters {λ1, λ2, …, λN} by Eq. (6).
-
Step 7:
Update the region descriptor u as \( \widehat{u} \) in Eq. (8).
-
Step 8:
Check the convergence condition. Stop the process until the convergence condition has been attained or maximum number of iterations reached. If not, continue from step 2.
The variables m, u and w are updated based on fixed value of other two variables which are estimated in the previous iteration. So, it is necessary to initialize two out of three variables initially. The convergence criterion in step 8 is specified as|Z(n) − Z(n − 1)| < ε, where Z(n)the region descriptor vector updated in nth iteration of step 7 and ε is the convergence threshold.
4 Implementation
The implementation of AWFRM algorithm is segmenting the brain MR images into three regions, namely GM, WM and CSF. The parameters employed in this experiment are initialized as follows. The different degrees of polynomials used in this experiment are set to 3 and 4. The size of the window is set to be 4. The fuzzifier q is initialized with 2. Here ε is set to 0.001. There are 20 polynomials are used for approximating the linear combinations.
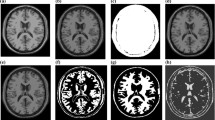
The proposed AWFRM algorithm is analysed using simulated Brain dataset (BrainWeb) [3]. It provides three dimensional brain MR image dataset of T1, T2 and PD weighted dataset. It also provides brain images with various levels of intensity non-uniformity, variety of slice thicknesses and noise levels. The proposed algorithm is applied to all the sets of T1 and T2 weighted brain datasets among variety of thicknesses, different levels of intensity non-uniformity, and noise levels. The three different random initializations of region descriptors u1, u2, …, uN and the membership functions m1, m2, …, mN yield the same segmentation results are shown in Fig. 1 with negligible difference in calculated bias field. The proposed algorithm is applied to T1 weighted, 1 mm slice thickness and 20% INU image. The energy E(m, u, w) is computed and it is plotted. The robustness of the initialization in the proposed algorithm is established in that the energy curve is decreased rapidly and converged quickly for different initializations.
To demonstrate the initialization robustness of the proposed algorithm with higher noise level and slice thickness, it is applied to T2 weighted, 9 mm slice thickness, 40% intensity non-uniformity and 9% noise image. In this assessment, the energy curve maintains the same results and converges quickly for different initializations. The same are shown in Fig. 2.
The next test is performed in T1 weighted brain image dataset among various form of noises and variety of intensity non-uniformity as shown in Fig. 3. It also gives calculated bias, bias corrected images and segmented regions. The ground truth provided by BrainWeb dataset is compared with the result of the proposed segmentation algorithm. The result of this comparison shows that the brain MR image tissue segmentation is consistent with that of ground truth.
Next, the proposed algorithm is tested with simulated T2 weighted brain dataset containing noise with 3% and three different INU as depicted in Fig. 4. It is also shows that the calculated bias, bias corrected images and segmented images. It exposes the outcomes of the AWFRM algorithm are reliable with those of the four different intensity non-uniformity and noise levels, and various thicknesses of brain MR images.
5 Results and discussion
The performance of AWFRM algorithm is evaluated with respect to intensity non-uniformity correction, a comparison is made with the well known methods namely N3 and MICO. It has been assessed by computing the intensity non-uniformity of the bias corrected brain MR images by means of Coefficient of Variation (CV) and Coefficient of Joint Variation (CJV). The CV is described for every brain tissue T (WM, GM or CSF) is as follows
The CV is calculated by the statistical information such as mean μ(T) and standard deviation σ(T) derived from the intensities in the tissue T and CJV is described as
The proposed is applied and the results are compared with MICO and N3 methods in 30 different images with variety of noises and non-uniformity in intensity obtained from T1 and T2 weighted simulated brain dataset. The proposed algorithm is effectively handles the bias estimation and correction. The CV and CJV results are plotted and shown in Fig. 5. It shows that the smaller CV and CJV values of the AWFRM are the sign of greater bias field correction results.
To illustrate the ability of proposed algorithm with respect to intensity non-uniformity correction and segmentation, a comparison is made with the methods developed by Zexuan Ji et al., Chuming Li et al. and Kaihua Zhang et al. GRFCM, MICO and Locally Statistical Level Set Method (LSLSM) respectively. The results are quantitatively evaluated using the Jaccard Similarity (JS) index devised as
where S1 is the segmented region of the proposed algorithm S2 is the region acquired from the ground truth. The segmentation outcomes of various techniques and ground truth of simulated T1-weighted brain image with 1 mm thickness, 3% noise, 40% intensity non-uniformity is shown in Fig. 6.
BrainWeb dataset provides the ground truth for WM, GM and CSF, that is used as reference region to estimate JS index. Figure 7 reveals the JS values of the 18 T1-weighted brain images with three different intensity non-uniformity and six different noise levels with respect to ground truth. The proposed algorithm is effectively addresses the issues stated in the different state of art methods and the robustness of the proposed approach with respect to initialization sensitivity, INU estimation and correction, and accurate segmentation. The higher JS value indicates the sign of greater segmentation accuracy. The JS values plotted by means of box plot are shown in Fig. 7 reveals that the AWFRM provides better performance than LSLSM, MICO and GRFCM algorithms by means of segmentation accuracy and robustness.
Further, the T2-weighted brain dataset with 1 mm slice thickness, 3% noise and 40% intensity non-uniformity is used for analyzing the performance of above said four methods. The ground truth and segmentation results of four methods are shown in Fig. 8.
Figure 9 illustrate the evaluation of JS values of the 18 T2-weighted brain MR images with three different intensity non-uniformity and six different noise levels with respect to ground truth. The JS values are plotted using box plot is shown in Fig. 9 reveals that the AWFRM again provides better performance than LSLSM, MICO and GRFCM algorithms by means of segmentation accuracy and robustness even for the higher contrast and thicknesses of the brain MR images.
The effectiveness of the four methods to segment T1-weighted MR images into GM and WM with different noise levels is examined and the quantitative outcomes are tabulated as follows.
The JS outcomes are measured and the segmentation results of GM and WM for four algorithms applied to T1-weighted brain dataset with different form of noises are plotted and shown in Fig. 10. The results prove that the AWFRM algorithm yields higher segmentation accurateness than the other algorithms.
The performance ability of the four methods in segmenting T2-weighted input brain dataset with different forms of noise is tested and the quantitative results are tabulated as follows. The results plotted are revealed in Fig. 11. The results prove that the AWFRM algorithm is more efficacious than the other methods in segmentation accuracy.
6 Conclusion
The AWFRM algorithm is proposed as the combination of adaptive weighted fuzzy rough set approach with energy minimization formulation. The existing locally statistical level set method approximates the bias field by considering forward difference in temporal derivatives. Thus it is not a reliable process to estimate and correct intensity non-uniformity which in turn will affect the results of the segmentation accuracy. The proposed algorithm effectively overcomes the drawbacks of the existing methods. The robustness of initialization, INU, noise level, and the parameters used in the proposed method are established by quantitative assessment and comparison with simulated brain MR images. The outcomes show that this algorithm is robust to noise, intensity non-uniformity, parameters used, and initialization. In sum, the proposed algorithm can be reliably used for producing accurate brain MR image segmentation.
References
Chang H, Huang W, Wu C, Huang S, Guan C, Sekar S, Bhakoo KK, Duan Y (2016) A new Variational method for Bias correction and its applications to rodent brain extraction. IEEE Trans Med Imaging 13(9):1–14
Chen Y, Zhang J, Wang S, Zheng Y (2012) Brain magnetic resonance image segmentation based on an adapted non-local fuzzy c-means method. IET Comput Vis 6(6):610–625
Cocosco CA, Kollokian V, Kwan RK-S, Evans AC (1997) BrainWeb: online Interface to a 3D MRI simulated brain database. NeuroImage 5(4, part 2/4):S425 Proceedings of 3rd International Conference on Functional Mapping of the Human Brain, Copenhagen
Hou Z (2006) A review on MR image intensity inhomogeneity correction. Int J Biomed Imaging 2006:1–11
Ji Z, Sun Q, Xia Y, Chen Q, Xia D, Feng D (2012) Generalized rough fuzzy c-means algorithm for brain MR image segmentation. Comput Methods Prog Biomed Elsevier 108:644–655
Jijun REN, Yachong Zhang, Jun Che, Tao Wang, Long Shi, Liang Zhang (2012) A Novel Multiphase Level Set Method to Image Segmentation Combined with Edge Link method. Proc IEEE Int Conf Indust Info (INDIN)
Krinidis S, Chatzis V (2010) A robust fuzzy local information C-means clustering algorithm. IEEE Trans Image Proc 19(5):1328–1337
Kumar S, Ray SK, Tewari P (2012) A hybrid approach for image segmentation using fuzzy clustering and level set method. Int J Image Graph Signal Proc 6:1–7
Kuo WF, Lin CY, Hsu WY (2011) Medical image segmentation using the combination of watershed and FCM clustering algorithms. Int J Innov Comput Info Control 7(9):5255–5267
Li C, Huang R, Zhaohua D, Chris Gatenby J, Metaxas DN, Gore JC (2011) A level set method for image segmentation in the presence of intensity Inhomogeneities with application to MRI. IEEE Trans Image Process 20(7):2007–2015
Li C, Gore JC, Davatzikos C (2014) Multiplicative intrinsic component optimization (MICO) for MRI bias field estimation and tissue segmentation. Magnet Reson Imaging Elsevier 32:913–923
Likar B, Viergever M, Pernus F (2001) Retrospective correction of MR intensity inhomogeneity by information minimization. IEEE Trans Med Imaging 20(12):1398–1410
Nathan Lowry, Rami Mangoubi, Mukund Desai, Youssef Marzouk and Paul Sammak (2011) A Unified Approach to Expectation-Maximization and Level Set Segmentation Applied To Stem Cell and Brain MRI Images. Proc IEEE Int Sympos Bio Med Imaging: Nano Macro
Zhang Shi, She Lihuang, Lu Li, Zhong Hua (2013) A Modified Fuzzy C-Means for Bias Field Estimation and Segmentation of Brain MR Image. Proc IEEE Conf Control Dec Conf (CCDC)
Sled JG, Zijdenbos AP, Evans AC (1998) A nonparametric method for automatic correction of intensity non-uniformity in MRI data. IEEE Trans Med Imaging 17(1):87–97
Tustison N, Avants B, Cook P, Zheng Y, Egan A, Yushkevich P, Gee J (2010) N4ITK: improved N3 Bias correction. IEEE Trans Med Imaging 29(6):1310–1320
Vovk U, Pernus F, Likar B (2007) A review of methods for correction of intensity inhomogeneity in MRI. IEEE Trans Med Imaging 26(3):405–420
Wang L, Li C, Sun Q, Xia D, Kao C-Y (2009) Active contours driven by local and global intensity fitting energy with application to brain MR image segmentation. Comput Med Imaging Graph 33:520–531
Wang L, Shi F, Yap P-T, Lin W, Gilmore JH, Shen D (2013) Longitudinally guided level sets for consistent tissue segmentation of neonates. Human Brain Mapping Wiley 34(4):956–972
Wang Y, Xiang S, Pan C, Wang L, Meng G (2013) Level set evolution with locally linear classification for image segmentation. Patt Recog Elsevier 46:1734–1746
Bei Yan, Mei Xie, Jing-Jing Gao, Wei Zhao (2010) A Fuzzy C-means based algorithm for Bias Field Estimation and Segmentation of MR Images. Proc IEEE Int Conf Apperceiving Comput Intel Anal (ICACIA)
Yang X, Fei B (2011) A multiscale and multiblock fuzzy C-means classification method for brain MR images. Med Phys Euro PMC 38(6):2879–2891
Kaihua Zhang, Lei Zhang and Su Zhang (2010) A Variational Multiphase Level Set Approach to Simultaneous Segmentation and Bias Correction. Proceedings of IEEE International conference on Image processing
Zhang H, Ye X, Chen Y (2013) An efficient algorithm for multiphase image segmentation with intensity Bias correction. IEEE Trans Image Process 22(10):3842–3851
Zhang K, Liu Q, Song H, Li X (2015) A Variational approach to simultaneous image segmentation and Bias correction. IEEE Trans Cybernet 45(8):1426–1437
Zhao F, Jiao L, Liu H (2013) Kernel generalized fuzzy c-means clustering with spatial information for image segmentation. Digit Signal Proc Elsevier 23:184–199
Zheng Q, Dong E, Cao Z, Sun W, Li Z (2014) Active contour model driven by linear speed function for local segmentation with robust initialization and applications in MR brain images. Signal Proc Elsevier 97:117–133
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Arulanandam, S., Selvarasu, S. Adaptive weighted fuzzy region based optimization for brain MR image segmentation. Multimed Tools Appl 79, 3603–3621 (2020). https://doi.org/10.1007/s11042-018-6215-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-018-6215-y