Abstract
Feature representations extracted from hippocampus in magnetic resonance (MR) images are widely used in computer-aided Alzheimer’s disease (AD) diagnosis, and thus accurate segmentation for the hippocampus has been remaining an active research topic. Previous studies for hippocampus segmentation require either human annotation which is tedious and error-prone or pre-processing MR images via time-consuming non-linear registration. Although many automatic segmentation approaches have been proposed, their performance is often limited by the small size of hippocampus and complex confounding information around the hippocampus. In particular, human-engineered features extracted from segmented hippocampus regions (e.g., the volume of the hippocampus) are essential for brain disease diagnosis, while these features are independent of diagnosis models, leading to sub-optimal performance. To address these issues, we propose a multi-task deep learning (MDL) method for joint hippocampus segmentation and clinical score regression using MR images. The prominent advantages of our MDL method lie on that we don’t need any time-consuming non-linear registration for pre-processing MR images, and features generated by MDL are consistent with subsequent diagnosis models. Specifically, we first align all MR images onto a standard template, followed by a patch extraction process to approximately locate hippocampus regions in the template space. Using image patches as input data, we develop a multi-task convolutional neural network (CNN) for joint hippocampus segmentation and clinical score regression. The proposed CNN network contains two subnetworks, including 1) a U-Net with a Dice-like loss function for hippocampus segmentation, and 2) a convolutional neural network with a mean squared loss function for clinical regression. Note that these two subnetworks share a part of network parameters, to exploit the inherent association between these two tasks. We evaluate the proposed method on 407 subjects with MRI data from baseline Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. The experimental results suggest that our MDL method achieves promising results in both tasks of hippocampus segmentation and clinical score regression, compared with several state-of-the-art methods.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Hippocampus segmentation in magnetic resonance (MR) images has attracted increasing scientific attention since the morphological analysis of the hippocampus is of vital importance to monitor and diagnose clinical conditions of the brain [14, 15, 32, 46, 47]. In human brains, the hippocampus locates in the medial temporal lobe (the site of functional and structural pathologies in mental illnesses) [37]. It is well recognized that the changes in shape and size of the hippocampus are closed related to the Alzheimer’s disease (AD) and other brain diseases [19, 23]. In recent years, various approaches have been proposed for compter-aided AD diagnosis based on features extracted from the hippocampus [2, 13, 21, 33]. Thus, accurate segmentation of hippocampus has been remaining an active topic.
The most straightforward way for hippocampus segmentation is the manual annotation in MRI. However, manual segmentation is highly demanding, since we have to identify the hippocampus in each slice of MRI, and thus such process is not only tedious but also error-prone [41, 44]. Many algorithms for hippocampus segmentation have used atlas-based (e.g., single-atlas or multi-atlas) and deformable models techniques [8]. However, the performance of these methods highly relies on supplementary techniques such as classifiers, optimizers, and thresholding strategies. Also, multi-atlas based methods are usually time-consuming, because of non-linear registration from multiple atlases onto a target image [6, 12, 18]. Particularly, because of irregular shape and blurred edges of the hippocampus, atlas-based methods usually generate sub-optimal segmentation performance. In recent years, many learning-based methods (e.g., random forest based regression method [10, 41, 43, 45, 48, 49]) have been proposed and shown good results in brain structure segmentation using MRI data. The primary disadvantage of these methods is that they usually require human-engineered features for MR images, and such features may be not consistent with specific learning models, which could degrade the segmentation performance.
On the other hand, since hippocampus has been proven to be closely related to many kinds of brain diseases (e.g., AD and Parkinson disease), many previous studies have proposed computer-aided disease diagnosis systems based on feature representations extracted from hippocampus in MRI. There are at least two disadvantages in these methods. First, diagnosis performance is usually highly dependent on the accurate segmentation for the hippocampus, while it is very challenging to achieve accurate segmentation results using current approaches. Second, feature representations extracted from hippocampus regions are usually prep-defined by human beings, without considering the heterogeneous characteristics between feature and subsequent diagnosis models. In such a case, these methods usually yield sub-optimal performance in computer-aided brain disease diagnosis.
To address these issues, we propose a multi-task deep learning (MDL) method for joint hippocampus segmentation and clinical score regression, without using any time-consuming non-linear registration for pre-processing MRI and pre-defining features for the hippocampus. The flowchart of our proposed MLD method is illustrated in Fig. 1. The intuitive idea here is to exploit the underlying association between the task of hippocampus segmentation and the task of brain disease diagnosis. Specifically, we first linearly align all studied MR images onto a common template space. To speed up the learning process, in the template space, we then extract image patches by approximately define a bounding box for hippocampus regions. Based on those image patches, we then design a multi-task convolutional neural network to simultaneously perform hippocampus segmentation and clinical score regression. It is worth noting that our proposed network consists of two subnetworks designed for two learning tasks, respectively. The first subnetwork follows a U-Net architecture with a Dice-like loss function, for hippocampus segmentation. The second one is a conventional CNN architecture with a mean squared loss function for clinical score regression. These two networks share a part of network parameters, which are used to exploit the underlying association between two tasks.
Illustration of the proposed multi-task deep learning (MDL) framework for joint hippocampus segmentation and clinical score regression. There are four main elements in MDL: 1) MR image processing via linear registration, 2) patch extraction, and 3) multi-task neural network for joint segmentation and regression. The input data are MR images, while the output contain both segmented hippocampus and Mini-mental state examination (MMSE) scores for subjects
The major contributions of this study can be summarized in the following. First, we propose to jointly perform hippocampus segmentation and clinical score regression via a multi-task neural network. This network contains two subnetworks, with each one corresponding to a particular task. And these two subnetworks share a part of parameters to underlying the inherent association of these two tasks. Second, we develop an automatic hippocampus segmentation methods, without using any manual annotation. In addtion, we develop an automatic feature extraction method for brain disease diagnosis, without defining hand-crafted features in hippocampus.
The remainder of this paper is organized as follows. In Section 2, we briefly introduce relevant studies. In Section 3, we describe materials used in this study and introduce the proposed method. Section 4 presents experimental settings and experimental results. We finally conclude this paper in Section 6.
2 Related work
2.1 Hippocampus segmentation
Currently, there are various methods developed for the segmentation of hippocampus [15, 38], including 1) manual annotation, 2) atlas-based methods [17, 20, 28], and 3) learning-based methods [10, 17, 31, 39]. In the first category, one has to identify the hippocampus in each slice of MRI, and usually take up to 2 hours to finish the annotation task [8]. Thus, manual segmentation for the hippocampus is a highly repetitive task. Besides, due to the high intra-annotator and inter-annotator variability, the manual segmentation for the hippocampus may be inconsistent and inaccurate.
As a traditional automated method, the atlas-based approach for the segmentation of hippocampus has attracted much attention, which can the subjectivity and increasing segmentation accuracy compared to manual annotation. Atlas-based methods can also be further divided into two categories, i.e., single-atlas based and multi-atlas based methods. For instance, Barnew et al. [4] proposed a single-atlas based method, by registering a single atlas onto a target image. They first performed affine registration to generate an ROI that corresponds to the hippocampus, and then adopt a different affine registration on the ROIs defined in the previous stage. Recently, Kwak et al. [24] proposed a graph-cuts algorithm based single-atlas method, by optimizing the output results of the initial atlas-based registration. However, the use of single atlas for segmentation can affect the segmentation accuracy, because of the difference between the atlas and the target image. Then, many multi-atlas based methods [12, 18, 30] have been proposed, where multiple atlases are selected and registered on the target image individually. Then, different label fusion methods are employed to obtain the final segmentation, e.g., majority voting, minimizing an energy function with intensity and prior terms, and simultaneous truth and performance level estimation (STAPLE). For instance, Heckemann et al. [18] select 30 MR images from 30 normal control subjects as atlases, and assign the class with the greatest occurrence among those 30 atlases to each voxel in the target image. They achieved results with an accuracy comparable to manual segmentation results. Although many new label fusion algorithms have been developed to improve the performance of multi-atlas based methods, it remains an open problem in determining how to choose those multiple atlases. Liu et al. [30] proposed a clustering algorithm based method for multi-atlas selection, and reported promising results in brain disease diagnosis. However, previous multi-atlas based methods are also highly dependent on the selected segmentation algorithm and the preprocessing steps [7, 11].
In recent years, many learning-based methods have been proposed for the automated segmentation of the hippocampus, by using advanced machine learning algorithms, such as hierarchical classification [34], random forest regression [27, 41], and support vector machine (SVM) based classification [17]. For instance, Pohl et al. [34] proposed a hierarchical classification method by first evaluating the probabilities of each voxel in the atlas, and then aggregated its neighboring voxels to form higher levels that represent larger portions of the segmented structures. In [41], Zhang et al. developed a partially joint random forest regression model to separately improve the reliability of segmentation, and achieved promising results. They incorporated the prior knowledge into a conventional random forest regression model and learned a non-linear mapping between a voxel’s local appearance and its 3D displacements to landmarks in the target image using Haar-like features to describe the local appearance of voxels. Hao et al. [17] proposed to a support vector machine to learn a classifier for each of the target image voxels from its neighboring voxels in the atlases based on both image intensity and texture features.
However, previous learning based methods require human-engineered feature representations to describe the local appearance of MR images [26]. In particular, these methods generally treat the process of feature extraction and regression/classification model learning as two standalone tasks, without considering the possible heterogeneous characteristics among features and models. Intuitively, incorporating feature learning and model training into a unified framework is expected to generate better performance.
2.2 Hippocampus-based brain disease diagnosis
Since the changes in shape and size of the hippocampus are closed related to the Alzheimer’s disease (AD) and its prodrome (i.e., mild cognitive impairment, MCI) [19], various compter-aided brain disease diagnosis approaches have been proposed, based on features extracted from the hippocampus and machine learning techniques. In [13], a classification method for AD diagnosis was developed based on the distinction of particular atrophic patterns of the hippocampus and entorhinal cortex. One of the features they used is the volume of the hippocampus, and quadratic discriminant analysis was performed for AD classification. Ahmed et al. [2] proposed to first extract local features from the hippocampus and posterior cingulate cortex in each slice of MRI, and then quantized those features using the Bag-of-Visual-Words approach to generate a histogram of quantized features. They finally adopt principal component analysis (PCA) to perform feature dimension reduction, followed by an SVM classifier to identify three classes of subjects: normal controls (NC), mild cognitive impairment (MCI) and Alzheimer’s disease (AD). Moodley et al. [33] investigated the correlation between AD in its earliest stages and hippocampal volume and cortical thickness of the precuneus and posterior cingulate gyrus, and demonstrated that 4 Mountains Test (4MT) was useful in the diagnosis of pre-dementia due to AD. In general, these methods usually require accurate segmentation for the hippocampus, and pre-defined human-engineered features representations for the hippocampus to facilitate the learning of classification/regression models [25]. However, those human-engineered features may be not consistent with the following learning models, leading to poor diagnosis performance.
In this paper, we propose a joint learning framework for hippocampus segmentation and clinical score regression. In our method, we do not require any non-linear registration process and any human-engineered features for the hippocampus. Experiments on 797 subjects from the baseline ADNI database demonstrate the effectiveness of our proposed methods (Table 1).
3 Materials and methods
3.1 Materials
We adopt the public Alzheimer’s Disease Neuroimaging Initiative (ADNI-1) database [22] in this study. Specifically, the baseline ADNI-1 database contains 797 subjects with 1.5T T1-weighted structural MRI data, including 181 AD, 165 progressive MCI (pMCI), 225 stable MCI (sMCI), and 226 normal control (NC) subjects. The definitions for these four categories can be found online.Footnote 1 One types of clinical scores are employed for all studied subjects in ADNI-1, i.e., Mini–Mental State Examination (MMSE) [16]. The MMSE is a test for the evaluation of efficiency intellectual disorders and presence of deteriorating cognitive and is often used as a screening tool in the investigation of patients with dementia and neuropsychological syndromes of different nature. The total MMSE score is between a minimum of 0 and a maximum of 30 points. A score equal to or less than 18 is indicative of a severe impairment of cognitive abilities; a score between 18 and 24 is an indication of a compromised moderate to mild, a score of 25 is considered borderline, from 26 to 30 is an index of cognitive normality. It has been widely used for evaluating cognitive levels of subjects in the diagnosis of AD and MCI.
3.2 Image pre-processing and patch extraction
We pre-process all studied MR images using a standard pipeline. To be specific, we first perform For all studied MR images, we pre-process them using a standard pipeline. Specifically, we first anterior commissure (AC)-posterior commissure (PC) correction using MIPA.Footnote 2 Then, we resample all images to have the same resolution of 256 × 256 × 256, followed by intensity inhomogeneity correction via N3 algorithm [36]. Finally, we linearly align all images onto a template image. Note that, in this study, we do not need any skull stripping or cerebellum removal process. Also, no nonlinear registration is required in image pre-processing.
After pre-processing, all MR images are aligned onto a common template space. In this common space, we then define a bounding cube for the hippocampus and extract an image patch from this box with the size of 72 × 72 × 72, with the axises informatin is (x=,y=,z=). Because of the relatively small size of the hippocampus in the brain, such process helps us discarding many confounding background information. Otherwise, the number of voxels in the background (i.e., negative samples) will be much larger than that of voxels in the hippocampus region (i.e., positive samples), leading to a severe class-imbalance problem. Giving a new testing MRI, we first pre-process it and align it onto the template space. Given the 3D axises of the pre-defined bounding cube, we can directly extract and image patch from this new testing MRI. For image patches extracted from the right hippocampus, we flip these patches to make them consistent with the direction of those from the left hippocampus. After segmentation, we then flip them into their own directions.
To generate the ground truth of segmentation, we first performed hippocampus segmentation using a nonlinear registration based method (i.e., Dartel in the SPM tool box [3]) to get the rough segmentations of the hippocampus, based on whole MR images. Then, three radiologists manually edited the rough segmentations to complete the annotation of the hippocampus for all studied subjects.
3.3 Multi-task convolutional neural network
We develop a multi-task convolutional neural network for joint learning of hippocampus segmentation and MMSE score regression. The schematic diagram of the proposed network is illustrated in Fig. 2. As we can see from this figure, the input data of the proposed network are image patches extracted from MRI, while the output data contain both segmented hippocampus regions and estimated MMSE scores for subjects. Particular, there are two subnetworks, i.e., Subnetwork 1 and Subnetwork 2, which are developed to perform clinical score regression and hippocampus segmentation, respectively. Note that these two subnetworks share a part of parameters (see left part of Fig. 2), which is expected to exploit the underlying association of those two tasks. In the following, we introduce the proposed network in detail.
Illustration of the proposed multi-task convolutional neural network for joint hippocampus segmentation and clinical score regression. The input data of this network are cropped MR image patches (72 × 72 × 72). And the output include both probability maps for segmented hippocampus and MMSE scores, estimated by a Dice-like loss function and a mean squared error loss function, respectively. Two subnetworks are included in the proposed neural network, i.e., Subnetwork 1 and Subnetwork 2, desingned for clinical score regression and hippocampus segmenation, respectively. Also, these two subnetworks share a part of common parameters
As shown in Fig. 2, the first step of Subnetwork 1 contains two 3 × 3 × 3 convolutional layers, followed by a rectified linear unit (ReLU) and a 2 × 2 × 2 max pooling operation. And the stride is 2 for down-sampling in the max pooling operation. The second and third steps of Subnetwork 1 include three 3 × 3 × 3 convolutional layers with ReLU and 2 × 2 × 2 max pooling operations. The output of the third step contains 128 kernels, followed by three fully connected (FC) layers. The numbers of elements in these FC layers are 128, 64, and 32, respectively. Then, we adopt a mean squared error loss function to optimize the network parameters, and the output is the estimated MMSE score for a particular subject.
Besides, the Subnetwork 2 adopts a U-Net architecture [35] to capture both the global and the local structural information of input image patches. To be specific, there are a contracting path and an expanding path in FCN1. The contracting path follows the typical architecture of a CNN, and share the same parameters of the subnetwork 1. Different from the Subnetwork 1, this subnetwork includes 2 additional convolutions in the fourth step. Also, each step in the expanding path consists of a 3 × 3 × 3 up-convolution, followed by a concatenation with the corresponding feature map from the contracting path, and two 3 × 3 × 3 convolutions. Similarly, each convolution is followed by a ReLU function. Due to the contracting path and the expanding path, Subnetwork 2 can grasp a large image area. Thus, even using small kernel sizes, it still can keep high localization accuracy [35]. The output of the last layer in Subnetwork 2 is normalized into [− 1;1]. For the purpose of segmentation, we use a Dice-like loss function in the Subnetwork 2.
Denote \(\mathcal {X}=\{\textbf {X}_{n}\}_{n = 1}^{N}\) as the training data in a batch, and Xn represents the n-th subject. Denote MMSE scores for subjects as \(\textbf {Y}=\{Y_{n}\}_{n = 1}^{N}\). Given V voxels in Xn, we denote the class label of the v-th voxel as Zn,v (n = 1,⋯ ,N;v = 1,⋯ ,V). Specifically, Zn,v = 1 if the v-th voxel locates in the hippocampus, and 0, otherwise. In this study, both class label and MMSE scores are used in a back-propagation procedure to update the network weights in the convolutional layers and learn the most relevant features in the FC layers. The proposed Subnetwork 1 aims to learn a non-linear mapping \({\Psi } : \mathcal {X} \to \textbf {Y}\) from the input space to the class label, with the objective function defined as follows:
which is the mean squared loss for regression to evaluate the difference between the estimated MMSE score f(Xn;W) and the true MMSE score Yn. To perform segmentation, we adopt a Dice-like loss function listed as follows:
where g(Xn,v;W) is the estimated probability map by using the network coefficients W, and Zn,v is the ground truth. In (2), the second term is the dice similarity coefficient (DSC) [50] used to evaluate the overall segmentation performance. That is, we adopt the Dice-like loss function to evaluate the capabilities of our model in detecting voxels in the hippocampus and in discarding confounding voxels in the background.
With the proposed neural network, we can not only jointly perform hippocampus segmentation and clinical score estimation, but also automatically learn local-to-glocal feature representations from MR images for both tasks. That is, we do not require any human-engineered features for MRI, and the learned features from data are consistent with subsequent regression and segmentation models. The optimization of the network parameters are performed via a stochastic gradient descent (SGD) approach [5] and a backpropagation algorithm to compute the network gradients. Specifically, we empirically set the momentum coefficient and learning rate for SGD as 0.9 and 10− 2, respectively. Besides, we implement the network based on the platform of Tensorflow [1] and a computer with a single GPU (i.e., NVIDIA GTX TITAN 12GB).
4 Experiments
4.1 Methods for comparison
For segmentation results, we compare the proposed MDL method with two conventional segmentation methods, including 1) random forest (RF) regression based method [41], and 2) multi-atlas (MA) based method [44]. For clinical score regression results, we compare our MDL method with a hippocampus volume based method (VBM) [13] using the volume based method. It is worth noting that our MDL method simultaneous perform tasks of segmentation and regression, RF and MA can only be used to perform the task of hippocampus segmentation, and VBM can only be employed to perform the task of clinical score regression, respectively. These three methods are briefly introduced in the following.
-
1)
Random forest (RF) based method with local energy pattern (LEP) features [40]. In this method, we learn a non-linear mapping between a local patch and the label of the center of this patch via a random forest classification model. For random forest construction, we adopt 20 trees, and the depth of each tree is empirically set as 25. For a fair comparison, RF share a same immage patch pool as our proposed MDL method. That is, image patches extracted (via the method presented in Section 3.2) from MR image are used as in both RF and MDL. In RF method, we extract the Harr-like features from each image patch [7], and fed such features into the subsequent segmentation model via random forest based regression.
-
2)
Mutli-atlas (MA) based method. In the experiments, we use the whole image for image registration, and we transfer the labeled regions from multi-atlas images to the target image using the majority voting strategy [28]. The detailed implementation can be found in [44].
-
3)
Volume based method (VBM) [13]. In VBM method, we use the normalized volume of the hippocampus as the feature for the linear regression of clinical score. The support vector regressor (SVM) is used as the regression model, which is implemented in LibSVM toolbox [9].
4.2 Experimental settings
We conduct two types of tasks, including hippocampus segmentation and clinical score regression. In the experiments, we adopt a 5 fold cross validation strategy [29, 42] using the subjects from ADNI-1. Specifically, we first randomly partition the whole dataset into 5 subsets, where each subset has roughly equal number of subjects. Then, we treat each subset as the testing set, while the remaining 4 subsets are combined to the training set. This process is repeated until all subsets have been used as the testing set. We finally record the mean and standard deviation of results in segmentation and regression tasks.
For the segmentation results, we use the criteria of Dice similarity coefficient (DSC), sensitivity (SEN), and positive predicted value (PPV). Denote true positives (TP) as predicted hippocampus voxels inside positive regions in ground-truth, false positives (FP) as predicted hippocampus voxels outside positive regions in ground-truth, true negatives (TN) as predicted background voxels outside positive regions in ground-truth, and false negatives (FN) as predicted background voxels inside positive regions in ground-truth. Then, these three measurements are defined as: DSC=\(\frac {2TP}{2TP+FP+FN}\), SEN =\(\frac {TP}{TP+FN}\), and PPV=\(\frac {TP}{TP+FP}\). Note that the final segmentation results are achieved by averaging the results for the left and the right hippocampus regions. For the task of clinical score regression, we use two evaluation criteria, including correlation coefficient (CC) and root mean square error (RMSE) between the predicted clinical scores and ground-truth clinical scores.
4.3 Results and analysis
4.3.1 Clinical score regression
Table 2 shows the MMSE score regression results achieved by our MDL method and VBM. From Table 2, we notice that our MDL method significantly improves the regression performance compared with VBM regarding CC and RMSE. There are two conclusions that can be obtained from the results. First, it demonstrates that clinical scores of subjects are somewhat correlated to the volume of the hippocampus since both MLD and VBM achieve reasonable performance in clinical score regression by using the volume of the hippocampus as feature representation for MRI. Second, besides the volume of hippocampus, shape or texture information of the hippocampus may also have the association to clinical scores, since our method (using features extracted from MRI) can achieve better regression performance.
For clarity, we further illustrate the scatter plots of the estimated MMSE scores versus the true MMSE scores achieved by MDL and VBM in Fig. 3. As shown Fig. 3, our predictions are more related to the ground-truth scores, since much higher correlation coefficients (i.e., CC) are achieved by MDL.
4.3.2 Hippocampus segmentation
In Table 3 and Fig. 4, we report the segmentation results achieved by our MDL method and two competing methods (i.e., RF, and MA) regarding DSC, SEN, and PPV, respectively. Note that the segmentation result of a particular method reported in Table 3 and Fig. 4 is the average of segmentation results for the left and the right hippocampus. From the table, we can observe that MDL achieves the best performance regarding three evaluation criteria. Specifically, RF achieves relatively lower performance in DSC because of the lower SEN value, MA obtains more stable results (small standard deviation), and MDL yields the best DSC (i.e., 0.893) which is much better than the second best DSC (i.e., 0.870) achieved by MA. Furthermore, our MDL method yields much better PPV value (0.890) in hippocampus segmentation. This implies that our method can effectively identify hippocampus regions from those confounding background regions, which is particularly useful in practical applications.
Furthermore, we also illustrate several typical segmentation results qualitatively in Fig. 5. As shown in the figure, the two competing methods (i.e., RF, and MA) cannot remove some confounding voxels very well. For instance, some regions can not be well connected in the segmentation results obtained by these two methods. On the contrary, the segmentation results achieved by our MDL methods are more smooth and accurate compared with RF and MA.
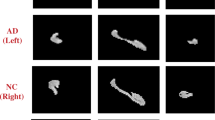
Also, several more segmentation results of our method are shown in Fig. 6. Figure 6a shows the original image, and we can notice that the hippocampus regions are difficult to be distinguished from neighboring tissues. It demonstrates that the accurate segmentation for the hippocampus is a very challenging task. The probability maps in Fig. 6b indicate that our method can describe the boundary of hippocampus accurately, and some confounding areas can be assigned very small probabilities. Therefore, smooth and accurate segmentation results can be achieved by our method. Besides, we also show the 3D rendering of the segmentation results in Fig. 6c. From Fig. 6c, we can see that the segmented hippocampus generated by our MDL method is smooth, implying that the global segmentation performance of our method is reasonable.
5 Discussion
In this section, we first investage the influence of the proposed joint learning strategy. We then study the the influence of our proposed averaging strategy for segmentation results of the left and the right hippocampus.
5.1 Comparision with single-task variants
In this group of experiments, we investigate the influence of our proposed joint learning strategy. Specifically, we compare our proposed MDL method with its two variants that only perform sing-task, including 1) MDL-1 that can conduct clinical score regression, and 2) MDL-2 that can only perform hippocampus segmentation. That is, MDL-1 uses the same Subnetwork 1 as our proposed network (see Fig. 2), while MDL-2 simply perform hippocampus segmentation using the Subnetwork 2. We report the experimental results achieved by three different methods (i.e., MDL-1, MDL-2, and MDL) in Table 4 and Fig. 7.
Scatter plots of the estimated MMSE scores vs. the real MMSE scores achieved by the proposed MDL method and its single-task variant MDL-1. Note that in MDL-1, we only adopt the Subnetwork-1 in Fig. 2 to perform clinical score regression. CC: Correlation Coefficient
From Table 4 and Fig. 7, we can make the following observations. First, in both tasks of regression and segmentation, the proposed joint learning model MDL is consistently superior to single-task models (i.e., MDL-1, and MDL-2). For instance, in the regression task for MMSE scores, the CC value obtained by MDL (0.559) is much higher than that obtained by MDL-1 (0.524). In the segmentation task, MDL achieves a DSC values of 0.893 which is better than that of MDL-2 (0.876). Second, from Fig. 7, we can observe that in clinical score regression task, MDL usually achieves higher CC value compared with its single-task variant (i.e., MDL-1).
5.2 Results using Single-side Hippocampus
In the above-mentioned experiments, for the hippocampus segmentation task, we report the results averaging the results for the left and the right hippocampus regions. Now we investigate the segmentation results for the left and the right hippocampus individually. Table 5 shows the results of methods using the left hippocampus (denoted as MDLl) and the right hippocampus (denoted as MDLr) separately. As shown in Table 5, the use of single-side hippocampus achieves comparable segmentation results and slightly lower performance in MMSE score regression, compared with our MDL method using the average segmentation results of the left and the right hippocampus. The results suggest that our methods using two-side hippocampus can achieve stable and accurate prediction.
6 Conclusions
In this paper, we proposed a multi-task deep learning (MDL) framework for joint hippocampus segmentation and clinical score regression based on convolutional neural networks. The prominent advantages of our MDL method lie on that we don’t need any time-consuming non-linear registration for pre-processing MR images, and features generated by MDL are consistent with subsequent learning models. The experimental results suggested that the proposed joint learning strategy can boost the performances of hippocampus segmentation and MMSE score regression. In the current work, we only use the MMSE score in the clinical score regression experiments. As a future work, we plan to incorporate more clinical scores into the proposed framework, to further improve the performance of our method.
References
Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M et al (2016) Tensorflow: A system for large-scale machine learning. In: Proceedings of the 12th USENIX symposium on operating systems design and implementation
Ahmed O B, Mizotin M, Benois-Pineau J, Allard M, Catheline G, Amar C B, Initiative A D N et al (2015) Alzheimer’s disease diagnosis on structural MR images using circular harmonic functions descriptors on hippocampus and posterior cingulate cortex. Comput Med Imaging Graph 44:13–25
Ashburner J (2007) A fast diffeomorphic image registration algorithm. NeuroImage 38(1):95–113
Barnes J, Boyes R, Lewis E, Schott J, Frost C, Scahill R, Fox N (2007) Automatic calculation of hippocampal atrophy rates using a hippocampal template and the boundary shift integral. Neurobiol Aging 28(11):1657–1663
Boyd S, Vandenberghe L (2004) Convex optimization. Cambridge University Press, Cambridge
Cao X, Gao Y, Yang J, Wu G, Shen D (2016) Learning-based multimodal image registration for prostate cancer radiation therapy. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, pp 1–9
Cao X, Yang J, Gao Y, Guo Y, Wu G, Shen D (2017) Dual-core steered non-rigid registration for multi-modal images via bi-directional image synthesis. Medical Image Analysis
Carmichael O T, Aizenstein H A, Davis S W, Becker J T, Thompson P M, Meltzer C C, Liu Y (2005) Atlas-based hippocampus segmentation in Alzheimer’s disease and mild cognitive impairment. NeuroImage 27(4):979–990
Chang C C, Lin C J (2011) Libsvm: A library for support vector machines. ACM Trans Intell Syst Technol 2(3):27
Chincarini A, Bosco P, Calvini P, Gemme G, Esposito M, Olivieri C, Rei L, Squarcia S, Rodriguez G, Bellotti R et al (2011) Local MRI analysis approach in the diagnosis of early and prodromal Alzheimer’s disease. NeuroImage 58 (2):469–480
Clark K A, Woods R P, Rottenberg D A, Toga A W, Mazziotta J C (2006) Impact of acquisition protocols and processing streams on tissue segmentation of T1 weighted MR images. NeuroImage 29(1):185–202
Coupé P, Manjón J V, Fonov V, Pruessner J, Robles M, Collins D L (2011) Patch-based segmentation using expert priors: Application to hippocampus and ventricle segmentation. NeuroImage 54(2):940–954
Coupé P, Eskildsen S F, Manjón J V, Fonov V S, Collins D L (2012) Simultaneous segmentation and grading of anatomical structures for patient’s classification: Application to Alzheimer’s disease. NeuroImage 59(4):3736–3747
Dagher A, Owen A M, Boecker H, Brooks D J (2001) The role of the striatum and hippocampus in planning: A PET activation study in Parkinson’s disease. Brain 124(5):1020–1032
Dill V, Franco A R, Pinho M S (2015) Automated methods for hippocampus segmentation: The evolution and a review of the state of the art. Neuroinformatics 13 (2):133
Folstein M F, Folstein S E, McHugh P R (1975) “mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 12(3):189–198
Hao Y, Wang T, Zhang X, Duan Y, Yu C, Jiang T, Fan Y (2014) Local label learning (LLL) for subcortical structure segmentation: Application to hippocampus segmentation. Hum Brain Mapp 35 (6):2674–2697
Heckemann R A, Hajnal J V, Aljabar P, Rueckert D, Hammers A (2006) Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. NeuroImage 33(1):115–126
Hyman B T, Van Hoesen G W, Damasio A R, Barnes C L (1984) Alzheimer’s disease: Cell-specific pathology isolates the hippocampal formation. Science 225:1168–1171
Iglesias J E, Sabuncu M R (2015) Multi-atlas segmentation of biomedical images: A survey. Med Image Anal 24(1):205–219
Jack C R, Petersen R C, O’Brien P C, Tangalos E G (1992) MR-based hippocampal volumetry in the diagnosis of Alzheimer’s disease. Neurology 42(1):183–183
Jack C R, Bernstein M A, Fox N C, Thompson P, Alexander G, Harvey D, Borowski B, Britson P J, L Whitwell J, Ward C (2008) The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J Magn Reson Imaging 27(4):685–691
Jin K, Peel A L, Mao X O, Xie L, Cottrell B A, Henshall D C, Greenberg D A (2004) Increased hippocampal neurogenesis in Alzheimer’s disease. Proc Natl Acad Sci 101(1):343–347
Kwak K, Yoon U, Lee D K, Kim G H, Seo S W, Na D L, Shim H J, Lee J M (2013) Fully-automated approach to hippocampus segmentation using a graph-cuts algorithm combined with atlas-based segmentation and morphological opening. Magn Reson Imaging 31(7):1190–1196
Lian C, Ruan S, Denoeux T (2015) An evidential classifier based on feature selection and two-step classification strategy. Pattern Recogn 48(7):2318–2327
Lian C, Ruan S, Denœux T, Jardin F, Vera P (2016) Selecting radiomic features from FDG-PET images for cancer treatment outcome prediction. Med Image Anal 32:257–268
Lindner C, Thiagarajah S, Wilkinson J, Consortium T, Wallis G, Cootes T (2013) Fully automatic segmentation of the proximal femur using random forest regression voting. IEEE Trans Med Imaging 32(8):1462–1472
Liu M, Zhang D, Shen D (2015) View-centralized multi-atlas classification for Alzheimer’s disease diagnosis. Hum Brain Mapp 36(5):1847–1865
Liu M, Zhang D, Chen S, Xue H (2016) Joint binary classifier learning for ECOC-based multi-class classification. IEEE Trans Pattern Anal Mach Intell 38 (11):2335–2341
Liu M, Zhang D, Shen D (2016) Relationship induced multi-template learning for diagnosis of Alzheimer’s disease and mild cognitive impairment. IEEE Trans Med Imaging 35(6):1463–1474
Liu M, Zhang J, Yap P T, Shen D (2017) View-aligned hypergraph learning for alzheimer’s disease diagnosis with incomplete multi-modality data. Med Image Anal 36:123–134
Liu M, Zhang J, Adeli E, Shen D (2018) Landmark-based deep multi-instance learning for brain disease diagnosis. Med Image Anal 43:157–168
Moodley K, Minati L, Contarino V, Prioni S, Wood R, Cooper R, D’incerti L, Tagliavini F, Chan D (2015) Diagnostic differentiation of mild cognitive impairment due to Alzheimer’s disease using a hippocampus-dependent test of spatial memory. Hippocampus 25(8):939–951
Pohl K M, Bouix S, Nakamura M, Rohlfing T, McCarley R W, Kikinis R, Grimson W E L, Shenton M E, Wells W M (2007) A hierarchical algorithm for MR brain image parcellation. IEEE Trans Med Imaging 26(9):1201–1212
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. arXiv:150504597
Sled J G, Zijdenbos A P, Evans A C (1998) A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 17(1):87–97
Tulving E, Markowitsch H J (1998) Episodic and declarative memory: Role of the hippocampus. Hippocampus 8(3):198–204
Zandifar A, Fonov V, Coupé P, Pruessner J, Collins DL, Initiative ADN et al (2017) A comparison of accurate automatic hippocampal segmentation methods. NeuroImage
Zarpalas D, Gkontra P, Daras P, Maglaveras N (2014) Accurate and fully automatic hippocampus segmentation using subject-specific 3D optimal local maps into a hybrid active contour model. IEEE J Trans Eng Health Med 2:1–16
Zhang J, Liang J, Zhao H (2013) Local energy pattern for texture classification using self-adaptive quantization thresholds. IEEE Trans Image Process 22(1):31–42
Zhang J, Gao Y, Wang L, Tang Z, Xia J J, Shen D (2016) Automatic craniomaxillofacial landmark digitization via segmentation-guided partially-joint regression forest model and multiscale statistical features. IEEE Trans Biomed Eng 63(9):1820–1829
Zhang J, Gao Y, Park SH, Zong X, Lin W, Shen D (2017) Structured learning for 3D perivascular spaces segmentation using vascular features. IEEE Transactions on Biomedical Engineering
Zhang J, Liu M, An L, Gao Y, Shen D (2017) Alzheimer’s disease diagnosis using landmark-based features from longitudinal structural MR images. IEEE Journal of Biomedical and Health Informatics
Zhang J, Liu M, Shen D (2017) Detecting anatomical landmarks from limited medical imaging data using two-stage task-oriented deep neural networks. IEEE Trans Image Process 26(10):4753–4764
Zhu X, Suk H I, Shen D (2014) A novel matrix-similarity based loss function for joint regression and classification in AD diagnosis. NeuroImage 100:91–105
Zhu X, Suk H I, Wang L, Lee SW, Shen D (2015) A novel relational regularization feature selection method for joint regression and classification in AD diagnosis. Medical Image analysis
Zhu Y, Zhu X, Kim M, Shen D, Wu G (2016) Early diagnosis of Alzheimer’s disease by joint feature selection and classification on temporally structured support vector machine. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, pp 264–272
Zhu Y, Zhu X, Zhang H, Gao W, Shen D, Wu G (2016) Reveal consistent spatial-temporal patterns from dynamic functional connectivity for autism spectrum disorder identification. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, pp 106–114
Zhu Y, Zhu X, Kim M, Kaufer D, Wu G (2017) A novel dynamic hyper-graph inference framework for computer assisted diagnosis of neuro-diseases. In: International Conference on Information Processing in Medical Imaging, Springer, pp 158–169
Zou K H, Warfield S K, Bharatha A, Tempany C M, Kaus M R, Haker S J, Wells W M, Jolesz F A, Kikinis R (2004) Statistical validation of image segmentation quality based on a spatial overlap index 1: Scientific reports. Acad Radiol 11(2):178–189
Acknowledgments
This study was supported by National Natural Science Foundation of China (Nos. 61703301, 61573023) one more support grant and University Science and Technology Project of Shandong Province (No. J17KA086).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cao, L., Li, L., Zheng, J. et al. Multi-task neural networks for joint hippocampus segmentation and clinical score regression. Multimed Tools Appl 77, 29669–29686 (2018). https://doi.org/10.1007/s11042-017-5581-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-017-5581-1