Abstract
Vitexin (apigenin-8-C-d-glucopyranoside) is a flavonoid isolated from natural sources. It has been employed as an anti-oxidant, anti-inflammatory, and anti-cancer agent, and is used as a traditional Chinese medicine to treat a variety of illnesses. The present study investigated the effect of vitexin on osteoblast differentiation of C3H10T1/2 mesenchymal stem cells, MC3T3-E1 preosteoblast, mouse calvarial primary cells, and primary bone marrow stem cells (BMSCs). RT-PCR and quantitative PCR demonstrated that vitexin increased mRNA expression of the osteogenic genes distal-less homeobox 5 (Dlx5) and Runxt-related transcription factor 2 (Runx2). Vitexin also increased the Dlx5 and Runx2 protein levels, Smad1/5/9 phosphorylation, and alkaline phosphatase (ALP) activity. In addition, vitexin increased Runx2-luciferase activity. Moreover, knockdown of Runx2 attenuated the increase in ALP activity induced by vitexin. These results demonstrate that vitexin enhances osteoblast differentiation via Runx2.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bone is a dynamic tissue that is remodeled via the actions of bone-forming osteoblasts and bone-resorbing osteoclasts [1]. An imbalance in these actions can lead to various diseases such as osteoporosis, bone metastasis, and tumor-induced bone destruction, rheumatoid arthritis, and osteosclerosis [2, 3]. Therefore, bone remodeling is very important. Osteoblasts are derived from mesenchymal stem cells (MSCs), which are pluripotent cells that can differentiate into osteoblasts, adipocytes, myocytes, and chondrocyte [4,5,6].
Runx2 is a member of the runt domain gene family, also known as Cbfa1. Runx2 has first demonstrated in 1997 that the role of it was essential in osteoblast differentiation [7]. Runx2 role was important for the cell cycle and maturation stages during osteoblast differentiation [8]. Runx2 is a transcription factor that increases alkaline phosphatase (ALP) activity and matrix mineralization in vitro [9,10,11]. Previous reports indicate that Runx2 is a transcription factor integral to osteoblast differentiation.
Vitexin (apigenin-8-C-d-glucopyranoside) is a flavonoid isolated from plants [12]. It has been employed as an anti-oxidant, anti-inflammatory, and anti-cancer agent, and is used as a traditional Chinese medicine to treat a variety of illnesses [12,13,14,15]. Vitexin elicits anti-inflammatory effects by inhibiting secretion of tumor necrosis factor-α [16] and exerts anti-cancer effects by upregulating p53 [12]. A recent study reported that vitexin suppresses osteoclastogenesis and reduces lipopolysaccharide-induced osteolysis [17]. However, the effect of vitexin alone on osteogenic differentiation has not been investigated at the mRNA level.
The present study investigated whether vitexin enhances osteogenic differentiation of C3H10T1/2 cells. We show that vitexin induced Smad 1/5/9 phosphorylation and enhanced osteogenic differentiation via Runx2.
Materials and methods
Animals
ICR mice used to isolate primary cells were maintained following the institutional guidelines of the Committee for Laboratory Animal Care and Use of Daegu University (DUIACC-12020/4-0313-006). Animals were maintained at 20–22 °C, 50–60% humidity, and 12-h-dark/light cycle conditions.
Primary mouse calvarial cells isolation
Isolated from calvaria in 2 days ICR mice. Isolated calvaria was incubated for 20 min in 5 mL PBS containing 0.1% collagenase (Gibco, Grand Island, NY, USA) and 0.2% dispase (Gibco, Grand Island, NY, USA) with shanking. And then, the supernatant was collected using for 8 min at 1800 RPM. This step was repeated 5 times. Primary mouse calvarial cells were maintained in Minimum Essential Medium Eagle-alpha modification (Gibco, Grand Island, NY, USA) with 10% fetal bovine serum (FBS) (Atlas Biologicals, Fort Collins, CO, USA), 100 units/mL penicillin, and 100 μg/mL streptomycin in humidified air containing 5% CO2 at 37 °C.
Primary mouse bone marrow stromal cells (BMSCs) isolation
BMSCs were isolated from femur in 10 weeks ICR mice. Isolated femur is centrifuged at 10,000×g in a collection tube made using 200 mL pipette tips and e-tube to extract bone marrow. Isolated bone marrow cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco, Grand Island, NY, USA) containing 10% FBS (Atlas Biologicals), 100 units/mL penicillin, and 100 μg/mL streptomycin in humidified air containing 5% CO2 at 37 °C [18].
Cell culture
C3H10T1/2 mouse MSCs (ATCC, Manassas, VA, USA) were maintained in DMEM (Gibco) containing 10% FBS (Atlas Biologicals), 100 units/ml penicillin, and 100 μg/mL streptomycin in humidified air containing 5% CO2 at 37 °C. To evaluate the effects of vitexin, cells were cultured in medium containing 10 μM vitexin. The medium with vitexin was replaced every 2 days.
MTT assay
C3H10T1/2 cells were seeded into 48-well-plates at a density of 3 × 104 cells per well and treated with 1–100 μM vitexin (Cayman Chemical, Ann Arbor, MI, USA) for 4 days. Cell viability was monitored by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide (MTT; Sigma-Aldrich, St.Louis, MO, USA) assay. Cells were incubated with MTT solution for an additional 2 h, and then 200 μL of dimethyl sulfoxide was added to each well. Finally, absorbance at 570 nm was measured using a multi-plate reader (Infinite M 200 Pro; Tecan Austria GmbH, Grödig, Austria).
RT-PCR and quantitative PCR (Q-PCR)
Total RNA was isolated from cells using Tri-Solution (Bioscience Technology, Gyeongsan, Korea) according to the manufacturer’s instructions. RT-PCR was performed using 5 µg of total RNA. The reaction conditions were as follows: an initial denaturation step at 94 °C for 1 min followed by 25–30 cycles of denaturation at 94 °C for 30 s, annealing at the optimal temperature for each primer pair for 30 s, and extension at 72 °C for 30 s, with a final extension at 72 °C for 5 min. The primer sequences were as follows: β-actin forward 5′-TTCTTTGCAGCTCCTTCGTTGCCG-3′ and reverse 5′-TGGATGGCTACGTACATGGCTGGG-3′ (25 cycles), Dlx5 forward 5′-CAGAAGAGTCCCAAGCATCC-3′ and reverse 5′-GAGCGCTTTGCCATAAGAAG-3′ (30 cycles), Runx2 forward 5′-CCGCACGACAACCGCACCAT-3′ and reverse 5′-CGCTCCGGCCCACAAATCTC-3′ (30 cycles), ALP forward 5′-ATCTTTGGTCTCGCTCCCATG-3′ and reverse 5′-TTTCCCGTTCACCGTCCAC-3′ (30 cycles).
ALP staining
C3H10T1/2 cells were cultured with vitexin (10 μM) for 8 days. ALP staining was performed using standard protocols. Cultured cells were fixed with 10% formalin, rinsed twice with deionized water, incubated with BCIP®/NBT solution (Sigma-Aldrich) for 15 min, washed again.
Western blotting
Total cells were harvested using an EzRIPA Lysis Kit (ATTO Technology, Tokyo, Japan) and centrifuged at 12,000 g for 10 min at 4 °C. Total proteins in the lysates were quantified by the Bradford assay, separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to a polyvinylidene fluoride membrane. After blocking with 5% skim milk prepared in Tris-buffered saline containing Tween 20, the membrane was incubated with specific primary antibodies (1:1000). Signals were detected using enhanced chemiluminescence reagents (Advansta, Menlo Park, CA, USA). Densitometric analysis of the membrane was performed using a FUSION Solo analyzer system (Vilber Lourmat, Eberhardzell, Germany).
Runx2 silencing and Runx2-luciferase assay
Deprotected and annealed Runx2-targeting small interfering RNAs (siRunx2) were chemically synthesized (Bioneer, Daejeon, Korea) and transfected according to the manufacturer’s instructions. C3H10T1/2 cells were transfected with siRunx2 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The sequences were as follows: siRunx2(1) sense 5′-GAGCUAUUAAAGUGACAGU-3′ and antisense 5′-ACUGUCACUUUAAUAGCUC-3′, siRunx2(2) sense 5′-UGAUGACUCUAAACCUAGU-3′ and antisense 5′-ACUAGGUUUAGAGUCAUCA-3′, siRunx2(3) sense 5′-GAGGAUGUACUGUGAUCAU-3′ and antisense 5′-AUGAUCACAGUACAUCCUC-3′. Luciferase activity was measured using a luciferase reporter assay system (Promega, Madison, WI, USA) and a luminometer according to the manufacturer’s instructions.
Statistical analysis
All experiments were repeated at least three times. Statistical analyses were performed using the Student’s t-test or an analysis of variance, followed by Duncan’s multiple comparison test. P values less than 0.05 were considered significant. Results are presented as the mean ± SEM of triplicate independent samples.
Results
Effect of vitexin on the viability of C3H10T1/2 cells
The chemical structure of vitexin is shown in Fig. 1a. Vitexin was soluble in dimethyl sulfoxide. The effects of various concentrations of vitexin the cell viability of C3H10T1/2 cells were determined by the MTT assay (Fig. 1b). Cells were treated with 1, 5, 10, 50, and 100 μM vitexin for 4 days. Treatment with up to 50 μM vitexin did not elicit cytotoxic effect; however, treatment with 100 μM vitexin decreased the viability of C3H10T1/2 cells. Therefore, 10 μM vitexin was used in subsequent experiments.
Effect of vitexin on the viability of C3H10T1/2 cells. a Chemical structure of vitexin. b C3H10T1/2 cells were treated with vitexin for 4 days, and then their viability was measured by the MTT assay. Data are the mean ± SEM of three individual experiments (*p < 0.05 compared with untreated control cells)
Effect of vitexin on osteogenic differentiation of C3H10T1/2 cells via Smad phosphorylation
The effect of vitexin on mRNA expression of osteogenic genes (Dlx5 and Runx2) was determined by RT-PCR and Q-PCR. Vitexin significantly increased mRNA expression of Dlx5 and Runx2 (Fig. 2a, b). Moreover, vitexin increased phosphorylation of Smad-1/5/9 after 1 h and the maximal protein levels of Dlx5 and Runx2 at 4 h. The phosphorylation of Smad-2/3 increased from 1 h, whereas Smad-4 increased at 6 h and Smad6/7 was not expressed (Fig. 2c). Moreover, ALP staining revealed that treatment with vitexin for 8 days significantly increased the activity of ALP (Fig. 2d). Furthermore, the induction of osteogenic gene by vitexin showed an additional effect when treated with BMP2 (Fig. 2e, f). These results indicate that vitexin induces osteogenic differentiation of C3H10T1/2 cells.
Vitexin induces osteogenic differentiation of C3H10T1/2 cells. (a and b) C3H10T1/2 cells were treated with 10 μM vitexin for 0–6 h harvested. mRNA expression of osteogenic marker genes was measured by RT-PCR (a) and real-time PCR (b). c C3H10T1/2 cells were treated with vitexin for 0–6 h, and then the levels of osteogenic proteins were determined by western blotting. d C3H10T1/2 cells were treated with vitexin for 8 days and then ALP staining was performed to determine ALP activity. e and f C3H10T1/2 cells were treated with 250 ng/ml BMP2 and/or 10 μM vitexin. mRNA expression and protein level of osteogenic marker genes was measured by RT-PCR (e) western blot analysis (f). (*p < 0.05, and **p < 0.01 compared to the untreated control)
Effect of vitexin on osteogenic differentiation of MC3T3-E1 cells via Smad phosphorylation
Vitexin significantly increased mRNA expression of Dlx5 and Runx2 in MC3T3-E1 cells (Fig. 3a, b). In addition, vitexin also increased phosphorylation of Smad-1/5/9 after 1 h and the protein levels of Dlx5 and Runx2 were increased. The phosphorylation of Smad-2/3 decreased, whereas Smad-4 increased and Smad6/7 was decreased (Fig. 3c). Moreover, ALP staining revealed that treatment with vitexin significantly increased the activity of ALP (Fig. 3d). Furthermore, the vitexin-induced osteogenic gene expression showed an additional effect when treated with BMP2 in MC3T3-E1 cells (Fig. 3e, f). These results indicate that vitexin induces osteogenic differentiation of MC3T3-E1 cells.
Vitexin induces osteogenic differentiation of MC3T3-E1 cells. a and b MC3T3-E1 cells were treated with 10 μM vitexin for 0–6 h harvested. mRNA expression of osteogenic marker genes was measured by RT-PCR (a) and real-time PCR (b). c MC3T3-E1 cells were treated with vitexin for 0–6 h, and then the levels of osteogenic proteins were determined by western blotting. (d) MC3T3-E1 cells were treated with vitexin for 8 days and then ALP staining was performed to determine ALP activity. e and f MC3T3-e1 cells were treated with 250 ng/mL BMP2 and/or 10 μM vitexin. mRNA expression and protein level of osteogenic marker genes was measured by RT-PCR (e) western blot analysis (f). (*p < 0.05, and **p < 0.01 compared to the untreated control)
The Knockdown of Runx2 attenuates the effects of vitexin
We confirmed whether vitexin directly regulates Runx2 promoter activity. Vitexin increased Runx2 P1-luciferase activity in both C3H10T1/2 and MC3T3-E1 cells (Fig. 4a, d). And then, we knocked down Runx2 expression in C3H10T1/2 and MC3T3-E1 cells using siRunx2 respectively. We confirmed that siRunx2 depleted mRNA levels of Runx2 and ALP (Fig. 4b, e). In addition, Knockdown of Runx2 markedly reduced ALP staining in vitexin-treated cells (Fig. 4c, f). Taken together, these results demonstrate that vitexin induces osteogenic differentiation through Runx2.
The knockdown of Runx2 attenuates the effects of vitexin on osteoblast differentiation. a and d C3H10T1/2 and MC3T3-E1 cells were transfected with Runx2 P1-luciferase (0.4 μg) and then treated with vitexin (+; 10 μM) Relative luciferase activity was measured. b and e C3H10T1/2 and MC3T3-E1 cells were transfected with siRunx2 (1), (2), or (3) for 6 h and then treated with vitexin (+; 10 μM) for 4 h. Total RNA was harvested, and the expression of osteogenic genes was assessed by real-time PCR. c and f C3H10T1/2 and MC3T3-E1 cells were transfected with siRunx2 (1), (2), or (3) for 6 h and then incubated in the absence or presence of vitexin for 8 days. The graph has shown using ImageJ. Data are the mean ± SEM of three individual experiments (*p < 0.05, and **p < 0.01 compared to the untreated control, #p < 0.05, ##p < 0.01, and ###p < 0.005 compared with the vitexin treated group)
Effect of vitexin on primary mouse calvarial cells and bone marrow stromal cells
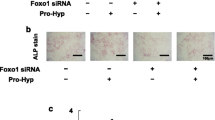
To examine whether vitexin regulates osteoblast differentiation in primary calvarial osteoblasts and bone marrow stromal cells, cells were cultured with 10 μM vitexin for 0, 1, 2, 4, and 6 h. Western blot analysis showed that vitexin significantly increased the Dlx5 levels at 4 h (Fig. 5a). To confirm the stimulation of ALP activity by Vitexin further, cells were treated with 10 μM vitexin for 8 days. Vitexin markedly increased ALP activity (Fig. 5b). Furthermore, Runx2 silencing also reduced vitexin-induced ALP activity in primary mouse calvarial cells (Fig. 5c). In bone marrow stromal cells (BMSCs), vitexin increased the Dlx5 protein levels at 2 and 4 h (Fig. 5d). ALP staining results showed that vitexin increased ALP activity and Runx2 silencing decreased vitexin-induced ALP activity in BMSCs (Fig. 5e, f). Taken together, these results suggest that vitexin induce osteoblast differentiation on ex vivo such as in vitro.
Effect of vitexin on osteogenic differentiation of primary mouse calvarial cells and BMSCs. a Primary calvarial osteoblasts were determined by western blotting after treated with vitexin for 0–6 h. b ALP protein expression in primary calvarial cells cultured with vitexin (10 μM) for 8 days (lower panel) and densitometry analysis was performed by the ImageJ program (upper panel). c In primary cells, ALP staining in transfected with siRunx2 (1), (2), or (3) for 6 h and then incubated in the absence or presence of vitexin for 8 days was measured. d Primary BMSCs were determined by western blotting after treated with vitexin for 0–6 h. e ALP protein expression in primary BMSCs cultured with vitexin (10 μM) for 8 days (lower panel) and densitometry analysis was performed by the ImageJ program (upper panel). f In primary BMSCs, ALP staining in transfected with siRunx2 (1), (2), or (3) for 6 h and then incubated in the absence or presence of vitexin for 8 days was measured. The graph has shown using ImageJ. Data are the mean ± SEM of three individual experiments (**p < 0.01, and ***p < 0.005 compared to the untreated control, #p < 0.05, ##p < 0.01, and ###p < 0.005 compared with the vitexin treated group)
Discussion
The balance between osteoclasts and osteoblasts is very important. Anim this balance in the actions of these cells typically causes osteoporosis and bone diseases [2, 3]. Phosphorylation of Smad-1/5/8/9 and increased expression of Runx2 and ALP induce differentiation of osteoblasts, which are involved in bone formation [19,20,21]. The current study demonstrated that vitexin positively regulates osteoblast differentiation by increasing expression of Runx2 and phosphorylation-Smad (Fig. 2). In addition, a recent study reported that vitexin inhibits the effects of osteolysis induced by RANKL-induced osteoclastogenesis and inflammation, thereby strengthening bones [17]. Our results were in agreement with this previous report (Fig. 6).
Cell survival and proliferation must be maintained as osteoblast differentiation progresses because osteoblasts migrate to the bone surface and proliferate prior to bone formation [22]. The MTT assay confirmed that treatment with vitexin at concentrations of up to 50 μM did not reduce the viability of C3H10T1/2 cells. A previous study also reported that treatment with vitexin at concentrations of up to 50 μM dose did not elicit cytotoxic effect on osteoblasts [17]. Therefore, we treated cells with 10 μM vitexin to ensure that they survived (Fig. 1b). Expression of specific marker genes increases during osteoblast differentiation. We showed that treatment with 10 μM vitexin increased expression of osteoblast differentiation marker genes (Fig. 2a, b).
In conclusion, we demonstrated that vitexin enhances osteoblast differentiation via phosphorylation of Smad1/5/9 and expression of Runx2 at invitro and ex vivo. This study provides evidence that vitexin can be used as a bone-modifying agent.
Abbreviations
- MSC:
-
Mesenchymal stem cell
- BMSC:
-
Bone marrow stromal cell
- BMP:
-
Bone morphogenetic proteins
- Runx2:
-
Runt-related transcription factor 2
- TNF-α:
-
Tumour necrosis factor-α
- ALP:
-
Alkaline phosphatase
- DMEM:
-
Dulbecco's Modified Eagle's Medium
- Dlx5:
-
Distal-less homeobox 5
- DMSO:
-
Dimethyl sulfoxide
- Smad:
-
Small mothers against
- FBS:
-
Fetal bovine serum
References
Olsen BR, Reginato AM, Wang W (2000) Bone development. Annu Rev Cell Dev Biol 16:191–220. https://doi.org/10.1146/annurev.cellbio.16.1.191
Abu-Amer Y, Darwech I, Clohisy JC (2007) Aseptic loosening of total joint replacements: mechanisms underlying osteolysis and potential therapies. Arthrit Res Ther 9(Suppl 1):S6. https://doi.org/10.1186/ar2170
Mbalaviele G, Novack DV, Schett G, Teitelbaum SL (2017) Inflammatory osteolysis: a conspiracy against bone. J Clin Investig 127(6):2030–2039. https://doi.org/10.1172/JCI93356
Ducy P, Karsenty G (1995) Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol 15(4):1858–1869. https://doi.org/10.1128/mcb.15.4.1858
Grigoriadis AE, Heersche JN, Aubin JE (1988) Differentiation of muscle, fat, cartilage, and bone from progenitor cells present in a bone-derived clonal cell population: effect of dexamethasone. J Cell Biol 106(6):2139–2151. https://doi.org/10.1083/jcb.106.6.2139
Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR (1999) Multilineage potential of adult human mesenchymal stem cells. Science 284(5411):143–147
Li Y, Ge C, Long JP, Begun DL, Rodriguez JA, Goldstein SA, Franceschi RT (2012) Biomechanical stimulation of osteoblast gene expression requires phosphorylation of the RUNX2 transcription factor. J Bone Miner Res 27(6):1263–1274. https://doi.org/10.1002/jbmr.1574
Stein GS, Lian JB, van Wijnen AJ, Stein JL, Montecino M, Javed A, Zaidi SK, Young DW, Choi JY, Pockwinse SM (2004) Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene 23(24):4315–4329. https://doi.org/10.1038/sj.onc.1207676
Banerjee C, McCabe LR, Choi JY, Hiebert SW, Stein JL, Stein GS, Lian JB (1997) Runt homology domain proteins in osteoblast differentiation: AML3/CBFA1 is a major component of a bone-specific complex. J Cell Biochem 66(1):1–8
Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89(5):747–754. https://doi.org/10.1016/s0092-8674(00)80257-3
Harada H, Tagashira S, Fujiwara M, Ogawa S, Katsumata T, Yamaguchi A, Komori T, Nakatsuka M (1999) Cbfa1 isoforms exert functional differences in osteoblast differentiation. J Biol Chem 274(11):6972–6978. https://doi.org/10.1074/jbc.274.11.6972
He M, Min JW, Kong WL, He XH, Li JX, Peng BW (2016) A review on the pharmacological effects of vitexin and isovitexin. Fitoterapia 115:74–85. https://doi.org/10.1016/j.fitote.2016.09.011
Ganesan K, Xu B (2017) Molecular targets of vitexin and isovitexin in cancer therapy: a critical review. Ann N Y Acad Sci 1401(1):102–113. https://doi.org/10.1111/nyas.13446
Min JW, Hu JJ, He M, Sanchez RM, Huang WX, Liu YQ, Bsoul NB, Han S, Yin J, Liu WH, He XH, Peng BW (2015) Vitexin reduces hypoxia-ischemia neonatal brain injury by the inhibition of HIF-1alpha in a rat pup model. Neuropharmacology 99:38–50. https://doi.org/10.1016/j.neuropharm.2015.07.007
Zhang Y, Bao B, Lu B, Ren Y, Tie X, Zhang Y (2005) Determination of flavone C-glucosides in antioxidant of bamboo leaves (AOB) fortified foods by reversed-phase high-performance liquid chromatography with ultraviolet diode array detection. J Chromatogr A 1065(2):177–185
Borghi SM, Carvalho TT, Staurengo-Ferrari L, Hohmann MS, Pinge-Filho P, Casagrande R, Verri WA Jr (2013) Vitexin inhibits inflammatory pain in mice by targeting TRPV1, oxidative stress, and cytokines. J Nat Prod 76(6):1141–1149. https://doi.org/10.1021/np400222v
Jiang J, Jia Y, Lu X, Zhang T, Zhao K, Fu Z, Pang C, Qian Y (2019) Vitexin suppresses RANKL-induced osteoclastogenesis and prevents lipopolysaccharide (LPS)-induced osteolysis. J Cell Physiol. https://doi.org/10.1002/jcp.28378
Maridas DE, Rendina-Ruedy E, Le PT, Rosen CJ (2018) Isolation, culture, and differentiation of bone marrow stromal cells and osteoclast progenitors from mice. J Vis Exp. https://doi.org/10.3791/56750
Erceg I, Tadic T, Kronenberg MS, Marijanovic I, Lichtler AC (2003) Dlx5 regulation of mouse osteoblast differentiation mediated by avian retrovirus vector. Croat Med J 44(4):407–411
Lee MH, Kim YJ, Kim HJ, Park HD, Kang AR, Kyung HM, Sung JH, Wozney JM, Kim HJ, Ryoo HM (2003) BMP-2-induced Runx2 expression is mediated by Dlx5, and TGF-beta 1 opposes the BMP-2-induced osteoblast differentiation by suppression of Dlx5 expression. J Biol Chem 278(36):34387–34394. https://doi.org/10.1074/jbc.M211386200
Wu M, Chen G, Li YP (2016) TGF-beta and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res 4:16009. https://doi.org/10.1038/boneres.2016.9
Rodan GA, Noda M (1991) Gene expression in osteoblastic cells. Crit Rev Eukaryot Gene Expr 1(2):85–98
Acknowledgements
This research was supported by Daegu University Research Grant 2018.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None of the authors have a conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kim, KM., Son, HE., Min, HY. et al. Vitexin enhances osteoblast differentiation through phosphorylation of Smad and expression of Runx2 at in vitro and ex vivo. Mol Biol Rep 47, 8809–8817 (2020). https://doi.org/10.1007/s11033-020-05929-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05929-y