Abstract
Hydrogen peroxide (H2O2) is known to accumulate in plants during abiotic stress conditions and also acts as a signalling molecule. In this study, Arabidopsis thaliana transgenics overexpressing cytosolic CuZn-superoxide dismutase (PaSOD) from poly-extremophile high-altitude Himalayan plant Potentilla atrosanguinea, cytosolic ascorbate peroxidase (RaAPX) from Rheum australe and dual transgenics overexpressing both the genes were developed and analyzed under salt stress. In comparison to wild-type (WT) or single transgenics, the performance of dual transgenics under salt stress was better with higher biomass accumulation and cellulose content. We identified genes involved in cell wall biosynthesis, including nine cellulose synthases (CesA), seven cellulose synthase-like proteins together with other wall-related genes. RNA-seq analysis and qPCR revealed differential regulation of genes (CesA 4, 7 and 8) and transcription factors (MYB46 and 83) involved in secondary cell wall cellulose biosynthesis, amongst which most of the cellulose biosynthesis gene showed upregulation in single (PaSOD line) and dual transgenics at 100 mM salt stress. A positive correlation between cellulose content and H2O2 accumulation was observed in these transgenic lines. Further, cellulose content was 1.6–2 folds significantly higher in PaSOD and dual transgenic lines, 1.4 fold higher in RaAPX lines as compared to WT plants under stress conditions. Additionally, transgenics overexpressing PaSOD and RaAPX also displayed higher amounts of phenolics as compared to WT. The novelty of present study is that H2O2 apart from its role in signalling, it also provides mechanical strength to plants and aid in plant biomass production during salt stress by transcriptional activation of cellulose biosynthesis pathway. This modulation of the cellulose biosynthetic machinery in plants has the potential to provide insight into plant growth, morphogenesis and to create plants with enhanced cellulose content for biofuel use.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salinity stress is one of the major environmental factors that severely affect plant growth and productivity [1]. Abiotic stresses lead to excessive production of reactive oxygen species (ROS) such as, free radicals, hydrogen peroxide (H2O2) and singlet oxygen (1O2). At high concentration, these ROS molecules cause damage to cells by lipid peroxidation, oxidation of proteins and damage to nucleic acids, while at low concentration these molecules act as signalling molecules, thereby regulating plant response to stress [2, 3]. As a signal, ROS molecules regulate a large network of genes, which play important role in acclimation against abiotic stress [4]. Among enzymes, involved in ROS production and scavenging, superoxide dismutase (SOD) plays a crucial role, as its enzymatic action results in the formation of H2O2 which is then detoxified into water and oxygen by ascorbate peroxidase (APX) [5]. H2O2 is considered to be more stable and freely diffusible molecule within the cell compartments than other ROS molecules [6].
During abiotic stress, ultrastructural properties of plant cell wall give protection and act as the first line of defence against any kind of detrimental effects on plants [7]. In plant cell wall, cellulose constitutes the major component of both, primary and secondary cell walls and represents a major sink for photosynthesis products [8]. Cellulose synthases (CesA) is the major enzyme involved in cellulose synthesis and there are about 10 CESA genes in the Arabidopsis genome [9]. Among different CESAs, CESA1, CESA3, and CESA6 are involved in primary cell wall and CESA4, CESA7, and CESA8 for secondary cell wall depositions [10]. Cellulose microfibrils not only determine cell shape and patterns of development, but are also involved in stress response [11]. Salinity for a prolonged period in barley roots can regulate various genes involved in cellulose biosynthesis [12]. A similar effect has been reported in the cell wall of petioles of an aspen hybrid, where the increased rigidity acts as a barrier for salt permeation [13]. To survive under abiotic stress, improvement of secondary cell wall thickening by increased deposition of hemicellulose, cellulose [13] and through enhanced lignin content [14,15,16] in the cell wall have been reported previously. In Arabidopsis, several CesA genes showed implication in salt stress response [17], but the detailed experimental investigation is still lacking.
In addition to the role of H2O2 as a signalling molecule, its involvement in other crucial functions has been reported, as well. Role of H2O2 signalling in cellulose synthase subunit dimerization and secondary cell wall cellulose biosynthesis was first observed in cotton fibers [18]. Further, Kim and Barbara [19] reported that H2O2 generated by extracellular SOD and peroxidases in developing cotton fibers is involved in the biogenesis of primary and secondary cell wall. Role of H2O2 in increasing cell expansion and root diameter in rice by enhanced accumulation of pectin was shown recently by Xiong et al. [20]. We have also depicted the role of H2O2 in enhancing secondary cell wall thickening by enhanced lignification to mitigate salt stress [15, 21].
To understand the changes in the metabolic pool in response to salinity stress, an extensive study of cell wall-related genes is obligatory. Transgenic plants that overexpress genes involved in scavenging ROS have become valuable tools in determining the physiological functions of these genes. Based on our previous work [14, 15], where we have reported that the simultaneous expression of PaSOD and RaAPX induced downstream transcription factors and genes which are specifically involved in lignin deposition under salt stress. Since, secondary cell wall includes cellulose, hemicellulose, and lignin, we tried to find out the effect of PaSOD and RaAPX over-expression on cellulose biosynthesis. Thus, the aim of the present study was to provide a detailed analysis of secondary cell wall cellulose-related gene expression in response to salt stress in WT and transgenic lines. Using RNA-seq approach, we analysed expression of nine cellulose synthases, along with the genes involved in cell wall biogenesis (namely sucrose synthase, SS; cellulose synthase interactive protein, CSI; cellulose micro-fibril organisation protein, COBRA; cellulose synthase-like gene, CSLA; cellulose biosynthesis gene, KOB; β-1,4-glucanase, KORIGAN) and the specific transcription factors (WRKY, MYB, NAC), which are directly or indirectly involved in cellulose biosynthesis in secondary cell wall. The present work stands out from the previous work in terms of major focus on cellulose biosynthesis pathway genes and transcription factors. We present a mechanism in which the production of H2O2 promotes cellulose biosynthesis by increased CesA activity to promote biomass production under salt stress.
Materials and methods
Development of transgenic plants
Full-length cDNAs of PaSOD, (GenBank accession EU532614.1) and RaAPX, (GenBank accession DQ078123.2) from Potentilla atrosanguinea and Rheum australe, respectively were overexpressed in Arabidopsis thaliana (Ecotype Columbia 0) as described earlier by Gill et al. [22]. The seeds were collected and screened on Murashige and Skoog [23] medium supplemented with 20 µg ml− 1 hygromycin. Homozygous (T3 generation) transgenic PaCuZn-SOD and RaAPX plants were crossed to obtain dual transgenics. Putative transgenics were screened by PCR using gene-specific primers (Supplementary Table S1). Single copy gene insertion of each of Cu/Zn-SOD and APX, in transgenics, was determined in the T2 Generation. The results were statistically analyzed by χ2 Test [14].
Gene-specific semi-quantitative RT-PCR
Total RNA was isolated from transgenics and WT plants using iRIS solution [24]. One microgram of total RNA was used for oligo (dT) primed first-strand cDNA synthesis in 20 µl reaction using Superscript III reverse transcriptase (Invitrogen). This cDNA was used in 27 cycle PCR using gene-specific primers for PaSOD and RaAPX genes. All the experiments were conducted in three biological replicates.
Plant growth and salt stress treatment
Seeds of transgenic lines (T3) over-expressing PaSOD (S26, S15), RaAPX (APX20, APX2), both genes (18O, 19c) and WT were grown in Murashige and Skoog (MS) medium for 10 days and transplanted thereafter to the soil mixture of vermiculite: peat moss: perlite (1:1:1) with MS medium and in the greenhouse under a 16 h light and 8 h dark cycle at 20 ± 1 °C and light intensity of 60–70 µmol PPFD m− 2 s− 1. For stress treatment, 21d old seedlings of WT, and transgenics were supplemented with the desired concentration of NaCl (0, 50, 100 and 150 mM). Three biological replicates were collected from each sample at respective time points after salt stress.
Assays of antioxidant enzymes
Total enzyme activity of SOD and APX was estimated after 1 h and 24 h of salt stress. Total SOD activity was estimated as the intensity of NBT reduction using spectrophotometer as described earlier [25]. All the experiments were conducted in three biological replicates.
Determination of H2O2 content
The level of H2O2 was measured following Sonja et al. [26] method with some modifications [14].
Estimation of cellulose, proline and total soluble sugars content
The sample was cut into small segments and resuspended in 500 µl of an acetic acid/nitric acid/water solution (8: 1: 2, v: v: v), as described by Updegraff [27]. The samples were boiled in a water bath for 2 h. The acid insoluble material was recovered by centrifugation at 6700 g for 10 min. The pellets were washed with water and acetone and dried at 30 °C overnight. The dried samples were resuspended in 67% (v: v) of sulfuric acid. Cellulose was quantified colorimetrically using the phenol–sulfuric acid method [28]. Cellulose content was calculated as µg glucose in cellulose per mg of dry weight. Total Soluble sugar (TSS) content was determined by the anthrone method. Free proline content was estimated using the acid ninhydrin method [29]. All the experiments were conducted in three biological replicates.
In-situ ROS and H2O2 detection in plants
In situ ROS detection was based on the principle of NBT (nitroblue tetrazolium) reduction to blue formazan by O2−. and H2O2 was visually detected in the leaves of plants by using 3,3-diaminobenzidine (DAB) as substrate as described by Orozco-Cardenas and Ryan [30] with some modifications.
Transcriptome sequencing, assembly, and annotation
RNA-Seq libraries were made from 2 µg RNA from each of these samples using TruSeq RNA sample prep kit (Illumina) as per manufacturer’s instruction. All the experiments were conducted in three biological replicates and two-time points were taken (1 h and 24 h) after stress treatment. Rest of the method was done as per Shafi et al. [14].
Quantitative real-time PCR analysis (qRT-PCR)
Total RNA was pretreated with RNase-free DNase I (Invitrogen, USA) followed by first strand cDNA synthesis with 2 µg of total RNA using superscript III (Invitrogen, USA) according to the manufacturer’s instructions. Gene-specific primers (Supplementary Table S1) were designed using Primer 3 software [31] and a β-actin gene (AT3G18780) was used as the endogenous control. Gene expression was performed on a Stratagene Mx3000P system (Agilent Technologies, Germany) using 2× Brilliant III SYBR@ Green qPCR Master Mix (Agilent Technologies, Germany). All qPCRs assays were run with biological triplicates with a no-template control to check for contamination.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank. Transcriptomic data were deposited into the Sequence Read Archive Handbook (http://www.ncbi.nlm.nih.gov/books/NBK47529/#SRA_Quick_Sub_BK.BioProject) under SRA Accession SRP029598 and Bio-project Id: PRJNA217812.
Cell wall regeneration studies from protoplasts
Protoplasts were isolated from Arabidopsis cotyledons excised from 1 to 2 week-old plants [32]. Aliquots of protoplast medium were taken at various time points (0, 3, 6, 12, 18, 24, 36, 48, and 72 h) and observed using fluorescence microscopy with staining of the cellulose by 0.001% Calcofluor White M2R (Sigma, St Louis, MO, USA) at 25 °C for 5 min [33]. Both ordinary light images and fluorescent images for individual specimens were observed under a fluorescence microscope (Zeiss AX10, fluorescent Imager. M1, Germany) equipped with a UV fluorescence filter set (excitation filter, 350 nm; barrier filter, 430 nm). The images were then photographed with a digital camera (Axio Cam Hc Zeiss, Germany).
Microscopy analysis
For light microscopy, developed stems were used to prepare sections by microtome and it was stained with safranin-fast green, then observed and photographed under a light microscope. For scanning electron microscopy (SEM), segments from the apical 1 cm of WT and transgenic lines at 0, 50, 100 and 150 mM NaCl and at 1 h and 24 h of salt stress stem cross-sections were fixed in two steps: first with a mixture of 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 mol/l cacodylate buffer, pH 7.4 for 1 h and then with 1% OsO4 in 0.1 mol/l cacodylate buffer, pH 7.4 for 30 min. After critical point drying, the samples were sputter-coated with gold, and the coated samples were viewed with a Hitachi S-3400N field emission SEM using an accelerating voltage of 30 kV.
Metabolite profiling
Metabolite profiling was performed using LC-MS. All the solvents used were of HPLC grade and procured from J.T. Baker (USA). The reference standard, kaempferol 3,7-di-rhamnoside (kaempferitrin) (ChomaDex™, USA). A stock solution of kaempferitrin standard (2.0 mg/10 ml) was prepared in acetonitrile: water (1:1, v/v) in a volumetric flask and different amounts of standard (3.125-100 µg/ml) were injected for UPLC analysis. The stems and leaves (100 mg) of both WT and transgenic lines were extracted three times with 20 ml of methanol: water (8:2, v/v) at 40 °C for 15 min/cycle under ultrasonication. The extracts were filtered and dried in a rotary evaporator (45 °C). The extract was re-dissolved in 2 ml of acetonitrile: water (1:1, v/v) and filtered through a membrane filter (0.20 µm) for UPLC analysis. All the experiments were conducted in three biological replicates.
Statistical analysis
All experiments were conducted with at least three independent repetitions in triplicates (biological triplicates). All values are shown as the mean ± the standard deviation. The statistical analysis was performed using Statistica software (v.7). The statistical significance of the mean values was assessed by analysis of variance (ANOVA) applying Duncan’s Multiple Range Test (DMRT). A probability level of P ≤ 0.05 was considered significant.
Results
Molecular analysis of transgenic Arabidopsis overexpressing PaSOD and RaAPX
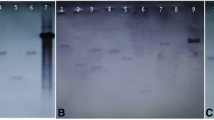
To confirm the expression of transgenes in the transgenic Arabidopsis, semi-quantitative PCR reactions were carried out with PaSOD and RaAPX gene-specific primers, individually (Fig. 1a, b). Dual transgenic plants (18O) were generated by crossing homozygous transgenic lines S26 (for PaSOD) and APX20 (for RaAPX) and were selected for subsequent investigation. Double transgenic plants were also confirmed for expression of both the genes using reverse transcriptase PCR, respectively (Fig. 1c). Further to confirm gene expression observation at transcriptional levels, the expression of PaSOD and RaAPX were analyzed by qPCR assay (Fig. 1d, e). PaSOD expression level was significantly higher in PaSOD and dual transgenic lines (Fig. 1d) and RaAPX expression level were significantly higher in PaSOD and dual transgenic lines compared to WT (Fig. 1e) under salinity stress.
(a) PCR confirmation of PaSOD and RaAPX in transgenic lines of Arabidopsis. PCR amplification of cDNA with Cu/Zn-SOD specific primer of single copy inserts lines, Lane 1—100 bp DNA ladder, Lane 2–5 amplification of 459 bp PaSOD transgenic lines (S15,S26,S7,S31), Lane 7—water (non-template control), Lane 8—plasmid as positive control (b), PCR amplification of cDNA with APX reverse primer of single copy inserts lines, Lane 1—100 bp DNA ladder, Lane 2–5 amplification of 858 bp APX single copy insert transgenic lines (APX2, APX10, APX18 and APX20), lane 6 is the WT control and lane 7 is a positive control (c), PCR amplification of PaSOD (459 bp, lower band) and RaAPX (858 bp, upper band) in double transgenic lines Lane 1—100 bp ladder, Lane 2–6—double transgenic lines 18O and 19C (d), (e) Transcript level of SOD and APX genes in WT and transgenic lines
Physiological, biochemical and molecular response of PaSOD and RaAPX transgenic plants to salinity stress
To elucidate the molecular and physiological mechanism underlying the enhanced stress tolerance, enzymatic activity assays of antioxidant system in transgenic and WT plants were performed under salt stress. As predicted, higher SOD and APX activities were observed transgenic lines than WT plants. However, SOD and APX activities in transgenic lines were further increased with increase in the magnitude of salt stress (Fig. 2). The SOD activity change was obviously little different between WT and transgenic lines, as shown in Fig. 1, prior to salt treatment. After stress, enzyme activities changed and twofold higher SOD activity was in PaSOD lines and nearly threefold in dual transgenic lines as compared to WT, at 100 mM NaCl treatment, while SOD activity in the WT plants showed an only slight increase (Fig. 2a). The APX activity showed a similar increasing pattern in transgenic lines with the stress; however, the APX activity in WT plants reduced slightly compared to what observed before treatment. APX activity was 3–5 fold higher in RaAPX lines and twofold in dual transgenic lines as compared to WT (Fig. 2b). Before salt stress, mRNA levels of PaSOD and RaAPX showed no significant difference in transgenic lines and WT (Fig. 1d, e). After salt treatment, the mRNA abundance of the transgenic lines demonstrated the higher transcription level of PaSOD and RaAPX, whereas their transcriptions showed no change in WT plants.
Biochemical parameters (a) SOD enzyme activity. (b) APX enzyme activity. (c) TSS. (d) Proline content. (e) Cellulose. (f) Glycoside content in WT and transgenic lines (SOD lines: S15, S26, APX lines: APX2 and APX20, Dual lines: 18O and 19c) at 0, 50, 100 and 150 mM salt stress. Data is Mean ± S.E of three biological replicates
Effect of PaSOD and RaAPX overexpression on biomass and osmoprotectant accumulation in transgenic lines and WT under salt stress
In order to investigate the effects of salinity stress on growth and biomass parameters of WT and transgenic plants at the whole-plant scale were evaluated. Under control conditions, the growth of the PaSOD, RaAPX and dual transgenic lines were comparable to that of WT, with no major indication of transgene effect on biomass and yield parameters (Fig. 3). Under saline conditions, improved performance in several biomass factors such as shoot and root biomass along with yields such as the number of siliques/plant, number of seeds/silique, seed weight and total seeds were higher in transgenic lines than WT (Fig. 3). The transgenic lines showed an increase in the shoot biomass only at 100 mM NaCl (Fig. 3a) in comparison to WT, with highest in dual transgenic lines (nearly 9–10%), PaSOD lines (nearly 8–9%) and RaAPX lines (nearly 7–8%), with further increase in the NaCl stress, the shoot biomass declined at 150 mM (Fig. 3a). Biomass accumulation for WT was observed to be less than that of the transgenics at all levels of salt stress. Thereafter, the difference in performance of the transgenics over the WT showed a significant (p ≤ 0.05) increase with the increase in the NaCl stress level (Fig. 3a). Root biomass was also studied at all levels of NaCl (0, 50, 100 and 150 mM) and in all the transgenic lines along with wild-type (Fig. 3b). Nevertheless, the difference in the root biomass between the transgenic lines and the WT was observed after imposition of NaCl stress at various concentrations (Fig. 3b). A marked decrease in root biomass accumulation was observed in all the transgenic lines with the increase in salt stress, but at100 mM reduction was less compared to 50 and 150 mM NaCl stress conditions. Among transgenic lines, double transgenic, 18O, and 19c showed higher biomass, followed by SOD (S26, S15) and APX (APX2, APX20) lines, although the root biomass content was higher than that of the WT at all levels of NaCl stress. Similarly, yield parameters (seed weight, number of siliqua, number of seeds, total seed weight) showed a similar trend in transgenic lines with a higher yield than WT under salt stress conditions (Fig. 3 c–f).
Effect of salt stress on Growth and yield-attributing traits. Shoot (a) and root (b) biomass accumulation, (c) seed weight (mg), (d) number of siliqua/plant, (e) number of seeds/siliqua and (f) total seed yield in WT and transgenic lines (SOD lines: S15, S26, APX lines: APX2 and APX20, Dual lines: 18O and 19c) at 0, 50, 100 and 150 mM salt stress. Data is Mean ± S.E of three biological replicates. (Color figure online)
Effect of salt stress on ROS accumulation and H2O2 content
To test whether overexpression of antioxidant enzyme transgene (PaSOD and RaAPX) and how the salt stress affected the ROS and H2O2 accumulation in transgenic and WT, NBT staining and ROS estimation were done. Blue colouration intensity indicates the production of ROS inside the tissue. Before salt stress treatment ROS accumulation was insignificant in all the lines as less blue colouration was observed (Fig. 4a), however, following stress treatment, the accumulation of ROS was strongly induced but was notably lower in transgenic lines and evident in WT, where blue intensity increased with the stress levels (Fig. 4a, b). Further, to study the possible relationships between the overexpression of antioxidant genes and H2O2 accumulation, we assessed the H2O2 levels both quantitatively and qualitatively in transgenics and WT under salinity stress (Fig. 4c, d). The intensity and presence of brown colour indicate H2O2 accumulation. Under 0 mM conditions, significant change between control and transgenic plants were not observed (Fig. 4c, d). However, significantly lower H2O2 content was detected in transgenic lines in response to salt stress except in S26 line which showed increased H2O2 production (Fig. 4d). Thus, H2O2 accumulation corresponded to the time of salinity stress treatment in transgenics (S26 and 18O) and declined afterwards. The action of the introduced RaAPX in addition to the native APX is also evident from the higher scavenging of H2O2 as shown by a less H2O2 accumulation in transgenic lines APX20 and 18O as compared to the control plants (WT) (Fig. 4d).
In situ ROS assay (a), superoxide content (b) DAB assay (c) and H2O2 content (d) in WT and transgenic lines (S15, S26, APX2, APX20, 18O and 19C) under control and salt stress. The assay was performed after four weeks of growth on respective NaCl concentration. Values represent the means ± SE of three biological replicates. Values marked with different letters are significantly different at p ≤ 0.05. (Color figure online)
Induction pattern of secondary cell wall cellulose genes and transcription factors in transgenic lines upon salt stress
Based on the previous finding that PaSOD and RaAPX are able to affect the lignin deposition in transgenic lines under salt stress [14, 15] and preliminary experiments done on cellulosic changes (data unpublished) prompted us to explore entire secondary wall biosynthetic program in transgenic Arabidopsis, particularly cellulose biosynthesis. In silico analysis of salt stress data led to the identification of some changes in expression pattern in transcripts of genes involved in cell wall biosynthesis (Table S2), where the majority of genes related to cellulose syntheses such as cellulose synthase, glycosyl hydrolase, 3-ketoacyl-CoA synthase, UDP-D-glucose/UDP-D-galactose 4-epimerase1, pectin methylesterase, xyloglucan endo trans-glucosylase/hydrolase, Glycosyl hydrolase 9A2, and starch synthase etc. (data unpublished) were upregulated in S26 and 18O lines after 24 h salinity stress (Fig. 5). Variation in the expression pattern in response to expression pattern can be observed among the different cell wall-related genes. These genes related to secondary cell wall cellulose biosynthesis exhibit significant upregulation in 18O and S26 line followed by APX line after 24 h of salt stress in comparison with WT (Table S2). Whereas in the case of WT, expression of most of the genes was found to be either unchanged or downregulated (Table S2). On the basis of gene IDs and associated FPKM values, 46 genes belonging to cellulose (primary and secondary wall) biosynthesis pathway were recognized and their expression level was also validated using a qRT-PCR assay (Fig. 5). From the Heat Map visualisation, it is possible to discern the degree of up-regulation of genes particularly those which are involved in secondary cellulose deposition (CESA4, CESA7, CESA8, KOBI, KOR1, COBRA4 and COBRA6) by salt stress were much higher in transgenic (S26 and 18O) than those that in the WT plants at least at one time point (24 h of stress) than 1 h time point (Fig. 5). In this study, most of the exclusive genes linked to secondary cell wall cellulose synthesis showed noteworthy induction; these were: cellulose synthase CESA’s (2–6 fold-up), which catalyzes the synthesis of cellulose, cellulose micro-fibril organisation protein, COBRA (2–5 fold-up); cellulose biosynthesis gene, KOB1 (1.7-2 fold-up); β-1,4-glucanase, KORIGAN (3–5 fold-up). Upregulation of various MYB related transcription factors occur during abiotic stress, thus the transcription factors which are actively involved in secondary cell wall cellulose biosynthesis (SND2, SND3, MYB83, MYB85, MYB 103 and MYB46) under abiotic stress were also validated by qPCR. A similar trend in expression pattern was observed (Fig. 6), where most of the transcription factors showed differential expression pattern under 100 and 150 mM salt stress, higher levels (2–3 fold) were observed in transgenic lines (S26, APX20 and 18O) in compared to WT under similar conditions. No significant change in expression levels of cellulose biosynthesis genes and transcription factors were observed in WT (Figs. 5, 6), even after 24 h salinity stress at any of the given stress concentrations.
Heat map showing differential expression of cellulose biosynthesis genes involved in cell wall biosynthesis under control and salt stress. Heat map represents the relative expression ratio of each gene under control and salt stress treatment with respect to WT of three biological replicates. The bar indicates the relative expression ratio whereby blue, white and red colours represent upregulation, no change and downregulation, respectively. (Color figure online)
Heat map showing differential expression of transcription factors involved in cell wall biosynthesis under control and salt stress. Heat map represents the relative expression ratio of each gene under control and salt stress treatment with respect to WT of three biological replicates. The bar indicates the relative expression ratio whereby blue, white and red colours represent upregulation, no change and downregulation, respectively. (Color figure online)
Cellulose content, anatomic and ultra-structural changes of transgenic lines
To examine transgenic lines (S26, APX20 and 18O) and WT under the stressed and non-stressed condition, some biochemical and anatomical aspects was done. Since, cellulose is an important component of the cell wall that is associated with mechanical strength. Thus, we measured the levels of cellulose in WT and transgenic lines under control and stressed conditions. Cellulose content was significantly enhanced in all transgenic lines with the salinity stress as compared to WT, indicating overexpression of PaSOD and RaAPX not only changed the cellulose transcriptome level but also leads to increased secondary cell wall cellulose deposition (Fig. 2e). The cellulose content was nearly threefold higher in S26 line and 2.5 fold higher in 18O followed by 1.5 fold higher in APX line than in the WT under 100 mM NaCl (Fig. 2e). A positive correlation was observed between cellulose content and H2O2 accumulation (correlation coefficient ‘r’ = 0.45). Further, we tried to document the difference of tissue structure between the transgenic lines and the WT by light microscopy (Fig. 7). Morphology of vascular system in all lines and WT changed with the stress conditions. However, compared with the transgenic lines, the xylem deformations were evident in WT (Fig. 7). Meanwhile, all cells of the vascular system of the WT exhibited severe deformations of xylem cell morphology at 100 and 150 mM salt stress, which resulted in irregular or collapsed xylem phenotype but the transgenic lines showed normal anatomy with enlarged xylary cells (Fig. 8). We tried to validate the results of light microscopy with SEM, some significant differences were also observed between the scanning electron micrographs of WT and transgenic lines, where anatomical modifications in the inflorescence of transgenic lines under salinity stress were documented (Fig. 8). Cell wall measurements were also done with SEM for all the transgenic lines along with WT under different salt stress conditions. The results obtained from SEM were in accordance with the light micrographs, wherein electron micrographs showed that cell walls of vascular bundles in transgenic lines particularly in 18O and S26 transgenic lines were denser and enlarged compared to WT (Fig. 8). A significant difference in cell wall thickness could be observed (Fig. 8), where S26 and 18O showed 3 times cell wall thickness compared to WT under 100 mM stress conditions.
Scanning electron micrographs of the stem of WT and transgenic lines (PaSOD; S26, RaAPX; APX20, dual line: 19c) under control and salt stress of three biological replicates, red arrows show wall thickening. Note the gradient in the degree of secondary wall thickness from developing to mature secondary xylem. (Color figure online)
Comparison of cell wall formation in WT and 18O transgenic line
To further evaluate the effect on antioxidants genes on cellulose accumulation and based on the above findings, we used protoplast technology to evaluate the dynamics of cell wall formation in WT and 18O. Protoplasts isolated from cotyledons of WT and 18O were transferred to protoplast culture media. Aliquots were taken at various points and studied using light and fluorescence microscopy with and without staining by Calcofluor White, which preferentially stains cellulose (Fig. 9). Development of new cell wall by the protoplasts did not occur in at the same time; several discrete phases could be recognized. Representative microscopic images of protoplasts at eight different time points were shown in the present study (Fig. 9). At initial stages of culture, fresh protoplasts remained unstained (Fig. 9a, b, j, k), indicating degraded cell wall. In the primary stages of culture, Calcofluor White staining was not observed in protoplasts and cell wall formation appeared after 18 h of protoplast culture (Fig. 9e, n), when the cell wall formation was observed in bulk of the cells, but in case of 18O, cell wall regeneration was relatively faster, within 12 h of culture as compared to WT (Fig. 9m). After 18–24 h of cultivation over 50% of 18O transgenic protoplasts reconstructed their cell walls. These protoplasts continued to grow and first cell division appeared after a few hours of culturing.
Time course changes in the cell surface during cell wall regeneration from the protoplasts of Arabidopsis of three biological replicates. Protoplasts were stained with calcofluor white and subsequently studied with (A–R) normal light and (a–r) UV light. Samples were taken at different time points after transfer to the protoplast culture medium: 0 h (A, a, J, j), 3 h (B, b, K, k), 6 h (C, c, L, l), 12 h (D, d, M, m), 18 h (E, e, N, n), 24 h (F, f, O, o), 36 h (G, g, P, p), 42 h (H, h, Q, q) and 48 h (I, i, R, r). Red arrows showing cell wall formation which appeared earlier in 18O (i, m) as compared to WT (e). Bar = 15 µm. (Color figure online)
Overexpression of antioxidant genes affects the accumulation of soluble phenolic compounds in transgenic Arabidopsis
Plants contain large amounts of soluble phenolic substances that function in protection against abiotic and biotic stresses. During the RNAseq analysis, we have observed some changes in flavonoid pathway genes, where upregulation was seen in transgenic lines under stress conditions (Fig. S2). To confirm the results of enrichment analysis (Fig. S1), we performed UPLC analysis on extracts obtained from WT and transgenic lines grown under control and salinity stress. Comparison of UPLC-PDA profiles (Fig. S2) demonstrated the accumulation of several flavonols in WT and transgenic plants. Several components were recognized by the characteristic UV absorbance spectra of their aglycones, and from the mass spectra data, glycosides nature was inferred (Fig. S2). In the tissue extracts of all the lines, the major peak was detected at 270 nm, which matches to kaempferitrin. Apart from this peak, numerous peaks equivalent to glycosides of kaempferol and quercetin were identified. Based on chromatographic and spectral properties which were then equated with a known standard of kaempferitrin, the amount of this metabolite was similar in all the lines at control condition, but under 100 mM salt stress the amount of principal compound that is kaempferitrin (phenolic glycoside) was found to be twofold higher in 18O line as compared to WT (Fig. 2f).
Discussion
Salinity stress and plant response to the stress situations have been intensively studied but it has not reflected in the development of abiotic stress tolerant crops. A cascade of genes involved in ROS scavenging, osmotic adjustments and some transcription factors has been utilized to generate salinity stress tolerant crops [34,35,36]. However, the rate of production of these tolerant crops is very low, which can be due to inadequate knowledge about the detailed processes involved salt stress tolerance by plants. Therefore, the major goal for plant biologists is to reduce the influence of abiotic stress on crop yield by maintaining growth, overall architecture, and metabolism. Majority of studies involved in the manipulation of single genes in the improvement of salt tolerance was reported. Though, salt tolerance is associated with the interactions of many genes and thus co-overexpression strategy of multi-genes is a promising way to improve salt tolerance. Taken together, our results allowed us to propose that the co-expression of the PaSOD and RaAPX genes in transgenic lines was closely associated with changes at a metabolic level such as increasing cellulose biosynthesis and other factors which probably led to the better adaptation of these plants under salinity stress. In the present work, all the stress conditions were used to validate/crosscheck whether other stress conditions (50 and 150 mM) had any effect on the cellulose biosynthesis in transgenic. This study proved that not only lignin, but cellulose biosynthesis pathway got differentially regulated at 100 mM salt stress conditions. Further, under salt stress, PaSOD and RaAPX-overexpression plants grew better and were characterized with significantly increased seed yield, root biomass and length, better tolerance to salt stress, and improved cellulose content, as compared to WT, in agreement with our previous studies [14,15,16, 21]. The constant production of low levels of H2O2 stimulated the onset of secondary cell wall cellulose synthesis signal transduction pathway (perhaps by inducing expression of secondary cellulose biosynthesis genes and transcription factors).
Enhanced accumulation of ROS molecules due to salinity results in the serious damage to plants and to evade this damage plants are well fortified with strong antioxidant systems that can scavenge these ROS and protect plants from oxidative damage [15, 21]. It has been well documented that overexpression of antioxidant genes, such as superoxide dismutase (SOD), and ascorbate peroxidase (APX) enhance ROS scavenging activity and improve salt stress tolerance [15, 21, 37], which was reflected in our transgenic lines under salinity stress (Fig. 2a, b). Therefore the enhanced activities of SOD and APX were considered to be closely related to plant tolerance to abiotic stress. Similarly, we found that the transgenic plants overexpressing PaSOD and RaAPX genes have SOD and APX activities elevated by about 2–3 fold under stress conditions, compared with the WT plants. Among transgenic, dual line (18O) accumulated substantially more SOD and APX levels under stress than other transgenic lines and WT plants. Further, to support this observation the endogenous levels of ROS in PaSOD and RaAPX plants was lower compared with the WT, suggesting that the WT plants were impaired extremely by salinity. As SOD and APX combo act as a major defence against ROS damage their activities and the transcription in transgenic lines increased more than that of WT plants after salt treatment, indicating that PaSOD and RaAPX overexpression amplified the ROS scavenging ability. These observations indicated that the transgenic line with higher antioxidant scavenging capacity is attributed to their higher antioxidant enzymes activities. Plants grown under control condition (0 mM) showed low accumulation of superoxide free radicals in a localization study (Fig. 4). Dark brown and blue-colour accumulation appeared after DAB and NBT staining, which is an indication of free radicals accumulation and was higher in WT compared to transgenic lines under salinity stress (Fig. 4).
In order to retain osmotic homeostasis across cell membrane during stress conditions, cell synthesizes osmoprotectants such as proline and total soluble sugars, which are considered as biochemical markers involved in osmotic adjustment [38, 39]. In our study, transgenic lines overexpressing PaSOD and RaAPX gene accumulated significantly higher levels of proline and reducing sugar, total sugar (Fig. 2c, d). This accumulation of proline and soluble sugars can stabilize the macromolecules, membranes and has a central role in energy provision. This augmentation in soluble sugar and proline levels along with enhanced cellulose in transgenics confirmed that the cell not only changes its cell wall structure but also maintains the internal balance of osmoprotectants.
At the phenotypic level, exposure to high salinity results in morphological traits, growth parameters, and seed productivity variations. As described previously [35], photosynthetic rates and chlorophyll fluorescence have efficiently enhanced in tobacco plants overexpressing SOD and APX genes. Similar results can be inferred from our observations where improved yield and biomass parameters were recorded in transgenic lines under different salt stress conditions compared to WT plants (Fig. 3), which reveals that the combination of antioxidant genes (SOD + APX) can ease the detrimental effects of salinity stress. This advance in growth appears to depend on improved cellulose accumulation during stress in these transgenic lines, as CesA’s Arabidopsis mutants showed a reduction in cellulose content and biomass accumulation [40, 41]. Treatment with 150 mM NaCl resulted in a drastic decrease in biomass and yield parameters probably due to maximum tolerance level with which plant can withstand stress conditions.
A group of researchers [42, 43] have shown that morphological changes in plants are found to be linked with increased cell dimensions and its architecture. Histochemical staining and quantitative analysis of transgenic and WT plants under stress conditions showed morphological changes in the vascular bundle architecture, width and its cell size (Fig. 7). Xylem vascular bundles cell size examination showed that PaSOD and RaAPX overexpression induced the cell expansion (Fig. 8). A similar observation has been reported by Yokoyama and Nishitani [44], xylem secondary cell wall formation was enhanced by a group of peroxidases. As we have expected, the cell size and cell wall width of vascular bundles in PaSOD and RaAPX overexpressors were enlarged than WT (Fig. 8).
Cellulose, hemicelluloses, and lignin constitute secondary cell wall components, their biosynthesis occurs by coordinated expression of various genes and transcription factors (SND1, NSTs, and VNDs) [45,46,47,48,49]. High salinity usually induces the expression of secondary cell wall genes and transcription factors which regulate their expression, among which MYB and NAC transcription factors [14,15,16], are recognized to be involved in various tolerance and adaptation. Based on these reports and some preliminary experiments (un-published data), we documented that PaSOD and RaAPX overexpression have resulted in increased expression of secondary cell wall cellulose synthesis genes in transgenic lines under salinity stress. However, at 100 mM salt stress and after 24 h time point more pronounced effect was observed in dual transgenic line (18O) followed by PaSOD and RaAPX lines, indicating that this difference in expression of cellulose genes biosynthesis between WT and within the transgenic lines might be due to the presence of extra copy PaSOD and RaAPX genes. We have observed that the essential CesA4, CesA7, and CesA8 genes which are required for synthesizing secondary cell wall cellulose [50] were upregulated in PaSOD and dual transgenic lines at 100 mM NaCl stress condition (Fig. 5). Zhong et al. [49] have reported impaired secondary wall formation in the fibers of Arabidopsis dual mutants (SND1 and NST1), which included a lack of cellulose. Among MYB family of transcription factors, MYB46 and MYB83 act as downstream targets of NAC regulators [51, 52] and their overexpression enhanced secondary CESA genes (CesA4, CesA7, and CesA8) expression and induced secondary cellulose deposition [53]. Similarly, we observed a shift in the expression pattern of MYB46 and MYB83 in dual transgenic line and PaSOD plants under salt stress conditions, which was lacking in WT and RaAPX plants. Higher levels of MYB46 and MYB83 transcription factors might have influenced the expression of secondary CESA genes (CesA4, CesA7, and CesA8) and which in turn induced secondary cell wall cellulose deposition. Transcription factors SND2, SND3, MYB46 and MYB83 act as leading transcriptional switches to activate their common targets and consequently the secondary wall cellulose biosynthetic cascade and these transcription factors showed 2–6 fold upregulation in dual and PaSOD line (Fig. 6). Thus in the present study most of the abiotic stress responsive genes involved in secondary cellulose biosynthesis were found upregulated. Based on the previous observation of Shafi et al. [14], 24 h stress time point provided better results as compared to 1 h of salt stress treatment, which could be due to stress induction occurs after 24 h of salinity stress. The increased thickness of the cell wall in transgenic lines could be correlated with the increased cellulose content. However, in WT, the decreased cellulose level under salt stress may result in thinner cell walls, as well as the shorter height. Collectively, the data suggest that overexpression of PaSOD and RaAPX has stimulated the minimal levels of H2O2 (required for signalling), which further increased the level of secondary cell wall cellulose synthesis and thus biomass accumulation.
Role of SOD and APX genes in cell wall metabolism has been reported [54]. Several investigations on cell wall synthesis have used protoplasts undergoing cell wall regeneration [55]. The choice of this system in the present study was based on the expectation that strongly enhanced cell wall regeneration would derive from amplified anti-oxidant scavenging gene expression levels (SOD and APX). More than 90% of protoplasts regenerated a new cell wall within 12 h of culture in 18O line, which is an indication that apart from the alterations in the lignin depositions other cell wall components are also being affected in 18O line (Fig. 9). Most of the protoplasts regenerated cell wall within 12 h of culture in 18O line, indicating that cell wall components are affected in 18O line in comparison with WT (Fig. 9). Our results propose that cellulose synthesis and its deposition is a significant influencing factor in adaptations of plants to salinity stress.
During abiotic stress, ROS molecules such as H2O2 at low levels act as signalling molecules [56] and its presence near cell wall was also observed [57] with its significant role in cell wall development. This study primarily focused on measurement of H2O2 in all the lines under control and stress conditions, where H2O2 levels were elevated in S26 line followed by 18O and APX20 (Fig. 1g, h). Under stress conditions, a strong but transient increase in H2O2 accumulation was observed in S26 and 18O lines. We also detected a trend in cellulose deposition under stress condition in transgenic lines (Fig. 2e), which was similar to H2O2 accumulation (Fig. 4d). A positive correlation between cellulose content and H2O2 accumulation in transgenic lines is an indication of a possible link of H2O2 signalling in cellulose deposition under salinity stress. Karpinska et al. [58] localized a novel isoform of CuZn-SOD in the secondary cell wall of the xylem cells of Scot pine and was involved in the H2O2 production. Such a continually elevated concentration of H2O2 within the cell is likely to produce an accumulation of damage to vital cellular components and therefore may result in the activation of programmed cell death. However, H2O2 produced by SOD is regulated by APX to optimal levels, which then acts as a signalling molecule inducing a series of downstream molecules which then upregulate secondary cell wall cellulose biosynthesis genes. Enrichment analysis at 100 mM salt stress condition in 18O transgenic line also confirmed that metabolic and biosynthetic processes involved in the H2O2 production were extremely enhanced (Fig. S1). These observations provide a molecular link between oxidative stress, redox homeostasis, and secondary cell wall cellulose biosynthesis during salinity stress. These results further support the notion that H2O2 generation in plants is developmentally regulated.
Flavonoid biosynthesis is tightly regulated by various developmental and ecological factors [59]. These flavonoid compounds are localized in different cell compartments, indicating their multiple physiological roles [60]. Arabidopsis phenolic pool consists of flavonol glycosides (Kaempferol and quercetin derivatives). The major soluble phenolic compound recovered from the samples was Kaempferol glycoside (Kaempferitrin) (Fig. S2). Besides this, other soluble phenolics were quercetin glycoside and rhamnetin glycoside. Since Kaempferitrin was found to be marker phenolics, it was used as a standard to observe changes in phenolic content in all the lines under control and salt stress conditions. It was also interesting to note that compared to control group, 18O transgenic line had increased levels of Kaempferitrin under salt stress (Fig. 2f), which indicates that apart from perturbations in the cellulose biosynthetic pathway, there must be some metabolic flux towards other branches of phenylpropanoid pathway as well. Enrichment analysis also revealed that there was positive regulation of flavonoid biosynthetic process in 18O line at 100 mM stress (Fig. S1).
Conclusion
The present work constitutes that the SOD and APX genes from Himalayan plants Potentilla atrosanguinea and Rheum austral respectively were isolated and ectopic expression in Arabidopsis up-regulated downstream genes involved in antioxidant protection and secondary cell wall cellulose biosynthesis, resulting in improved salt tolerance. The present study provided the information about changes occurring precisely at cellulose biosynthesis genes and its transcription factors. Taken together, our results demonstrated that PaSOD and RaAPX overexpression enhanced the salt stress tolerance of transgenic plants, possibly due to ultrastructural changes at cell wall level with improved cell wall integrity and higher cellulose, reduced accumulation of ROS, increased activities of antioxidant enzymes, enhanced biomass, yield, and flavonoid accumulation. SOD and APX act as an important gene for salinity stress tolerance and for the development of value-added cultivars.
References
Qin H, Gu Q, Zhang J, Sun L, Kuppu S, Zhang Y et al (2011) Regulated expression of an isopentenyltransferase gene (IPT) in peanut significantly improves drought tolerance and increases yield under field conditions. Plant Cell Physiol 52:1904–1914
Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F (2011) ROS signaling: the new wave? Trends Plant Sci 16:300–309
Wrzaczek M, Brosche M, Kangasjarvi J (2013) ROS signaling loops: production, perception, regulation. Curr Opin Plant Biol 16:575–582
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9:490–498
Asada K (1999) The water-water cycle in chloroplasts: scavenging of active oxygen and dissipation of excess photons. Ann Rev Plant Physiol Plant Mol Biol 50:601–639
Bienert GP, Møller AL, Kristiansen KA et al (2007) Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J Biol Chem 282:1183–1192
Somerville C, Youngs H, Taylor C, Davis SC, Long SP (2010) Feedstocks for lignocellulosic biofuels. Science 329:790–792
Demura T, Ye ZH (2010) Regulation of plant biomass production. Curr Opin Plant Biol 13:299–304
Richmond TA, Somerville CR (2001) Integrative approaches to determining Csl function. Plant Mol Biol 47:131–143
McFarlane HE, Do¨ring A, Persson S (2014) The cell biology of cellulose synthesis. Annu Rev Plant Biol 65:69–94
Carpita NC, McCann M (2000) The cell wall. In: Buchanan B, Gruissem W, Jones RL (eds) Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville, pp 52–108
Ueda A, Yamamoto-Yamane Y, Takabe T (2007) Salt stress enhances proline utilization in the apical region of barley roots. Biochem Biophys Res Commun 355:61–66
Muszyńska A, Jarocka K, Kurczynska E (2014) Plasma membrane and cell wall properties of an aspen hybrid (Populus tremula × tremuloides) parenchyma cells under the influence of salt stress. Acta Physiol Plant 36:1155–1165
Shafi A, Chauhan R, Gill T, Swarnkar MK, Sreenivasulu Y, Kumar S, Kumar N, Shankar R, Ahuja PS, Singh AK (2015) Expression of SOD and APX genes positively regulates secondary cell wall biosynthesis and promotes plant growth and yield in Arabidopsis under salt stress. Plant Mol Biol 87:615–631
Shafi A, Gill T, Sreenivasulu Y, Kumar S, Ahuja PS, Singh AK (2015) Improved callus induction, shoot regeneration, and salt stress tolerance in Arabidopsis overexpressing superoxide dismutase from Potentilla atrosanguinea. Protoplasma 252:41–51
Shafi A, Pal AK, Sharma V, Kalia S, Kumar S, Ahuja PS, Singh AK (2017) Transgenic potato plants Overexpressing SOD and APX exhibit enhanced lignification and starch biosynthesis with improved salt stress tolerance. Plant Mol Biol Rep 35:504–518
Heyndrickx KS, Van de Velde J, Wang X, Weigel D, Vandepoele K (2014) A functional and evolutionary perspective on transcription factor binding in Arabidopsis thaliana. Plant Cell 26:3894–3910
Kurek I, Kawagoe Jacob-Wilk YD, Doblin Delmer MD (2002) Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc Natl Acad Sci USA 99:11109–11114
Kim HJ, Barbara T (2008) Involvement of extracellular Cu/Zn superoxide dismutase in cotton fiber primary and secondary cell wall biosynthesis. Plant Signaling Behavior 3:1119–1121
Xiong J, Yang Y, Fu G, Tao L (2015) Novel roles of hydrogen peroxide (H2O2) in regulating pectin synthesis and demethylesterification in the cell wall of rice (Oryza sativa) root tips. New Phytol 206:118–126
Shafi A, Dogra V, Gill T, Ahuja PS, Sreenivasulu Y (2014) Simultaneous over-expression of PaSOD and RaAPX in transgenic Arabidopsis thaliana confers cold stress tolerance through increase in vascular lignifications. PLoS One 9:e110302
Gill T, Kumar S, Ahuja PS, Sreenivasulu Y (2010) Over-expression of Potentilla superoxide dismutase improves salt stress tolerance during germination and growth in Arabidopsis thaliana. J Plant Genet Transgenics 1:1–10
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Ghawana S, Paul A, Kumar H, Kumar A, Singh H, Bhardwaj PK, Rani A, Singh RS, Raizada J, Singh K, Kumar S (2007) An RNA isolation system for plant tissues rich in secondary metabolites. BMC Research Notes 4:85
Gill T, Sreenivasulu Y, Kumar S, Ahuja PS (2010) Over-expression of superoxide dismutase exhibits lignification of vascular structures in Arabidopsis thaliana. J Plant Physiol 167:757–760
Sonja V, Noctor G, Foyer CH (2002) Are leaf hydrogen peroxide concentrations commonly overestimated? The potential influence of artefactual interference by tissue phenolics and ascorbate. Plant Physiol Bioch 40:501–507
Updegraff DM (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32:420–424
Dubois M, Gilles K, Hamilton JK, Rebers PA, Smith F (1956)) Colorimetric method for determination of sugars and related substances. Anal Chem 38:350–361
Bates L, Waldren R, Teare ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207
Orozco-Cárdenas ML, Ryan CA (1999) Hydrogen peroxide is generated systematically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc Natl Acad Sci USA 96:6553–6557
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Mathur J, Koncz C, Szabados L (2005) A simple method for isolation, liquid culture, transformation and regeneration of Arabidopsis thaliana protoplasts. Plant Cell Rep 14:221–226
Nagata T, Takebe I (1970) Cell wall regeneration and cell division in isolated tobacco mesophyll protoplasts. Planta 92:301–308
Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N et al (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486:228–232
Faize M, Burgos L, Faize L, Piqueras A, Nicolas E, Barba-Espin G, Clemente-Moreno MJ, Alcobendas R, Artlip T, Hernandez JA (2011) Involvement of cytosolic ascorbate peroxidase and Cu/Zn-superoxide dismutase for improved tolerance against drought. J Exp Bot 62:2599–2613
Bhaskaran S, Savithramma DL (2011) Co-expression of Pennisetum glaucum vacuolar Na+/H+antiporter and Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic tomato. J Exp Bot 62:5561–5570
Abogadallah GM, Serag MM, El-Katouny TM, Quick PW (2010) Salt tolerance at germination and vegetative growth involves different mechanisms in barnyard grass (Echinochloa crusgalli L.) mutants. Plant Growth Regul 60:1–12
Waditee R, Bhuiyan MNH, Rai V, Aoki K, Tanaka Y, Hibino T, Suzukim S, Takanom J, Jagendorf AT, Takabe T, Takabe T (2005) Genes for direct methylation of glycine provide high levels of glycinebetaine and abiotic-stress tolerance in Synechococcus and Arabidopsis. Proc Natl Acad Sci USA 102:1318–1323
Koster KL, Lynch DV (1992) Solute accumulation and compartmentation during the cold acclimation of puma rye. Plant Physiol 98:108–113
Harris DM, Corbin K, Wang T, Gutierrez R, Bertolo AL, Petti C, Smilgies DM, Estevez JM, Bonetta D, Urbanowicz BR et al (2012) Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903V and CESA3T942I of cellulose synthase. Proc Natl Acad Sci USA 109:4098–4103
Fujita M, Himmelspach R, Ward J, Whittington A, Hasenbein N, Liu C, Truong TT, Galway ME, Mansfield SD, Hocart CH et al (2013) The anisotropy1 D604N mutation in the Arabidopsis cellulose synthase1 catalytic domain reduces cell wall crystallinity and the velocity of cellulose synthase complexes. Plant Physiol 162:74–85
Wang G, Gao Y, Wang J, Yang L, Song R et al (2011) Overexpression of two cambium-abundant Chinese fir (Cunninghamia lanceolata) alpha-expansin genesClEXPA1 and ClEXPA2 affect growth and development in transgenic tobacco and increase the amount of cellulose in stem cell walls. Plant Biotechnol J 9:486–502
Zenoni S, Fasoli M, Tornielli GB, Dal Santo S, Sanson A et al (2011) Overexpression of PhEXPA1increases cell size, modifies cell wall polymer composition and affects the timing of axillary meristem development in Petunia hybrida. New Phytol 191:662–677
Yokoyama R, Nishitani K (2006) Identification and characterization of Arabidopsis thaliana genes involved in xylem secondary cell walls. J Plant Res 119:189–194
Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19:1855–1860
Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17:2993–3006
Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19:270–280
Zhong R, Demura T, Ye ZH (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18:3158–3170
Zhong R, Richardson EA, Ye ZH (2007) Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225:1603–1611
Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100:1450–1455
Zhong R, Ye ZH (2007) Regulation of cell wall biosynthesis. Curr Opin Plant Biol 10:564–572
Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH (2008) A battery of transcription factors Involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20:2763–2782
Kim WC, Ko JH, Kim JY, Kim JM, Bae HJ, Han KH (2012) MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J 73:26–36
Passardi F, Penel C, Dunand C (2004) Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci 9:534–540
Cosgrove DJ (2001) Wall structure and wall loosening: a look backwards and forwards. Plant Physiol 125:131–134
Gechev TS, Hille J (2005) Hydrogen peroxide as a signal controlling plant programmed cell death. J Cell Biol 168:17–20
Olson PD, Varner JE (1993) Hydrogen peroxide and lignification. Plant J 4:887–892
Karpinska B, Karlsson M, Schinkel H, Streller S, Suss KH, Melzer M, Wingsle G (2001) A novel superoxide dismutase with a high isoelectric point in higher plants: expression, regulation and protein localisation. J Plant Physiol 126:1668–1677
Saslowsky DE, Warek U, Winkel BSJ (2005) Nuclear localization of flavonoid enzymes in Arabidopsis. J Biol Chem 25:23735–23740
Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5:218–223
Acknowledgements
This research was supported by grants from the Council of Scientific and Industrial Research (CSIR), New Delhi, India under CSIR Network Projects BSC107 and BSC109.
Author information
Authors and Affiliations
Contributions
AKS, AS and PSA conceived and designed the experiments. AS and TG performed the experiments. PSA, YS, SK and AKS analyzed the data. IZ did statistical analysis. AS and AKS wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2019_4648_MOESM1_ESM.pdf
Hierarchical tree graph of over-represented GO terms in down-regulated genes by singular enrichment analysis generated by agriGO. Boxes in the graph show GO terms labelled by their GO ID, term definition and statistical information. The significant terms (adjusted p < 0.05) are marked with colour, while non-significant terms are shown as white boxes. The degree of colour saturation of a box correlates positively with the enrichment level of the term. Solid, dashed and dotted lines represent two, one and zero enriched terms at both ends connected by the line, respectively. The rank direction of the graph runs from top to bottom. (PDF 500 KB)
11033_2019_4648_MOESM2_ESM.pdf
UPLC Analysis of Soluble Phenolics in WT (a), S26 (b), APX20 (c) and 18O (d) lines of Arabidopsis thaliana. Phenolic profiles of transgenic (b, c, d) and wild type (a) lines were compared at 270 nm under control and stress conditions. Key to peak Peaks was confirmed by mass spectrometry. (PDF 267 KB)
11033_2019_4648_MOESM4_ESM.xlsx
RNA seq FPKM values for the genes and transcription factors involved in secondary cell wall cellulose biosynthesis. (XLSX 22 KB)
Rights and permissions
About this article
Cite this article
Shafi, A., Gill, T., Zahoor, I. et al. Ectopic expression of SOD and APX genes in Arabidopsis alters metabolic pools and genes related to secondary cell wall cellulose biosynthesis and improve salt tolerance. Mol Biol Rep 46, 1985–2002 (2019). https://doi.org/10.1007/s11033-019-04648-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-04648-3