Abstract
Phytases are enzymes that hydrolysis phytic acid and makes mineral phosphorus available to animals. Phytases face relatively extreme heating during food processing, thus thermostability plays an important role in industrial applicability of this enzyme. Herein, we report the design of a thermostable phytase with favorable biochemical properties and high enzymatic activity using molecular dynamics and rational design-based molecular engineering. Based on the crystal structure of E. coli phytase, bioinformatics analysis and docking binding energy measurement, S392F mutant was introduced by site-directed mutagenesis in order to improve thermostability of phytase through strengthen the bounding interactions. Wilde type and Mutated constructs were expressed in E. coli BL 21. The WT and manipulated phytase were purified; their biochemical and kinetic was investigated. Results revealed that recombinant WT and mutant phytase have optimum temperature of 50 °C with no significance change but optimum pH of WT and mutant was respectively 5 and 6 with a pH shift. Furthermore, S392F phytase catalytic efficiency values showed significant improve of 25.6%, compared with the WT. Analysis of the retained enzymatic activity at high temperatures, indicated that despite of phytase stability reduction at high temperatures but mutant phytase showed more stable behavior in compare with WT phytase, So that at 70 °C showed twice thermo stability and at 80 °C and 90 °C display respectively 74% and 78.4% improvement of thermostability compared to the wild-type. In conclusion, our results implied that the designed phytase could be a potential candidate for phytase manipulation research and industrial applications with improved thermostability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With a vital role in seed viability and germination, myo-inositol-(1,2,3,4,5,6)-hexakisphosphate (InsP6), commonly known as phytic acid (PA), is a major component of all plant seeds, constituting about 60–90% of the total phosphorus. Although found to have antioxidant properties, dietary PA has been associated with mineral deficiency in human by binding to essential minerals and preventing their absorption through the gastrointestinal tract [1]. Generally, phytates have a potential for binding multivalent cations such as calcium, magnesium, copper, iron, zinc, cobalt and manganese. The resulting chelate complexes are nutritionally less available and difficult to hydrolyze during digestion. This effect has been also detected on starch, positively charged proteins and amino acids [2]. Furthermore, emission of undigested PA by monogastric animals such as poultry, fish and swine due to inadequate levels of phytase in their digestive tracts, imposes substantial environmental pollution risks [3]. The concern of phytate phosphorus pollution is specifically highlighted in intense fish production. Thus, to address the abovementioned issues, enzymatic and non-enzymatic approaches are routinely applied. In the non-enzymatic approach, reduction of phytate level is achieved during food processing steps or through physical removal of phytate-rich parts of the plant seed. Alternatively, supplementation of animal feed with inorganic phosphate was proposed, which has been linked to further ecological problems including eutrophication [4].
Enzymatic hydrolysis of PA involves the addition of phytate-degrading enzymes. Phytases (EC 3.1.3.8) catalyze the sequential hydrolysis of PA to inorganic phosphate [5]. This class of histidine acid phosphatases has been detected in several species of bacteria, fungi, plants and in some animal tissues [1]. Using phytase as a feed supplement has proven to increase nutrition uptake and moderate phosphate excretion by 50%. Although, relatively low amount of this enzyme in wild-type origins, excessive cost of exogenous recombinant production in microbial hosts and conformational instability of the enzyme molecule in various pH and temperature conditions of processing and manufacturing procedures are some of the obstacles currently associated with the use of phytases [1, 6]. Additionally, most phytases are not heat-resistant and animal feed are generally pelleted at temperatures between 65 and 80 °C, hence, significant percentage of phytase activity will be lost under these processing conditions [7].
Recent advances in genetic engineering has provided an ideal platform for manipulating molecular characteristics of recombinant proteins. Typically, improving the unfolding temperature (Tm) of a protein entails multiple amino acid exchanges [8]. Each modification may slightly enhance the unfolding temperature of the polypeptide. However, it is critical to identify the effectual amino acid residues on thermostability of the molecule. Generally, such amino acids may have a role in the formation of hydrogen bonds, salt and disulfide bridges, increasing the hydrophobic stacking, enhancing α-helix and β-sheet stability, or stabilizing β-turns, flexible termini or loops [9]. Due to the lack of a precise and reliable procedure, many residues have to be mutated and individually analyzed for selecting a hyperthermostable mutant.
Previously, several efforts have been made for this purpose [10,11,12]. Lehman et al. increased the thermostability of Aspergillus niger phytase by around 20 °C through de novo constructing consensus enzymes using primary protein sequence comparisons of thirteen fungal phytases. Van den Burg et al. achieved similar results for a thermolysin-like Bacillus stearothermophilus protease by substituting eight amino acid residues [13]. These are generally conducted via two main approaches of rational protein engineering, where point mutations are inserted based on the predicted protein conformation, and directed evolution in which a set of mutations are introduced in line with the selection of improved mutants [14]. Bioinformatics analysis has the potential to significantly improve our perspective of protein structure–function correlation. Recent advances in molecular dynamics allow for reliable structural comparison between a reference protein and a proposed mutant, providing valuable data for rational design of mutant variants with favorable characteristics [15]. Knowledge of E. coli phytase crystal structure and other similar phytases, beside on protein sequences studies of native app phytase have enabled our rational design toward improving the enzyme efficiency.
In the present study, since structural and docking predictions require experimental verification, we introduced S392F mutation with highest binding affinity to phytate substrate by using a site-directed mutagenesis method.
*Thermostability: thermal stability and the maintenance of the enzyme activity at higher temperatures.
Methods and materials
Strains, media and reagents
Escherichia coli DH5α was used for cloning and amplification of the gene and BL21 (DE3) as host for gene expression. pUC57 and pET26b were used as cloning and expression vectors respectively. For growth of E. coli, the LB medium is also used. Preparation the Competent cells and Transfer of pUC57-ansB plasmid and Plasmid extraction and cutting with the enzyme were performed according to standard molecular Cloning methods. Taq DNA polymerase, T4 DNA ligase and restriction enzymes were purchased from Thermo Fermentase (USA). Oligonucleotides were synthesized and automatic DNA sequencing was performed at Macrogene Inc (South Korea). Sodium phytate and phytic acid were purchased from Sigma Aldrich Co., Ltd. (St Louis, MO, USA). The columns used for protein purification were obtained from Amersham Pharmacia Biotech (USA).
Construct preparation
The amino acid sequence of E. coli (K12) phytase (appA, accession number: P07102), with reportedly strong affinity for sodium phytate as its substrate, was obtained from the UniProt database. The first 22 amino acids coding for the N-terminal signal peptide were excluded and the rest was used to design the phytase gene, based on the optimal codon usage of the expression host. A total of 1230 bp, coding for a reading frame of 410 amino acids, were synthesized and subsequently cloned into pUC57. Competent E. coli DH5α cells were transformed with pUC57-Phy. Successfully-transformed clones were selected by colony PCR, using universal M13 primers (Clontech, USA) and employed for plasmid preparation and cloning steps. Restriction enzymes, NcoI and BamHI, and T4 DNA ligase (New England Biolabs, USA), were applied to subclone the extracted (CinnaPure DNA extraction kit, CinnaGen, Iran) phytase gene into pET26b (+) expression vector. The final construct was transformed into E. coli BL21 competent cells, following confirmation by BamH1–NcoI double digestion. Successfully-transformed clones were selected by colony PCR, using universal T7 primers (Clontech, USA).
Rational design of mutation
Three-dimensional structure of phytase was obtained from the PDB entry, 1DKL, having 100% sequence homology with our wild-type bacterial phytase. One subunit of the enzyme and other additional molecules were removed for easier structural analysis using the ConText software.
In order to evade destabilizing mutations, SRide software (http://webclu.bio.wzw.tum.de/stride).
was used, which predicts stabilizing residues in the protein structure [16]. Suggested as stabilizing residues, Val12, Ala21, Phe299, Val334 and Val 350 were not considered as mutation candidates. As a polar amino acid, serine was proposed to enhance protein stability, whereas structural analysis using visual molecular dynamics (VMD software version 1.9.3; http://www.ks.uiuc.edu/Research/vmd), indicated reverse effect when adjoining a non-polar amino acid such as tryptophan [17]. Thus, a non-polar amino acid was decided to replace this residue, potentially resulting in hydrophobic interactions between the two non-polar amino acids, which could in turn improve protein structural stability. To investigate the effect of this mutation, MUpro software version 1.1 was used. Based on the prediction of this software, the S392F mutation could lead into an increase in the protein stability. MAESTROweb predicted ΔΔGpred of the mutant protein to be − 0.314, implying stabilizing role of the imposed mutation. This prediction was also supported by iMutant v2.0 and iStability software [18].
Three dimensional structure of the mutant phytase was subsequently predicted to analyze the effect of the suggested mutation in this context. Molegro Virtual Docker v6.0 software was applied to replace the wild-type residue with phenylalanine in 3D structure of phytase, and conformation of the surrounding amino acids was optimized to better mimic their native structures. Next, the effect of the introduced mutation was inspected on the whole structure of the protein by performing clean geometry analysis using Discovery Studio Visualizer, and energy minimization was studied using SPDBV 4.1 software.
Site-directed mutagenesis
Site-directed mutagenesis of Bac–Phy gene was conducted using QuikChange Site Directed Mutagenesis Kit (Stratagene, CA, USA). The pET26b vector containing Bac–Phy gene (1230 bp) with optimized codon preference for expression in E. coli was utilized to produce the mutant. In this methods, double stranded, dam methylated pET26b DNA with Bac–Phy gene isolated from E. coli DH5α was used with pair of designated complementary primers containing the desired point mutation (Table 1). The mutagenesis was extended by pfu Turbo DNA polymerase by using thermo cycling condition of 95 °C for 3 min, 18 cycles at 95 °C for 30 s, 67 °C for 45 s and 68 °C for 1 min/kb of plasmid DNA length. The product was treated by DpnI (Fermentase) at 37 °C for 1 h and nicked plasmid DNA was transformed to E. coli BL21 by which the nick was repaired. The plasmids harboring desired mutation was identified by DNA sequence analysis.
Phytase expression
Overnight culture of transformants were inoculated in 250 mL of LB medium contain Kanamycin and cultured at 37 °C until OD600 reached 0.8–1. After induction with 1 mM isopropyl β-thiogalactopyranoside (IPTG), the induced cells were cultured for 18 h at 37 °C in a shaker incubator. Phytase activity was measured in an assay mixture containing 20 mM sodium phytate and 200 mM sodium acetate buffer (pH 5.5) [19]. Following the incubation of 30 min at 37 °C, the reaction was stopped by adding acetone-molybdate-acid (AMA), stopping reagent, and liberated phosphate was measured by ammonium molybdate method at 380 nm [20]. Additional, the protein concentration was measured by Bradford assay.
Purification and biochemical characterization of expressed phytases
Culture of WT Bac–Phy and transformant Bac–Phy mutant were centrifuged at 11,000 rpm for 20 min at 4 °C. The filtered supernatants were concentrated by using Amicon centrifugal filter device (Cut-off, 30,000). The concentrated supernatants were loaded on Superdex size Column for gel permeation chromatography (Amersham, Inc, USA), which has previously been equilibrated with 0.1 M sodium acetate buffer (pH 5.5), then eluted with 0.1 M sodium acetate containing 0.15 M NaCl (pH 5.5) with flow rate of 2.5 ml min−1 using AKTA Purifier Fast Protein Liquid Chromatography system (Amersham bioscience, USA). The fraction profiles of OD280 and phytase activity were analysed to determine the desired protein peaks. The peak fractions were pooled and concentrated by Amicon centrifugal filter device and stored at − 20 °C for further characterization.
The phytase pH profile was determined at 37 °C with different buffers; 0.2 M glycin–Hcl (pH 1.0, 2.0 and 2.5) 0.2 M sodium citrate (pH 3.0, 4.0, 5.0 and 5.5), and 0.2M Tris–Hcl (pH 6.0 to 8.0) [19]. The temperature profile of phytase was investigated with 200 mM sodium acetate buffer (pH 5) ranging from 20 to 80 °C with 5 °C intervals. To evaluate thermostability, purified phytases were incubated at temperatures ranging from 40 to 90 °C for 10 min and they were placed on ice for 30 min immediately after heat treatment to assay phytase residual activity at 37 °C (pH 5.5). To determine the half-life of the expressed phytases, the enzyme was incubated at 80 and 90 °C for 0–60 min and then placed on ice for 30 min before being tested at 37 °C as described above.
Evaluation of kinetic characteristics
Kinetic parameters of Km and Vmax, were evaluated for the mutant phytase at 37 °C and pH 5.5 using gradient concentrations of sodium phytate salts (ranging from 50 to 1000 µM) as the substrate in 0.2 M sodium acetate buffer for 10 min. Results of triplicate experiments on mutant and wild-type phytases were compared and data obtained were analyzed by creating a Michaelis–Menten plot of reaction rate versus substrate concentration using GraphPad Prism. The Vmax values were converted to Kcat value, by normalizing the enzyme concentrations by the molecular mass of the monomer. Finally, catalytic efficiency (Kcat/Km) of the phytases was measured.
Statistical analysis
Data were analyzed by Minitab (release 14; Minitab State College, PA, USA). The Student t test was used to compare mean differences. Significance was set at a P value of < 0.05.
Results
Analysis of 3D structure
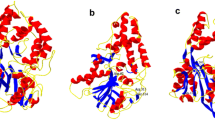
Based on the results obtained from bioinformatic evaluations, energy level of the protein structure was dramatically decreased from 15,990 to − 13,845 kJ/mol. TM-align algorithm was employed for comparing the structure of the mutant and wild-type molecules, and the root-mean-square deviation of atomic positions imposed by the mutation was measured around 0.37 Å, which inferred a high degree of structural similarity between the two molecules. This can be interpreted as estimation that the stabilized conformation of the protein may not change and the enzymatic activity of the phytase would be conserved following introduction of the mutation. Figure 1 depicts synchronized structure of the wild-type and mutant phytases. Therefore, it may be concluded that the active site and structural domains were not affected by the substitutions.
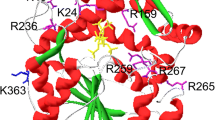
To study the effect of the mutation on the interactions of amino acids adjacent to the mutation point, LigPlot+v1.4.5 software was used and a comparison between the wild-type and mutant structures suggested a significant increase in hydrophobic interactions inflicted by the mutation. As shown in Fig. 2, the introduced mutation, S392F, forms two new hydrophobic interactions with the surrounding amino acids, which can lead to an increase in structural stability of the mutant phytase.
Expression analysis
Amino acid sequence of E. coli (K12) appA gene was translated into a codon-optimized nucleotide sequence and used for gene synthesis. The synthesized gene was cut out of the carrying plasmid, subcloned into pUC57 and transferred to E. coli DH5α cells (Fig. 3).
Expression of WT and mutant phytase in E. coli BL21 cells was analyzed on a 12% SDS-PAGE gel. Both proteins were overexpressed, concluded by 45-KD protein bands attributed as the molecular weight of the phytase enzyme. Purified phytases in the culture supernatants were also visualized by the same approach (Fig. 4). Based on the standard curve plotted using known concentrations of BSA, 0.351 mg/mL and 0.333 mg/mL of wild-type and mutant phytase was detected in their respective culture supernatants.
Effect of S392F mutation on pH and temperature Optima
To determine pH range of activity and optimum pH of phytase, E. coli expressed WT and mutant phytase activity were determined at different pH. The optimum pH of WT and mutant was 5 and 6 respectively with a significant pH shift. Furthermore mutant phytase showed 12.8%, 9.6% and 10.3% higher phytase activity at pH 6, 7 and 8, respectively in compare with WT (Fig. 5).
In order to determine WT and mutant phytase behavior at different temperature, purified recombinant samples were assayed for phytase activity at wide range of temperatures. It was discovered that WT and mutant phytase have optimum temperature of 50 °C (Fig. 6).
Culture supernatants pretreated at various temperatures were measured for their residual phytase activity and the results indicated fairly higher thermostability of the mutant phytase compared to its wild-type counterpart despite of phytase stability reduction at high temperatures. Analysis of the retained enzymatic activity of mutant phytase at high temperatures indicated twice thermo stability improvement at 70 °C and respectively 74% and 78.4% higher thermostability at 80 °C and 90 °C in compare with WT (Fig. 7).
Characterization of expressed phytases kinetic parameters
Kinetic analysis indicated that the apparent Km and Vmax of the WT was 19.45 µM and 98.5 ± 0.8 µmol min−1 mg−1, respectively. The Km value of mutant for sodium phytate was 20% lower than the WT (P < 0.05). Moreover, the catalytic efficiency values (Kcat/Km) of the mentioned mutant was improved about 25.6%, compared with the WT phytase which indicated a significant increase of kinetic efficiency for sodium phytate (Table 2).
Discussion
Based on the growing phytase market volume, developing efficient platforms for the production of manipulated phytases with favorable biochemical properties is irrefutable [21]. A wide range of bacterial, yeast and fungal hosts are employed for industrial production of phytase. On account of posttranslational modifications, host organism plays an important role in phytase activity and thermostability. Despite high expression level and increased specific activity of phytase in yeast, E. coli is the preferred host organism for this protein, due to its fast growth, non-chromosomal integration and high plasmid transformation efficiency [22].
Phytases are normally exposed to temperatures above 60 °C during feed pelleting applications, while most wild-type phytases undergo irreversible heat denaturation at this point. The unfolding temperature of phytase can be enhanced by protein engineering. Theoretically, any attempt toward increasing the number of intramolecular interactions may lead into a decrease in total free energy of the protein and consequently improves structural stability of the molecule [9]. This can be achieved through introduction of non-polar amino acids, including phenylalanine, glycine, alanine, proline, valine, leucine, isoleucine, methionine and tryptophan to the protein primary structure. Hydrophobic interaction between these residues naturally improves protein resistance against harsh conditions of heat and pH [23].
Particular guidelines for improving the thermostability of a target protein commonly evolve from recorded 3D structure data of the wild-type protein and its heat-resistant homologous variants, as well as general rules known about protein structures associated with thermostability [9]. Random mutagenesis is also applied to imitate the nature in developing and selecting improved variants with favorable physiochemical properties. The former approach was successfully used by Van den Burg et al. to enhance the heat resistance of Bacillus stearothermophilus thermolysin-like protease by 21 °C through substitution of eight amino acid residues [13]. Another study reported 20% improvement in phytase thermal tolerance by stabilizing local interactions using error-prone polymerase reaction. Chen et al. also reported 15% higher activity remained after heating at 75 °C for 10 min for the F43Y single mutant compared to the wild-type A. niger phytase [24]. Random mutagenesis includes a laborious and time-consuming process, therefore, prediction of the effect of mutations on the protein structure can be highly advantageous to rationalize specific modifications in silico [25]. Bioinformatic analysis represents a promising means for rational design of a molecule with favorable features. Through increasing knowledge on structural dynamics and rational design-based molecular engineering, mutations in protein sequence with potential to improve intended structural characteristics are identified and used for the prediction of primary, secondary and tertiary structure of the protein [26].
Generally, thermostability profile of a protein is governed by basic physiochemical forces between the constituting residues, including hydrogen bonding network, ionic and hydrophobic interactions, molecular flexibility in loop regions and surface charge–charge interactions. Additionally, other structural factors such as core and side chain packing, oligomerization, proline substitutions, helical content and helical propensities contribute to the thermal resistance of a protein [23].
In 1997, Pasamontes et al. developed the first thermostable recombinant phytase with 90% retained activity following 20 min heat treatment at 100 °C. Wang et al. increased phytase Tm by 7.5 °C via introduction of 10 point mutations, attributing this improvement to an enhancement of α-helix content and greater exposed hydrophobic surface [27]. An increase in glycosylation sites was also successfully employed to enhance phytase thermotolerance [28]. Fei et al. reported 39% increase in thermostability of phytase by generating a C-terminal deletion mutant [29]. Another study developed a phytase with 60% retained activity after heat treatment [30]. Kim et al. documented two phytase variants (K46E and K65E/K97M/S209G) with over 20% enhanced thermostability and 6–7 °C increased melting temperature, which was described to be caused by additional hydrogen bonds formed between the introduced amino acids with adjacent residues [24]. Double mutation, F89S and E226H, predicted by molecular dynamic analysis, resulted in improved stability of the engineered phytase against heat and proteases [31]. Few attempts have been made to improve phytase thermal resistance by incorporating hydrophobic interactions. Although rarely found in protein sequence, aromatic amino acids show thermostabilizing properties in most cases. Shivange et al. produced the phytase variant V298F with improved thermal and pH resistance, by introduction of additional hydrophobic interactions [32]. Based on the literature, 91.5% increase and 13.96% decrease in thermostability of E. coli phytase was observed by salt bridge addition or subtraction mutants, respectively. The amino acid residues, Glu31, E35, S42, K46, S51, F89, R168, E226, R248 and P257 were also previously associated with the thermostability of phytase [33]. Our observations suggested S392 as a novel key amino acid with significant influence on the thermal tolerance of phytase.
Generally, hydrophilic amino acids are found on the solvent exposed surface of the protein, whereas hydrophobic residues are buried inside of the protein folding. This hydrophobic effect stabilizes protein folding through interaction of side chains in the non-polar core of the protein, as well as hydrogen bonding network of nonpolar side chains on its surface. However, not all buried residues are nonpolar, which may provide an opportunity for protein engineering. Replacement of some polar amino acids in the folded protein interior may result in formation of new hydrophobic bonds and improvement of the protein stability [34].
Previously, we reported three double mutations (S205N-S206A, T314S-Q315R, T151A-V62N) with enhancing effects on the thermostability and catalytic efficiency of phytase, using bioinformatics tools [19]. In the present study, we used molecular dynamics to predict a rationalized thermostabilizing single mutation in E. coli phytase. In an effort to design a thermotolerant enzyme, several softwares were employed and 3D structure of the substituted phytase was compared to that of its wild-type counterpart. Many studies have identified the hydrophobic bond network as one of the most important stabilizing factors for thermostable proteins [19]. Based on the results of the molecular dynamics studies, the proposed mutation, S392F, forms two new hydrophobic bonds with the amino acid Trp347. This may justify the improved thermostability reported for the recombinant phytase. Furthermore, a dramatic decline in energy level of the protein structure to − 13,845 kJ/mol, may have contributed to conformational stabilization, resulting in reinforcement of the protein tertiary structure. Therefore, improvement of thermostability can be a result of minor global and local conformational changes, stabilizing local or long-rage interactions and reducing overall entropy of the molecule. An improvement of the thermodynamic stability of a protein increases the free-energy difference between the unfolded and the folded state or decreases the rate of unfolding by enhancing the free energy difference between the folded state and the transition state of unfolding [9].
Despite the significant change in the molecular free energy, data obtained from structural comparison using TM-align algorithm implied minimal deviation in atomic positions between the two phytases. Thus, a high degree of structural similarity might be an explanation for nearly intact enzymatic activity of the mutant phytase in comparison to the wild-type molecule. It is noteworthy that replacement of the mutated S392 residue, with three other hydrophobic amino acids of Trp, Val and Tyr was also suggested by our bioinformatics structural analysis to have thermostabilizing effects on the phytase structure, in the same manner.
Thermostability analysis indicated that culture supernatant containing the mutant phytase has higher remained specific activity comparing to the wild-type phytase. The introduced mutation had minimal effect on pH profile of the phytase, with only 10% improved activity at pH 6. Similarly, kinetic properties of the mutant phytase were slightly higher, while both enzymes displayed comparable phytase activity at different concentrations of the substrate.
In conclusion, our observations are an endorsement to the effectiveness of predictions based on bioinformatics analysis and the possibility of using information obtained from molecular dynamics studies to estimate 3D structure and physiochemical properties of engineered proteins. Such studies can help build a fairly realistic correlation between in silico and in vitro data, which may in turn lead into significant developments in protein engineering toward improved industrial applicability. Based on our results, the designed phytase possesses industrially favorable physiochemical and biochemical characteristics, signifying the introduction of a phytase with a potential to be used as an additive in animal livestock.
References
Vohra A, Satyanarayana T (2003) Phytases: microbial sources, production, purification, and potential biotechnological applications. Crit Rev Biotechnol 23:29–60
Noureddini H, Dang J (2009) Degradation of phytates in distillers’ grains and corn gluten feed by Aspergillus niger phytase. Appl Biochem Biotechnol 159:11–23
Greiner R, Farouk A-E, Carlsson N-G, Konietzny U (2007) myo-Inositol phosphate isomers generated by the action of a phytase from a Malaysian waste-water bacterium. Protein J 26:577–584
Bohn L, Meyer AS, Rasmussen SK (2008) Phytate: impact on environment and human nutrition. A challenge for molecular breeding. J Zhejiang Univ Sci B 9:165–191
Haros M, Bielecka M, Honke J, Sanz Y (2007) Myo-inositol hexakisphosphate degradation by Bifidobacterium infantis ATCC 15697. Int J Food Microbiol 117:76–84
Vats P, Banerjee UC (2004) Production studies and catalytic properties of phytases (myo-inositolhexakisphosphate phosphohydrolases): an overview. Enzyme Microb Technol 35:3–14
Elkhalil E, Männer K, Borriss R, Simon O (2007) In vitro and in vivo characteristics of bacterial phytases and their efficacy in broiler chickens. Br Poult Sci 48:64–70
Lehmann M, Pasamontes L, Lassen S, Wyss M (2000) The consensus concept for thermostability engineering of proteins. Biochim Biophys Acta 1543, 408–415
Lehmann M, Wyss M (2001) Engineering proteins for thermostability: the use of sequence alignments versus rational design and directed evolution. Curr Opin Biotechnol 12:371–375
Zhang W, Mullaney EJ, Lei XG (2007) Adopting selected hydrogen bonding and ionic interactions from Aspergillus fumigatus phytase structure improves the thermostability of Aspergillus niger PhyA phytase. Appl Environ Microbiol 73:3069–3076
Zhang W, Lei XG (2008) Cumulative improvements of thermostability and pH-activity profile of Aspergillus niger PhyA phytase by site-directed mutagenesis. Appl Microbiol Biotechnol 77:1033–1040
Lehmann M, Kostrewa D, Wyss M, Brugger R, D’Arcy A, Pasamontes L et al (2000) From DNA sequence to improved functionality: using protein sequence comparisons to rapidly design a thermostable consensus phytase. Protein Eng 13:49–57
Van den Burg B, Vriend G, Veltman OR, Venema G, Eijsink VG (1998) Engineering an enzyme to resist boiling. Proc Natl Acad Sci 95:2056–2060
Zhao Q, Liu H, Zhang Y, Zhang Y (2010) Engineering of protease-resistant phytase from Penicillium sp.: high thermal stability, low optimal temperature and pH. J Biosci Bioeng 110:638–645
Damborsky J, Brezovsky J (2014) Computational tools for designing and engineering enzymes. Curr Opin Chem Biol 19:8–16
Heinig M, Frishman D (2004) STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucl Acids Res 32:W500–W502
Humphrey W, Dalke A, Schulten K (1996) VMD—visual molecular dynamics. J Mol Graph 14:33–38
Laimer J, Hofer H, Fritz M, Wegenkittl S, Lackner P (2015) MAESTRO—multi agent stability prediction upon point mutations. BMC Bioinform 16:116. https://doi.org/10.1186/s12859-015-0548-6
Hesampour A, Siadat SER, Malboobi MA, Mohandesi N, Arab SS, Ghahremanpour MM (2015) Enhancement of thermostability and kinetic efficiency of Aspergillus niger PhyA phytase by site-directed mutagenesis. Appl Biochem Biotechnol 175:2528–2541
Hesampour A, Siadat SOR, Malboobi MA, Harati J, Mohandesi N (2014) Comparison of biochemical properties of recombinant phytase expression in the favorable methylotrophic platforms of Pichia pastoris and Hansenula polymorpha. Prog Biol Sci 4:97–111
Bhavsar K, Khire J (2014) Current research and future perspectives of phytase bioprocessing. RSC Adv 4:26677–26691
Shivange AV, Schwaneberg U (2017) Recent advances in directed phytase evolution and rational phytase engineering. In: Directed enzyme evolution: advances and applications. Springer, New York, pp 145–172
Kumar S, Tsai C-J, Nussinov R (2000) Factors enhancing protein thermostability. Protein Eng 13:179–191
Kim M-S, Lei XG (2008) Enhancing thermostability of Escherichia coli phytase AppA2 by error-prone PCR. Appl Microbiol Biotechnol 79:69–75
Lutz S (2010) Beyond directed evolution—semi-rational protein engineering and design. Curr Opin Biotechnol 21:734–743
Wu T-H, Chen C-C, Cheng Y-S, Ko T-P, Lin C-Y, Lai H-L et al (2014) Improving specific activity and thermostability of Escherichia coli phytase by structure-based rational design. J Biotechnol 175:1–6
Wang X, Yao M, Yang B, Fu Y, Hu F, Liang A (2015) Enzymology and thermal stability of phytase appA mutants. RSC Adv 5:43863–43872
Ushasree MV, Shyam K, Vidya J, Pandey A (2017) Microbial phytase: Impact of advances in genetic engineering in revolutionizing its properties and applications. Bioresour Technol 245:1790–1799
Fei B, Cao Y, Xu H, Li X, Song T, Fei Z et al (2013) AppA C-terminal plays an important role in its thermostability in Escherichia coli. Curr Microbiol 66:374–378
Zhang J, Liu Y, Gao S, Zhu L, Li W, Tian X et al (2016) Site-directed mutagenesis and thermal stability analysis of phytase from Escherichia coli. Biosci Biotechnol Res Commun 9:357–365
Niu C, Yang P, Luo H, Huang H, Wang Y, Yao B (2017) Engineering of yersinia phytases to improve pepsin and trypsin resistance and thermostability and application potential in the food and feed industry. J Agric Food Chem 65:7337–7344
Shivange AV, Dennig A, Schwaneberg U (2014) Multi-site saturation by OmniChange yields a pH-and thermally improved phytase. J Biotechnol 170:68–72
Rebello S, Jose L, Sindhu R, Aneesh EM (2017) Molecular advancements in the development of thermostable phytases. Appl Microbiol Biotechnol 101:2677–2689
Pace CN, Scholtz JM, Grimsley GR (2014) Forces stabilizing proteins. FEBS Lett 588:2177–2184
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fakhravar, A., Hesampour, A. Rational design-based engineering of a thermostable phytase by site-directed mutagenesis. Mol Biol Rep 45, 2053–2061 (2018). https://doi.org/10.1007/s11033-018-4362-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-018-4362-x