Abstract
Degenerative myelopathy (DM) is a late-onset, slowly progressive degeneration of spinal cord white matter which is reported primarily in large breed dogs. The missense mutation SOD1:c.118G>A is associated with this pathology in several dog breeds, including the German Shepherd Dog (GSD). The aims of the present study were to develop a tool for the rapid screening of the SOD1 mutation site in dogs and to evaluate the association of the polymorphism with DM in the German Shepherd breed. Two different techniques were compared: a minisequencing test and a real-time pcr allelic discrimination assay. Both approaches resulted effective and efficient. A sample of 47 dogs were examined. Ten subjects presented the symptoms of the illness; for one of them the diagnosis was confirmed by postmortem investigations and it resulted to be an A/A homozygote. In another clinically suspected dog, heterozygote A/G, the histopathological examination of the medulla showed moderate axon and myelin degenerative changes. GSD shows a frequency of the mutant allele equal to 0.17, quite high being a high-risk allele. Because canine DM has a late onset in adulthood and homozygous mutant dogs are likely as fertile as other genotypes, the natural selection is mild and the mutant allele may reach high frequencies. A diagnostic test, easy to implement, may contribute to control the gene diffusion in populations. The SOD1:c.118G>A mutation could be a useful marker for breeding strategies intending to reduce the incidence of DM.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Degenerative myelopathy (DM) is a late-onset, slowly progressive degeneration of the spinal cord white matter which is reported primarily in large-breed dogs. The condition has been described in German Shepherd Dog (GSD) as well as in other breeds like Siberian Husky, Miniature Poodle, Boxer, Pembroke Welsh Corgi, Chesapeake Bay Retriever, and Rhodesian Ridgeback [1]. Dogs with DM show an insidiously progressive ataxia and paresis of the pelvic limbs, ultimately leading to loss of the ability to ambulate within 6–12 months of the onset of signs. Definitive diagnosis of DM is confirmed by postmortem histological examination of the spinal cord [2]. There is no sex bias. Most dogs are at least 8 years old before the onset of clinical signs [1]. DM appears to be an autosomal, recessive, and incompletely penetrant disease [3].
Human amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by adult-onset, progressive loss of motor neurons in the motor cortex, brainstem and spinal cord (lower motor neurons), leading to muscle atrophy, generalized paralysis, respiratory depression, and inevitable death [4–7]. In 1993, mutations in superoxide dismutase 1 (SOD1) gene were associated with familial ALS [8]. So far, more than a hundred SOD1 different mutations have been identified in ALS patients. Mutations in other genes have been reported to account for a small number of cases of familial ALS [9].
Interestingly, relationship between the missense mutation SOD1:c.118G>A and canine DM has recently been reported [3].This mutation was associated with DM in several dog breeds, including the GSD. The related SNP could be a useful marker for breeding strategies intending to reduce the incidence of DM.
The first aim of the present study was to develop a tool for rapid screening of the SOD1 mutation site in dogs comparing two different molecular approaches: (1) minisequencing (or primer-extension reaction—PER) and (2) real-time pcr allelic discrimination TaqMan minor groove binding (MGB). The second aim was to evaluate the association of SOD1c.118G>A mutation with DM in GSD.
Materials and methods
Sample collection
Peripheral blood samples were collected from 47 GSD of both genders aged from 5 months to 14 years, including ten clinically suspected of having a progressive neurological disease. Dogs were patients from the Veterinary Hospital, University of Turin, and from several private veterinary practitioners. Blood samples were stored at −22 °C until analysed.
DNA extraction
Genomic DNA was obtained from blood using the NucleoSpin Blood Quick Pure kit (Macherey–Nagel GmbH & Co. KG, Düren, Germany), according to the manufacturer protocol, and stored at −20 °C until use. DNA purity was evaluated by absorbance readings using the NanoDrop-2000 (Thermo Fisher Scientific Inc., Waltham, Massachusetts).
Minisequencing test
The PER reaction is based on the addition of a [F]ddNTP to the 3′ end of an unlabeled oligonucleotide (sequencing primer) in the absence of dNTPs in the reaction. Each [F]ddNTP is labeled with a specific fluorescent dye and each sequencing primer is designed immediately adjacent to the diagnosis site.
Preliminary PCR
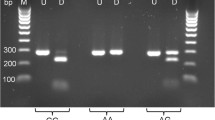
The PCR primers used in the preliminary PCR tests are shown in Fig. 1 (Eurofins MWG Operon, Ebersberg, Germany) [3], PCR reactions were performed in 50 μL volumes consisting of 0.04 U/μL HotStarTaq (Qiagen, Hilden, Germany), 0.2 mM each of dNTPs,0.5 μM of each primer, and 50–100 ng of DNA template. The PCR profile consisted of an initial activation step at 95 °C for 15 min, followed by 35 cycles of 95 °C denaturation for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. A final extension step of 72 °C for 7 min was added to all reactions. Amplifications were carried out using a GeneAmp PCR System 2,720 thermal cycler (Applied Biosystems by Life Technologies Italia, Monza, Italy). Amplicons were resolved on a 2.5 % agarose gel.
Primer-extension reaction (PER)
The PCR products acted as templates for PER after enzymatic clean up. In order to remove primers and unincorporated dNTPs, 2 μL of Exo-Sap (GE Healthcar Europe Gmbh, Glattbrugg, Schweiz) were added to 5 μL of PCR products. Tubes were incubated at 37 °C for 1 h and at 80 °C for 15 min.
Based on SOD1 sequence alignments, the sequencing primer was designed upstream from the SOD1 mutation site associated with DM (5′-GAACCATTACAGGGCTGACT-3′) (Fig. 1) PER reactions were performed following the SNaPshot multiplex Kit protocol (Applied Biosystems by Life Technologies Italia, Monza, Italy) with minor modifications: 2.5 μL of Ready Reaction Mix, 3 μL of purified amplicons from the preliminary PCR, diluted to obtain a range of 0.4–0.01 pmol and 0,2 μM of sequencing primer in a total volume of 10 μL. The reactions underwent 25 single base extension cycles of denaturation at 96 °C for 10 s, annealing at 50 °C for 5 s, and extension at 60 °C during 30 s. The reactions were carried out in a GeneAmp PCR System 2,720 thermal cycler (Applied Biosystems by Life Technologies Italia, Monza, Italy).
Subsequently, a post-extension treatment to remove 5′-phosphoryl group of the [F]ddNTPs helped to prevent unincorporated [F]ddNTPs co-migrating with the extended primers and producing a high background signal. For this purpose, the final volume (10 μL) was treated with 1 unit of Calf Intestine Alkaline Phosphatase (Invitrogen by Life Technologies Italia, Monza, Italy). The tubes were incubated at 37 °C for 1 h and at 65 °C for 15 min. Finally, samples were prepared by adding 1 μL of the post-PER product to 12 μL of formamide and 0.2 μL of GeneScan 120 LIZ size standard. Each sample was loaded on the ABI 310 Genetic Analyzer and analyzed using Pop4 polymer, a 47-cm capillary column, the ABI GeneScan E5 Run Module. Parameters for this analysis are the following: injection time 12 s and run time 15 min. Electropherograms were analysed using the GeneScan 3.1.2 software (Applied Biosystems by Life Technologies Italia, Monza, Italy).
Real-time PCR allelic discrimination test
In our study, we develop a real-time pcr allelic discrimination TaqMan MGB assay based on the SOD1:c.118G>A mutation analysis.
Recently, the real-time pcr allelic discrimination test has been largely used to genotype individual DNA at known muatation site [10].
During amplification, the fluorogenic TaqMan probes anneal specifically to complementary sequences between forward and reverse primer binding sites on the DNA template and they are then degraded owing to the 5′–3′ exonuclease activity of DNA polymerase. Once separated from the quencher, the reporter dye emits fluorescence, which is read by the real-team PCR system.
In the assay here described, the two probes are 100 % identical with their respective target sequences; only a single mismatch in the 5′ end is observed between them. The contemporary presence of the two probes allows the annealing of both regardless of the allele present because the single mismatch does not impair the probes’ annealing to heterologous DNA. During the extension, the 5′–3′ exonuclease activity of the DNA polymerase degrades the probe that is totally identical to the template with a consequent fluorescent emission. On the contrary, the probe that has a mismatch with the sequence is removed without degradation and devoid of fluorescent emission. The post-PCR analysis allows the instantaneous visualization of the results by mean of a specific plot.
Primer and probe design
The PCR primers [3] were used in conjunction with two MGB probes (Applied Biosystems by Life Technologies Italia, Monza, Italy). One probe was designed to specifically hybridize, with a 100 % of identity, to one allele (SOD1_ProbeA 5′-6FAM-CTGACTAAAGGCGAG-3′) and the other one to the second allele (SOD1_ProbeG 5′-VIC-CTGACTGAAGGCGAGC-3′) (Fig. 2).
Analysis of diagnostic site
Real-time PCR was performed in a 25 μL reaction mixture containing 900 nM of both forward and reverse primers, 200 nM of each probe, 12.5 μL of TaqMan Universal Master Mix (Applied Biosystems by Life Technologies Italia, Monza, Italy), and 30–50 ng of DNA template. Thermal cyclings were performed on a 7,300 real-time PCR system (Applied Biosystems by Life Technologies Italia, Monza, Italy) under the following conditions: 95 °C for 10 min, and 45 cycles of 95 °C for 15 s and 60 °C for 1 min. All PCR reactions were run in duplicate.
Results were obtained by the automatic calling feature of the allelic discrimination option in SDS v1.2.1 software. Alleles were assigned to samples by visual inspection of a plot of the fluorescence (Rn) from the A probe versus Rn from G probe generated from the post-PCR fluorescence read.
Sequencing
DNA sequencing was carried out as a confirmatory test. Amplified fragments of the preliminary PCR of a homozygous AA dog and a homozygous GG animal were cloned into the pDRIVE Cloning Vector using QIAGEN PCR Cloning Kit (GE Healthcar Europe Gmbh, Glattbrugg, Schweiz). Clones were subsequently submitted to RFLP analysis using Hae III to choose a clone that contained the diagnostic site. These cloned fragments were cycle sequenced on an ABI PRISM 310 Genetic Analyser (Applied Biosystems by Life Technologies Italia, Monza, Italy), using the ABI Prism BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems by Life Technologies Italia, Monza, Italy), version 1.1 by the dideoxy chain termination method with fluorescence dye terminators. The sequencing on both strands was performed using the two M13 vector primers. The resulting sequences were compared and aligned with ‘ClustalW’ program [11].
Results
Minisequencing test
The primers used for the preliminary PCR successfully amplified a 79 bp fragment. For homozygote dogs the PER analysis gave rise to a peak of the color that is specific to the nucleotide present on template, green for A and blue for G. In heterozygote animals both peaks, green and blue, were generated (Fig. 3). On 47 GSDs analysed by minisequencing, two homozygote individuals for the mutant allele (AA) were detected, 12 heterozygous GA, all other dogs were homozygote for the wild allele (GG).
Real-time PCR allelic discrimination test
Real-time pcr allelic discrimination test confirmed the genotypes obtained with PER. A representative plot used to genotype individuals is displayed in Fig 4. The real time test was applied to all 47 individuals. Two dogs were confirmed to be A/A, 33 dogs were G/G and 12 G/A. In the examined cohort the allele frequencies were 0.17 and 0.83 for A and G, respectively.
Sequencing
Ultimately, the sequencing of the amplicons obtained from the preliminary PCR confirmed the reliability of both techniques for genotyping the SOD1:c118G>A polymorphism in dog.
Association between DM and SOD1c.118G>A
Out of the 47 dogs examined in this study, ten were clinically suspected to have DM. Therefore, depending on availability of tissues and clinical information, diagnoses of DM were made on the basis of criteria of varying stringency. Only in one case the clinical diagnosis was confirmed by postmortem investigations (histopathology of the spinal cord tissue) (data not shown). It was a male, 8 years old, and it resulted to be an A/A homozygote. In another clinically suspected dog, a male 14 years old, heterozygote A/G, the histopathological examination of the medulla showed only moderate axon and myelin degenerative changes limited to the thoracic segments. One more A/A clinically suspected animal is still alive. For all the other clinically suspected dogs (1 A/G and 6 G/G) the diagnosis was not confirmed by histopathological examination of the medulla or unfortunately the necropsy was not carried out.
From the other 37 dogs not suspected to have DM, none was homozygote A/A, 27 were homozygotes G/G, and the other heterozygotes.
Discussion
In a previous study SOD1 in normal and DM affected dogs was sequenced and a G to A transition in exon 2, predicting an E40 K missense mutation, was observed [3]. Homozygosity for the A allele was highly associated with DM in five dog breeds (GSD, Pembroke Welsh Corgi, Boxer, Rhodesian ridgeback, and Chesapeake Bay retriever); therefore the A allele was recognized as a major genetic risk factor for the disease. Recently, two genotyping assays for the SOD1 mutation, using conventional and real time PCR methods have been proposed [12]. The aim of our study was to develop a test for rapid screening and identification of SOD1:c118G>A mutation carriers in dogs using two different approaches: a minisequencing test and a real-time pcr allelic discrimination test. Both approaches are effective and efficient, all genotypes were clearly determined without non-specific allelic amplification. Costs for consumables and reliability of results are similar. Real time allelic discrimination test provides several advantages because it does not require any post-amplification step and utilizes 96-well format plates that can be read in about 5 min, enabling a rapid diagnosis of large numbers of samples. On the other hand, the reads of the minisequencing test have a precision comparable to that of the sequencing. This technique can be suitable for automation and high-throughput testing for laboratories equipped with an ABI sequencer. In comparison with sequencing, the minisequencing has the advantage of being quicker, allowing an immediate interpretation of the results. Moreover, it may be implemented for the multiplex analysis of several mutation sites. Recently, a novel SOD1 missense mutation, associated with DM, has been described in the Bernese mountain dog breed [13]. It would be interesting to implement a minisequencing test for the concurrent genotyping of multiple SNPs. Ultimately, the choice of the more suitable approach depends on laboratory disposable equipment.
Representatives of 217 different dog breeds were genotyped at SOD1:c.118G>A and 115 of them were found to segregate the A allele [14].The wire fox terrier had the highest SOD1:c.118A allele frequency (0.98). Likewise, in a survey on Pembroke Welsh Corgi, the mutant allele was found to be prevalent (0.68) [12]. In the present investigation, GSD shows a frequency of the mutant allele equal to 0.17, quite high being a high-risk allele. Because canine DM has a late onset in adulthood and homozygous mutant dogs are likely as fertile as other genotypes, the natural selection is mild and the mutant allele may reach high frequencies. A diagnostic test, easy to implement, may contribute to control the gene diffusion in populations.
According to the hypothesis of Awano [3], the DM seems to be an age-related, incompletely penetrant disease. As far as the gene action is concerned, a model of inheritance based on dominant allele, as in humans, may not be excluded. In fact, not only the homozygote dogs exhibit clinical signs, but also heterozygote animals develop the pathology, although very slowly. Therefore, the clinical signs become apparent solely in dogs surviving longer the usual canine life span.
References
Coates JR, Wininger FA (2010) Canine degenerative myelopathy. Vet Clin North Am Small Anim Pract 40:929–950
March PA, Coates JR, Abyad R et al (2009) Degenerative myelopathy in 18 Pembroke Welsh corgi dogs. Vet Pathol 46:241–250
Awano T, Johnson GS, Wade C et al (2009) Genome-wide association analysis reveals a SOD1 missense mutation canine degenerative myelopathy that resembles amyotrophic lateral sclerosis. Proc Natl Acad Sci 106:2794–2799
Jonsson PA, Ernhill K, Andersen PM et al (2004) Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain 127:73–88
Shaw BF, Durazo A, Nersissian AM et al (2006) Local unfolding in a destabilized, pathogenic variant of superoxide dismutase 1 observed with H/D exchange and mass spectrometry. J Biol Chem 281:18167–18176
Rakhit R, Chakrabartty A (2006) Structure, folding, and misfolding of Cu, Zn superoxide dismutase in amyotrophic lateral sclerosis. Biochim Biophys Acta 1762:1025–1037
Marucci G, Morandi L, Bartolomei I et al (2007) Amyotrophic lateral sclerosis with mutation of the Cu/Zn superoxide dismutase gene (SOD1) in a patient with Down syndrome. Neuromuscul Disord 17:673–676
Rosen DR, Siddique T, Patterson D et al (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362:59–62
Pasinelli P, Brown RH (2006) Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci 7:710–723
Stopińska K, Grzybowski T, Malyarchuk BA et al (2006) Optimization of the Y831C mutation detection in human DNA polymerase gamma by allelic discrimination assay. Acta Biochim Pol 53:591–595
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Chang HS, Kamishina H, Mizukami K, et al.(2013) Genotyping assays for the canine degenerative myelopathy-associated c.118G>A (p.E40 K) Mutation of the SOD1 gene using conventional and real-time PCR methods: a high prevalence in the Pembroke Welsh corgi breed in Japan. J Vet Med Sci (Epub ahead of print)
Wininger FA, Zeng R, Johnson GS et al (2011) Degenerative myelopathy in a Bernese mountain dog with a novel SOD1 missense mutation. J Vet Intern Med 25:1166–1170
Zeng R, Coates JR, Hansen L, et al.(2012) The distributions of two SOD1 missense mutations in the pet dog population and their association with canine degenerative myelopathy, a model for amyotrophic lateral sclerosis. American Society of Human Genetics (ASHG)’s 62nd Annual Meeting, 6–10, November 2012 San Francisco, CA
Acknowledgments
The authors thank Alessandra Sereno for technical support.
Conflict of interest
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Maria Teresa Capucchio and Veronica Spalenza have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Capucchio, M.T., Spalenza, V., Biasibetti, E. et al. Degenerative myelopathy in German Shepherd Dog: comparison of two molecular assays for the identification of the SOD1:c.118G>A mutation. Mol Biol Rep 41, 665–670 (2014). https://doi.org/10.1007/s11033-013-2904-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-013-2904-9