Abstract
As a highly conserved nuclear protein, death domain-associated protein (Daxx) plays an important role in transcriptional control, carcinogenesis, and resistance to virus infection and so on. In order to further investigate the mechanism of Daxx, the yeast two-hybrid technique was used to screen the intra-cellular proteins interacting with Daxx. And 13 positive colonies and three proteins interacting with Daxx were obtained. One of the candidate proteins was identified as ferritin, heavy polypeptide 1(FTH1). The interaction between Daxx and FTH1 was further supported by GST pull-down and co-immunoprecipitation respectively. Then Daxx was determined to induce apoptosis and FTH1 can inhibit Daxx-mediated apoptosis. Besides, it is found that Daxx mediated apoptosis through the Fas-Daxx-ASK1-JNK1 signaling pathway, while FTH1 can inhibit the activation of JNK signaling pathway. We present evidence to demonstrate the FTH1 and Daxx are able to participate in apoptosis pathway through JNK signal molecule and FTH1 can inhibit this pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
DAXX, a death-domain-associated protein, was first discovered through its cytoplasmic interaction with the classical death receptor FAS. It has been associated heterochromatin and PML-NBs (Promyelocytic Leukaemia nuclear bodies) and has been implicated in many nuclear processes including transcription and cell cycle regulation [1]. Daxx interacts with pro-apoptotic receptors such as FAS and TGFβ receptor [2]. The canonical FAS pathway for apoptosis is based on the recruitment of the FAS associated death domain (FADD) protein, which, in turn, activates caspase-8 and downstream caspases. Daxx has been suggested to function as a pro-apoptotic protein downstream of FAS through activation of the c-Jun-N terminal kinase (JNK) pathway in a FADD-independent manner [1, 3]. Despite the reports advocating a pro-cell-death function for Daxx, several studies have suggested a potential antiapoptotic function for Daxx [4]. The function of Daxx remains unknown. In this study, we designed a bait plasmid containing the full of Daxx sequence to screen the proteins interacting with Daxx in adult liver cDNA library by the yeast two-hybrid system and 13 kinds of Daxx binding proteins were screened out in hepatocytes. These proteins were important to make sure Daxx molecular mechanism in liver cell. And it is significant to study the Daxx binding proteins to show the mechanism of Daxx in pro-apoptotic and anti-apoptotic function.
Materials and methods
Bacteria, yeast strains and plasmids
ProQuest two hybrid system (CAT SERIES 10835) and human adult liver cDNA library (ProQuest pre-made cDNA library) were obtained from Invitrogen (Carlsbad, USA). Yeast expression vectors pDBleu and pPC86 (both from Invitrogen) are fusion vectors for the linkage of proteins to the Gal4 DNA binding domain and to the VP16 transactivation domain, respectively. Escherichia coli JM109 and HEK293 cells were kept in our laboratory.
Chemical agents and culture media
Taq DNA polymerase was purchased from Tiangen Co. (Beijing, China). T4 DNA ligase, Sal I and Not I restriction endonucleases were from TaKaRa Biotechnology Co. (Dalian China). Both c-Myc mAb and goat anti-mouse IgG conjugated with horseradish peroxidase were from Zhongshan Co. (Beijing, China). Lithium acetate, semi-sulfate adenine, acrylamide, and N, N′-bis-acrylamide were from Sigma–Aldrich (St. Louis, USA). TEMED was from Boehringer Mannheim Co. (Mannheim, Germany). Tryptone and yeast extracts were from Oxoid Co. (Hampshire, UK). X-α-gal and culture media (YPAD, SD/-Trp, SD/-Leu, SD/-Trp/-Leu, SD/-Trp/-Leu/-His, and SD/-Trp/-Leu/-His/-Ade) were from Clontech (San Jose, USA). Protein-Gagarose was from Roche (Indianapolis, USA). Other chemical agents were from Sigma–Aldrich.
Bait plasmid construction
Take the adult liver cDNA library as DNA template, Daxx was generated by PCR amplification.The sequences of the primers containing the Sal I and Not I restriction enzyme sites were the following: sense primer, 5′-GCGTCGACC ATGGCCACC GCTAACAGCATC-3′ and anti-sense primer, 5′-TTGCGGCCG CTAATCAG AGTCTGAGAGCA-3′; The 100 ng of template was added to a PCR mixture in a final volume of 50 μl containing 10 μl 5∓ bufer, 2.5 μmol of each Of dATP, dCTP, dGTP and dTTP, 10 pmol of each set of the sense/antisense primers and 0.6 U of Pyrobest polymerase. The PCR conditions were: predenature at 94°C for 4 min, then 94°C for 45 s, 64°C for 45 s, 72°C for 2 min, altogether for 30 cycles, 72°C for 7 min. The construction was verified by restriction enzyme digestion and sequencing.
Western blotting analysis
The bait plasmid was transformed into yeast strain AH109 by lithium acetate method, in which Daxx cDNAs was inserted in-frame with a flag-epitope. After transformed, a single colony (1–2 mm in diameter, not older than 5 days) of yeast MaV203 transformed with pDBLeu-Daxx was selected and the yeast fusion protein was extracted according to Urea/SDS method described in the Yeast Protocols Handbook of Clontech (PT302421). Protein extract (25 μg of protein) was resolved on 8% SDS–polyacrylamide gel and transferred onto nitrocellulose membrane [5]. Western blotting was performed to confirm the expression of the fusion protein using BD monoclonal antibody.The untransformed yeast Mav203 cells were used as the negative control.
Screening of the human adult liver cDNA library
The screening of the human adult liver cDNA library was performed as described in instruction manual of ProQuest two hybrid system. Plasmids were extracted from cDNA library in large scale and were transformed into the yeast cell containing pDBLeu-Daxx. The yeast cells were inoculated onto SD/-Leu/-Trp/-His+3AT 25 mM plate. After 6–15 days, colonies grew on plate were picked up and analyzed for His, Ura, and LacZ phenotype. Plasmids from the blue colonies were isolated and co-transfected into yeast cells with pDBLeu-Daxx to further confirm the interaction.
Confirmation of the true interaction in yeast
Yeast plasmid was isolated from positive yeast colonies, and thirteen positive plasmids were obtained and transformed into JM109. The structure of insert was confirmed by direct sequencing, then compared with GenBank to analyze the structure and function of the genes. In order to exclude false positives, the plasmids of positive colonies and pDBLeu-Daxx were retransformed into yeast strain MaV203. After mating, the diploids yeast were plated on SD/-Leu-Trp-His+3AT and then replica plated.
GST-pull down assay
The pGEX-4T-2-FTH1 plasmid for GST-pull down assay was prepared by cloning of SalI/NotI fragment of pPC86-FTH1. The pFLAG-CMV-2-Daxx (below described as pFLAG-Daxx) was prepared by cloning of SalI/NotI fragment of pDBLeu-Daxx. Expression and purification of recombinant protein GST-FTH1 were performed as described by Shi et al. [6]. Expression and collection of fusion protein from E. coli JM109 were carried out with the fusion protein expression vector pGEX-4T-2 (Pharmacia, Uppsala, Sweden) [7]. GST-FTH1 fusion protein was bound with glutathione-Sepharose 4B (Amersham, Uppsala, Sweden) beads in binding buffer (50 mM Tris–HCl, 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM PMSF and 10% glycerol, pH 8.0) at 4°C for 1 h. The bound glutathione-Sepharose 4B beads were incubated with the HEK 293 cell lysates containing pFLAG-Daxx fusion protein for 2 h at 4°C in binding buffer. Then detected by Western immunoblot analysis with anti-FLAG antibody [8, 9].
Coimmunoprecipitation assay
The pCMV-Myc-FTH1 (below described as pMyc-FTH1) plasmid for co-immunoprecipitation assay was prepared by cloning of SalI/NotI fragment of pPC86-FTH1. The pMyc-FTH1 and pFLAG-Daxx plasmids were cotransfected into HEK293 cells using the LipofetamineTM 2000 in a 1:2.5 ratio. The cell lysates were harvested, centrifuged at 14,000×g at 4°C for 30 min. The supernatant obtained was mixed with 2 μg anti-FLAG polyclonal antibody and placed on a shaker for overnight at 4°C. 40 μl mixture of protein A/G agarose beads suspension were added to it, rocked at 4°C for 8 h. The beads were washed five times with lysis buffer. 20 μl of 2∓ sample loading buffer was added to the beads, and the mixture was boiled for 5 min. The extracts were then separated by SDS–PAGE. The proteins were transferred to PVDF membranes which were probed by Western blotting. The pFLAG-Daxx and pCMV-myc plasmids, pFLAG-CMV-2 and pMyc-FTH1 plasmids were transfected to HEK293 respectively provided a negative control.
Cell cycle and apoptosis analyses
For cell cycle analysis, Hela cells were seeded into 60 mm plastic tissue culture dishes at a density of 5 × 104 cells/dish. Twenty-four hours later, cells were transfected with 4 μg pDaxx and pFTH1; cells were harvested after 72 h and washed in cold PBS followed by fixation in 70% alcohol for 30 min on ice. After washing in cold PBS three times, cells were resuspended in 0.8 ml of PBS solution with 40 μg of propidium iodide (Sigma) and 0.1 μg of RNase A (Qiagen) for 30 min at 37°C. Samples were analyzed for DNA content by flow cytometry immediately. To measure apoptosis, cells were transfected with 4 μg pDaxx, set another group which transfected with pDaxx + pFTH1; cells were harvested and stained. Cell cycle analysis and apoptosis rate were measured by flow cytometry.
Results
Screening Daxx binding protein with yeast two hybrid system
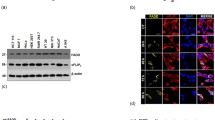
The full length sequence of Daxx generated by PCR amplification. The full length of Daxx was ligated into pDBLeu plasmid at the SalI/NotI site (Fig. 1a). Yeast strain MAV203 transformed with pDBLeu-Daxx could stably express the fusion protein at high level. The Daxx fusion protein was 120 kD (Fig. 1b). pDBLeu-Daxx was used as the bait for screening adult liver cDNA library. Thirteen positive clones grew in the SD/-Leu/-Trp/-His+3AT media. To confirm the true interaction with Daxx protein in yeast, the thirteen clones were transformed into MAV203 and were further tested for β-galactosidase assay, some blue colonies were picked. In Fig. 1c, 13 positive clones, which grew on SD/-Leu/-Trp/-His+3AT media, were lined on the media covered with X-Gal and all became blue. After confirmation of the true interaction with Daxx protein in yeast, 13 independent positive clones were identified and sequenced. Only three sequencing results of 13 clones revealed ORF in frame with the AD coding region. The presence of an open reading frame(ORF) fused to the GAL4 AD sequence was verified, and the sequence was compared with those in GenBank. The mRNA full length sequences were obtained with Vector NTI 8.0 and by searching BLAST database (http://www.ncbi.nlm.nih.gov/). These colonies were prescreened by PCR to make sure that only colonies with different products were subjected to sequencing. Summary of the data is presented in Table 1.
Screen Daxx binding protein in adult liver cDNA library. a Electrophoresis analysis of PCR production of Daxx cDNA; Electrophoresis analysis of Restriction enzyme digestion production of Daxx. b Expression of bait protein in yeast by Western blot analysis. c Reporter Gene Phenotypes of Yeast Strains. Yeast Strains were patched from isolated colonies onto an SC-Leu-Trp master plate and incubated for 18 h at 30°C.Cell from this master plate were replica plated onto SD/-Leu-Trp-His+3AT (25 mM) and a YPAD plate containing a nylon membrane for x-Gal Assay. a: master plate (SC-leu-Ttp); b: SD/-Leu-Trp-His+3AT (25 mM); c: X-Gal assay
Daxx directly binds to FTH1 in vitro
To verify the interaction between Daxx and FTH1 that was first screened in yeast, a GST pull-down assay was first performed (Fig. 2a) to test whether Daxx binds to FTH1 in vitro. The plasmid containing the fusion protein GST-FTH1 was transformed into E. coli JM109 cells and was overexpressed after IPTG induction (Fig. 2a).The interaction between FTH1 and Daxx was detected by GST-pull down assay. As shown in Fig. 2b, GST did not bind to Daxx, but GST-FTH1 was able to bind to the Daxx directly. It was identified that DAXX can interact with FTH1in vitro.
Daxx binds to FTH1 in vitro. a Expression of GST-FTH1 in JM109. Samples (0.5 μg) of affinity-purified GST-FTH1, as indicated, were examined by SDS–PAGE and Coomassie Blue staining. 1. protein marker; line 2. total bacterial protein without IPTG induction; 3. total bacterial protein induced by IPTG; 4. purified fusion prote of GST-FTH1 by glutathione-Sepharose 4B column. b GST pulldown assay. Purified GST-fused domains of FTH1, as indicated, immobilized on glutathione-Sepharose beads were incubated with purified Flag-Daxx(0.5 μg). Proteins trapped were examined by immunoblotting with anti-Daxx antibodies. Purified Flag-Daxx (lane 1) was used as a positive control
Interaction between Daxx and FTH1 in vivo
In order to confirm that these two proteins interacted with each other in vivo, co-immunoprecipitation assay was carried out. It was shown that the plasmid FLAG -Daxx, and Myc-FTH1 were expressed in HEK293 cells as revealed by Western blot analysis (Fig. 3, lane 1, 2, 3 and 7, 8, 9), and the lanes were 120 and 22 kDa respectively. The three groups of plasmids pFLAG-CMV-2 and pMyc-FTH1 (lane 4, 10), pFLAG-Daxx and pMyc-FTH1(lane 5, 11), pFLAG-Daxx and pCMV-Myc (lane 6, 12)were transfected into HEK293 cells. The protein were extracted and analyzed by immunoprecipitation assay. L means light chain which is 25 kDa and H means heavy chain which is 55 kDa. The cell lysates were precipitated with aMyc monoclonal antibody and determined by aFlag or aMyc antibodies (Fig. 3). It suggested that Daxx could interact with FTH1 in vivo. The pFLAG-Daxx and pCMV-myc plasmids, pFLAG-CMV-2 and pMyc-FTH1 plasmids were transfected to HEK293 respectively provided a negative control.
Daxx binds to FTH1 in vivo. Lane 1, 2, 3 and lane 7, 8, 9 are western blot analysis for whole-cell lysate. Lane 4, 5, 6 and lane 10, 11, 12 are western blot analysis for CO-IP precipitated with Myc monoclonal antibody. L means light chain, H means heavy chain. The remained arrowheads point to the aim protein. The IP antibody is a-myc in panel A and panel B, the IB antibody in panel A is a-Flag, the IB antibody in panel B is a-myc. Panel B is control
FTH1 inhibits Daxx-mediated apoptosis
To examine whether Daxx effect cell apoptosis, and whether FTH1 can also effect cell apoptosis through Daxx. We performed flow cytometry analyses. Firstly, we analysed cell cycle distribution after transfected pcDNA-3.1(−)-Daxx and pcDNA-3.1(−)-Daxx + pcDNA3.1(−)-FTH1 into Hela cells. It is showed the cell apoptotic rate increased and cell number in S phase reduced after transfected pcDNA-3.1(−)-Daxx. The cell apoptotic rate is 35.56%.The cell number in S phase is 14.53%. However, the cell apoptotic rate decreased and cell number in S phase increased after transfected pcDNA-3.1(−)-Daxx + pcDNA3.1(−)-FTH1 into Hela. The cell apoptotic rate is 16.51%.The cell number in S phase is 48.25%. The differences between treatment groups and control groups have statistical significance (P < 0.05, Fig. 4a, b).
Analysis on cell cycle distribution and cell apoptosis rate. a Cell cycle distribution Results. Data in b are Mean percentage of cells in S-phase ±SE for 3 independent experiments of Helas cells transfected with pDaxx and pDaxx + pFTH1 72 h before cell cycle analysis (P < 0.05). c Cell apoptosis rate Results. Data in d are mean ± SE for relative apoptosis normalized to control cells for 3 independent experiments of Helas cells transfected with pDaxx and pDaxx + pFTH1 72 h before flow cytometry analysis(P < 0.05)
Secondly, we analysed cell apoptotic rate through Annexin-V system of flow cytometry analyses. It is showed the cell apoptotic rate increased after transfected pcDNA-3.1(−)-Daxx. The cell early apoptotic rate is 21.82%.While the cell apoptotic rate decreased after transfected pcDNA-3.1(−)-Daxx + pcDNA3.1 (−)-FTH1 into Hela. The cell early apoptotic rate is 10.44%. The differences between treatment groups and control groups have statistical significance (P < 0.05, Fig. 4c, d).
FTH1 inhibits JNK activation in Daxx-mediated apoptosis
Then, in order to determine whether FTH1 can influence Daxx-ASK1-JNK apoptosis signal pathway, we detected p-JNK,JNK,ASK1 and caspase3 expression after transfected pcDNA-3.1(−)-Daxx and pcDNA-3.1(−)-Daxx + pcDNA3.1(−)-FTH1 into Hela cells. It is showed that phosphate JNK expression increased in time-depedent manner after transfected pcDNA-3.1(−)-Daxx in Hela cells, And phosphate JNK expression is up to the peak after transfected 48 h. Besides, ASK1 and Caspase3 expression were also increased after transfected pcDNA-3.1(−)-Daxx (Fig. 5a). However, phosphate JNK, ASK1 and Caspase3 expression reduced after transfected pcDNA-3.1(−)-Daxx + pcDNA3.1(−)-FTH1 in Hela cells. And these molecules expression were decreased in dose-dependent manner of pcDNA3.1(−)-FTH1(Fig. 5b).
It is showed that FTH1 can inhibit the activation of JNK signaling pathways. These data show that the FTH1 and Daxx are able to participate in apoptosis pathway.
Discussion
Apoptosis is important in the development and tissue homeostasis of multicellular organisms. Initiation of apoptosis can be activated through two signaling pathways via proteins that bind the death domain, the MAPK-JNK pathway mediated by Daxx and the caspase pathway mediated by FADD. Daxx was originally identified as a protein that specifically binds to the death domain of the trans-membrane death receptor FAS (also called CD95) in the cytoplasm and potentiates FAS-induced apoptosis. And a rather surprising property of Daxx is its anti-apoptotic function, But Daxx anti-apoptotic mechanism remain unknown [10, 11]. Daxx was originally cloned from a yeast two-hybrid screen using the intracellular tail of the Fas receptor as the bait [2]. DAXX is found in the cytoplasm, interacting with Fas-receptor or other cytoplasmic molecules, as well as in the nucleus where it is interacting with some subnuclear structures. Daxx accumulates in both the nucleus and the cytoplasm; in the nucleus, Daxx is found associated with the promyelocytic leukaemia (PML) nuclear body and with a-thalassemia/-mental retardation syndrome protein (ATRX)-positive heterochromatic regions. In the cytoplasm, Daxx has been reported to interact with various proteins involved in cell death regulation. Daxx exerts many reported functions that include mediating the signaling from FasL to apoptosis via activating JNK, induction and inhibition of apoptosis [10–13], and regulation of chromatin remodeling [14]. Daxx deletion mutant (aa501–625) has been known to be an inducer of apoptosis. Daxx fragment-induced activation of caspase-9 and caspase-3 was mediated through the apoptosis signal-regulating kinase1 (ASK1)-MEK-c-Jun-N-terminal kinase (JNK)/p38-Bax pathway [15]. Despite a significant number of studies attempting to determine Daxx function in apoptotic and non-apoptotic cell death, its precise role in this process is only partially understood. Therefore, the identification of the intracellular proteins directly interacting with Daxx is helpful to elucidate the possible pathway of Daxx signaling.
In our study, Yeast two-hybrid system provides a sensitive method for detecting relatively weak and transient protein–protein interactions. The bait plasmid pDBLeu-Daxx was transformed into yeast strain MAV203, then plasmids were extracted from cDNA library in large scale and were transformed into the yeast cell containing pDBLeu-Daxx, to select the diploid cells.After reintroduced into yeast strain MAV203, 13 positive colonies were obtained and verified.Finally we successfully screened out the protein FTH1 that interacts with Daxx. The interaction between Daxx and FTH1 were verified by GST pull-down in vitro and co-immunoprecipitation in vivo successfully.
In our screened proteins, FTH1 is ferritin, heavy polypeptide 1. This gene encodes the heavy subunit of ferritin, the major intracellular iron storage protein in prokaryotes and eukaryotes [2]. It is composed of 24 subunits of the heavy and light ferritin chains. A major function of ferritin is the storage of iron. Recent research found that FTH1 is not only an iron storage protein, but also a mediator of the NF-κB protective function [8]. FTH1 is induced downstream of NF-κB and is required to prevent sustained JNK activation and, thereby, apoptosis triggered by TNF [16, 17].
Our current study focuses on Daxx interacting with FTH1 in vitro and in vivo (Figs. 2, 3).Then we determined that Daxx can induce apoptosis and FTH1 can inhibit Daxx-mediated apoptosis (Fig. 4). Besides, it is found that Daxx mediated apoptosis through the Fas-Daxx-ASK1-JNK1 signaling pathway, while FTH1 can inhibit the activation of JNK signaling pathways (Fig. 5).These data show that the FTH1 and Daxx are able to participate in apoptosis pathway through JNK signal molecule.
This study also contributed to reveal the Daxx biological function, and brought some new clues for further exploration of the expressing and regulating mechanism of Daxx. It is of significance to clarify the Daxx gene transcriptional regulation mechanism, the connection between cis-acting element and its binding trans-acting factor within the Daxx regulation region.
References
Yang X et al (1997) Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell 89:1067–1076
Perlman R et al (2001) TGF-beta-induced apoptosis is mediated by the adapter protein Daxx that facilitates JNK activation. Nat Cell Biol 3:708–714
Chang HY et al (1998) Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science 281:1860–1863
Michaelson JS et al (1999) Loss of Daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes Dev 13:1918–1923
Arad U, Axelrod J, Ben-nun-Shaul O, Oppenheim A, Galun E (2004) Hepatitis B virus enhances transduction of human hepatocytes by SV40-based vectors. J Hepatol 40:520–526
Shi XB, Wei JM, Shen YK (2000) Using the yeast two-hybrid system to detect the interactions of ATP synthase subunits from Spinacia oleracea. Sci Sin (Ser C) 43:169–175
Shi XB, Wei JM, Shen YK (2001) Effects of sequential deletions of residues from the N- or C-terminus on the functions of epsilon subunit of the chloroplast ATP synthase. Biochemistry 40:10825–10831
Ni ZL, Wang DF, Wei JM (2002) Substitutions of the conserved Thr42 increased the roles of the ε-subunit of maize CF1 as CF1 inhibitor and proton gate. Photosynthetica 40:517–522
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, New York
Yang X, Khosrav F, Chang HY et al (1997) Daxx, a novel Fas-binding protein that activates JNK and apoptosis [J]. Cell 89(7):1067–1076
Wu S, Loke HN, Rehemtulla A (2002) Ultraviolet radiation induced apoptosis is mediated by Daxx. Neoplasia 4:486–492
Gostissa M, Morelli M, Mantovani F et al (2004) The transcriptional repressor hDaxx potentiates p53-dependent apoptosis. J Biol Chem 279:48013–48023
Chen LY, Chen JD (2003) Daxx silencing sensitizes cells to multiple apoptotic pathways. Mol Cell Biol 23:7108–7121
Li Q, Wang X, Wu X et al (2007) Daxx cooperates with the Axin/HIPK2/p53 complex to induce cell death. Cancer Res 67(1):66–74
Song JJ, Lee Y (2004) Daxx deletion mutant (amino acids 501–625) induced apoptosis occurs through the JNK/p38/Bax dependent mitochondrial pathway [J]. J Cell Biochem 92(6):1257–1270
Can GP, Concetta B, Francesca Z (2004) Ferritin heavy chain upregulation by NF-κB inhibits TNF-α-induced apoptosis by suppressing reactive oxygen species [J]. Cell 119(4):529–542
Pietsch EC, Chan JY, Torti FM et al (2003) Nrf2 mediates the induction of ferritin H in response to xenobiotics and cancer chemopreventive dithiolethiones [J]. J Biol Chem 278(4):2361–2369
Acknowledgments
This study was supported by National Natural Science Foundation (No. 30270510, 30700898) and Chongqing Science and Technology Commission (No. CSTC2009BA5082).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, F., Du, ZY., He, JL. et al. FTH1 binds to Daxx and inhibits Daxx-mediated cell apoptosis. Mol Biol Rep 39, 873–879 (2012). https://doi.org/10.1007/s11033-011-0811-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-0811-5