Abstract
In the present study, we investigated the diversity distributions of allelic frequencies of 15 short tandem repeats (STRs) loci in a sample of Chinese Hui ethnic group in the Ningxia Hui Autonomous Region. The allelic frequencies of the 15 STR loci (D8S1179, D21S11, D7S820, CSF1PO, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, vWA, TPOX, D18S51, D5S818 and FGA) were obtained from 2975 unrelated healthy Hui individuals. The STR genotyping data of all the samples were generated by DNA extraction, multiple amplification, GeneScan and genotype analysis. The genetic distances among different populations were calculated by using Nei’s method and a phylogenetic tree was constructed based on the allelic frequencies of the same 15 STR loci using the neighbor-joining method. A total of 185 alleles were observed in the Hui population, with the corresponding allelic frequencies ranging from 0.0002 to 0.5322. Chi-Square tests showed that all STR loci were in Hardy–Weinberg equilibrium. The forensic statistical parameters of all the loci showed high values. The population data in this study were compared with the previously published population data from other ethnics or areas. The Hui population showed significant differences from the Minnan Han, Uigur, Ewenki, Yi, Tibetan, Maonan and Malay ethnic minority groups in some loci, and from the South Morocco population and the Moroccan population in all the loci. Our results are valuable for human individual identification and paternity testing in the Chinese Hui population and are expected to enrich the genetic information resources of Chinese populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Hui ethnic group with a sizable population of over 9.81 million is the most widely distributed ethnic minority in China. The Hui ethnic group, typically distinguished by their practice of Islam, is one of the largest ethnic minorities in the country. Most of the Huis inhabit in the Ningxia Hui Autonomous Region, Gansu, Qinghai, Henan, Hebei, Shandong and Yunnan provinces and the Xinjiang Uygur Autonomous Region. The Huis speak Mandarin Chinese and write in Chinese characters although a number of Arabic and Persian words have remained in their daily interactions and religious activities. The Hui people living in the border areas of China often use the language of local ethnic minorities [1, 2].
The majority of the Hui people believe in Islamic religion, which has a deep influence on their daily life (http://www.china.org.cn/english/null/139573.htm). Usually a mosque is built and becomes the symbol of architecture in large Hui communities. The habitation of the Hui people is characterized by small scale concentration and wide distribution with mosques as the center of their social activities (http://www.chinahighlights.com/travelguide/nationality/hui.htm). Hui men usually wear Hui style hats, which are small black-colored and white-colored hats without brims. Most Hui men like wearing white hats while some do not wear hats and instead they wrap their heads with white towels or cloth. Women usually wear white round-edged hats and veils on their heads. Under the influence of Islamism, the Huis consume a large amount of beef and mutton in their diet and the meat of pig, dog, horse, donkey, mule and beasts of prey is forbidden [3, 4].
Qinghai province, where the samples for this study were collected, is located in the northeastern part of the Tibetan Plateau. The Yellow River originates in the middle of the province, while the Yangtze and Mekong have their sources in the southwestern part of the province. Qinghai is a multi-ethnic province inhabited by 44 ethnic groups, including the Han people (the China’s majority) and the Tibetan, Hui, Tu, Salar, Bonan, Yugur, Dongxiang, and Mongol ethnic groups. Minority people account for 44% of the total population in the province (http://www.china.org.cn/e-xibu/2JI/3JI/qinghai/qing-ban.htm).
Short tandem repeats (STRs) are the most ideal genetic markers because of their numerous allelic genes, their power for human identification, and their stable heredity in the human genome [5, 6]. STR typing is also suitable for analyzing degraded or outmoded human DNA and minute amounts of human DNA [7, 8]. Recent advances in study on polymorphic STRs as genetic markers have led to their wide application to population genetics, establishment of forensic DNA database, human individual identification, paternity testing, genetic mapping, disease linkage analysis, archaeology and potential inference of the ethnic origin of an individual. Previously published population data have shown that the allelic distributions of STR loci differ from one population to another or between the members of the same population living in different geographic areas [9–12]. In the present study, we investigated the distributions of the allelic frequencies and forensic statistical parameters of 15 STR loci in the Chinese Hui ethnic group living in Qinghai province of China, established a forensic DNA database and compared the study population with other neighboring populations. In addition, a phylogenic tree was constructed to illustrate the inter-population relationships based on allelic frequencies of 15 STR loci using neighbor-joining method.
Materials and methods
Sample collection and DNA extraction
After informed consent, bloodstain samples were obtained from 2,975 unrelated, healthy individuals from the Chinese Hui ethnic group living in Qinghai province of China. Donors were randomly chosen among the Hui individuals whose ancestors had lived in the region for at least three generations. Genomic DNA was extracted using the Chelex-100 protocol as described by Walsh et al. [13].
PCR amplification and DNA typing
PCR for 15 autosomal STRs was performed in a multiplex reaction using AmpFℓSTR Identifiler kit. 0.9 μl (2 ng/μl) genomic DNA samples were amplified in a total reaction volume of 10 μl consisting of 2.9 μl deionized water, 4 μl dNTP, 0.2 μl AmpliTaqGold DNA polymerase and 2.0 μl primer set. Thermal cycling was conducted under the following conditions: 95°C for 11 min; 28 cycles of 94°C for 60 s, 59°C for 60 s, 72°C for 60 s; a final extension of 60°C for 45 min. Detection and genotyping of all STR PCR products were carried out using ABI3130XL DNA Genetic Analyzer (Applied Biosystem, Foster City, CA, USA). Allele designations were determined by comparison of the sample PCR fragments with those of allelic ladders provided with the kit using Genemapper 3.2 (Applied Biosystem, Foster City, CA, USA). The alleles of all STR loci were named according to the number of repeat units present, as recommended by the DNA Commission of the International Society for Forensic Haemogenetics [14].
Quality control
All experimental steps were performed according to the laboratory internal control standards and kit controls.
Statistical and phylogenetic analysis
Allelic and genotypic frequencies were estimated by the direct counting method. The observed heterozygosity (HO), expected heterozygosity (HE), power of discrimination (DP), probability of paternity exclusion (EPP), polymorphism information content (PIC), probability values of exact tests of Hardy–Weinberg disequilibrium were calculated as previously reported [15, 16]. R × C contingency test was employed for pairwise interpopulation comparisons [17]. Our present data were compared with previously published data for the same 15 STR loci in samples from the Chinese Yi ethnic minority group living in Yunnan province [9], from the Chinese Dongxiang [10], Salar [10], Tu [11] and Tibetan [12] ethnic minority groups living in Qinghai province of China, from the Chinese Ewenki ethnic group in the Inner Mongolia Autonomous Region [18], from the Han population in Guangdong province [19], from the Minnan Han population in Southeast China [20], from the Chinese Uigur ethnic group in Xinjiang province [21], from the Chinese Maonan ethnic group in Guangxi [22], from the South morocco population [23], from the Malay ethnic group [24], and from the Moroccan population [25]. Excel program and SPSS 13.0 for windows software was used for analysis of the data. A phylogenetic tree showing inter-population relationships was constructed using genetic distances with hierarchical cluster method by SPSS 13.0 software.
Results and discussion
Allelic frequencies and forensic statistical parameters regarding the 15 STR loci of the Chinese Hui population living in Qinghai province of China are shown Table 1. The comparison results (P value) of the population data between the Hui ethnic group and the previously published ethnic groups are shown in Table 2.
As shown in Table 1, a total of 185 alleles were observed in the Hui population, with the corresponding allelic frequencies ranging from 0.0002 to 0.5322. The combined probabilities of exclusion and power of discrimination for all 15 STR loci were 0.999999, and 0.9999999999, respectively. Chi-Square tests show that all STR loci were in the Hardy–Weinberg equilibrium (P > 0.05).
The Hui population data were compared with the previously published population data of other ethnic groups for the same 15 STR loci by the method of the R × C contingency test. No differences were found in the comparison results between our study population and the Chinese Dongxiang, Salar and Tu ethnic minority groups living in Qinghai Province [10, 11] and the Chinese Han population living in Gongdong province [19] at all 15 STR loci. We found that the study population showed significant difference from the Uigur ethnic population [21] at the TH01 locus (P = 0.011), the Ewenki group [18] at D18S51 (P < 0.001) locus, the Minnan Han population in Southeast China [20] at D7S820 (P = 0.027), D8S1179 (P = 0.002) and D13S317 (P = 0.019) loci, the Chinese Yi ethnic minority group [9] at vWA (P < 0.001), FGA (P < 0.001), TH01 (P = 0.020), D2S1338 (P < 0.001), D3S1358 (P = 0.021) and D19S433 (P = 0.001) loci, the Chinese Tibetan ethnic minority group [12] at FGA (P = 0.001), TH01 (P = 0.001), D2S1338 (P = 0.002), D5S818 (P = 0.005), D13S317 (P = 0.013) and D18S51 (P < 0.001) loci, the Maonan ethnic group [22] at CSF1PO (P < 0.001), TPOX (P = 0.011), D13S317 (P < 0.001), D16S539 (P < 0.001), D8S1179 (P < 0.001), vWA (P < 0.001), D18S51 (P < 0.001) and FGA (P < 0.001) loci, the Malay ethnic group [24] at TH01 (P < 0.001), CSF1PO (P < 0.001), D3S1358 (P < 0.001), D2S1338 (P < 0.001), D7S820 (P = 0.012), D18S51 (P < 0.001), D5S818 (P < 0.001), D8S1179 (P = 0.027), D19S433 (P = 0.001), D21S11 (P = 0.018) and FGA (P < 0.001) loci, and the South morocco population [23] and the Moroccan population [25] at all loci.
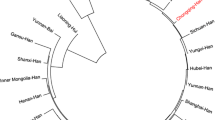
The genetic distances between the Hui population and the previously published populations are shown in Table 3. Short genetic distances were found between the Hui population and the Tu ethnic minority (0.050), the Dongxiang ethnic minority (0.073), the Salar ethnic minority (0.079) and the Ewenki ethnic group (0.081), but further distances were found between the Hui population and the Minnan Han population (0.115), the Yi ethnic minority group (0.150), the Uigur ethnic group (0.188), the Han population in Guangdong (0.253), the Maonan ethnic group (0.329), the Tibetan ethnic minority group (0.356), the Malay ethnic group (0.535), the southern Morocco population (0.622) and the Moroccan population (0.827). The results show that the genetic distances between the Hui population and the other populations living in the same area (in Qinghai province of China) was the nearest, much smaller than its distances to the Chinese populations living in other provinces. More genetic diversity existed between the Hui population and the foreign populations in other countries. A neighbor-joining phylogenetic tree (Fig. 1) was constructed based on the allelic frequencies of the 15 STR loci in the Hui population and other 13 populations [9–12, 18–25]. Cluster analysis shows that two main clusters existed within the 14 populations: the first-level cluster consisted of all the populations living in China, and the second-level cluster consisted of the foreign populations only (the Malay ethnic group, the southern Morocco population and Moroccan population). The Hui population clustered with the Tu, Dongxiang and Salar ethnic minority groups at the first level, followed in descending order by Ewenki, Minnan Han, Yi, Uigur, Guangdong Han, Maonan, and Tibetan. The results of the cluster analysis were consistent with those of the genetic distance analysis. The best explanation for the first-level clustering and the short genetic distances of the Hui and other ethnic groups in Qinghai province may be that there have been strong gene flows among these populations since they lived in the same area. Living together for a long time may help promote the gene communion and amalgamation between the Hui population and the other populations in the same area. Locations of the Hui ethnic group and other populations in China are shown in Fig. 2.
A phylogenetic tree constructed by the neighbor-joining method based on the allelic frequencies of all 15 STR loci. The tree shows the relationships between the Hui population and other 13 populations (1 Tu; 2 Uigur; 3 Dongxiang; 4 Ewenki; 5 Guangdong Han; 6 Salar; 7 Yi; 8 Hui; 9 South morocco; 10 Tibetan; 11 Malay; 12 Minnan Han; 13 Moroccan; 14 Maonan)
In summary, our results are valuable for human identification and paternity testing of the Chinese Hui population in Qinghai province of China. The study is expected to enrich Chinese population genetic information resources and provide biological evidence for the origin of the Hui population.
References
Li SX (2001) National Properties of Hui Language. J Ningxia Teach Univ 22:61–64 (in Chinese)
Jin XJ (1989) Language research of Hui nationality. China Natly 2:36–37 (in Chinese)
Ma ZJ (2001) Islamic law’s influence on the Huis’ moral concept and convention. J Ningxia Univ (Soc Sci Ed) 23:95–97 (in Chinese)
Li BP (2009) Dietary customs and cultural boundaries of Hui nationality. J Ningxia Teach Univ (Soc Sci Ed) 30:81–85 (in Chinese)
Edwards A, Civitello A, Hammond HA, Caskey CT (1991) DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am J Hum Genet 49:746–756
Ritter N (2007) Missing persons and unidentified remains: the nation’s silent mass disaster. NIJ Journal 256:7
Hammond HA, Jin L, Zhong Y, Caskey CT, Chakraborty R (1994) Evaluation of 13 short tandem repeat loci for use in personal identification applications. Am J Hum Genet 55:175–189
Zhu BF, Wang ZY, Yang CH, Li XS, Zhu J, Yang G, Huang P, Liu Y (2005) Y-chromosomal STR haplotypes in Chinese Uigur ethnic group. Int J Legal Med 119:306–309
Zhu BF, Shen CM, Wu QJ, Deng YJ (2008) Population data of 15 STR loci of Chinese Yi ethnic minority group. Leg Med (Tokyo) 10:220–224
Deng Y, Zhu B, Yu X, Li Y, Fang J, Xiong X, Mu H, Huang Y, Shi X (2007) Genetic polymorphisms of 15 STR loci of Chinese Dongxiang and Salar ethnic minority living in Qinghai province of China. Leg Med (Tokyo) 9:38–42
Zhu B, Yan J, Shen C, Li T, Li Y, Yu X, Xiong X, Mu H, Huang Y, Deng Y (2008) Population genetic analysis of 15 STR loci of Chinese Tu ethnic minority group. Forensic Sci Int 174:255–258
Yan J, Shen C, Li Y, Yu X, Xiong X, Mu H, Huang Y, Deng Y, Yu J (2007) Genetic analysis of 15 STR loci on Chinese Tibetan in Qinghai province. Forensic Sci Int 169:e3–e6
Walsh PS, Metzger DA, Higuchi R (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–513
Bär W, Brinkmann B, Budowle B, Carracedo A, Gill P, Lincoln P, Mayr W, Olaisen B (1997) DNA recommendations. Further report of the DNA commission of the ISFH regarding the use of short tandem repeat systems. Int J Leg Med 110:175–176
Nei M (1987) Molecular Evolutionary Genetics. Columbia University Press, New York
Ohno Y, Sebetan IM, Akaishi S (1982) A simple method for calculating the probability of excluding paternity with any number of codominant alleles. Forensic Sci Int 19:93–98
Nei M, Roychoudhury AK (1974) Sampling variances of heterozygosity and genetic distance. Genetics 76:379–390
Chen L, He Y, Li S (2006) Genetic analysis of 15 STR loci of Chinese Ewenki ethnic population. J Forensic Sci 51:1408–1409
Zhu J, Li J, Guo Y, Liu K, Zhu B, Liu Y (2005) Population data of 15 STR in Chinese Han population from north of Guangdong. J Forensic Sci 50:1510–1511
Hu SP, Wu DQ, Xu XH, Liu JW, Li B (2005) Genetic profile of 15 STR loci in the Min Nan population in Southeast China. Forensic Sci Int 152:263–265
Zhu B, Wang Z, Wu Q, Sedike ME, Zhu J, Huang P, Xu Y, Liu Y (2005) Genetic analysis of 15 STR loci of Chinese Uigur ethnic population. J Forensic Sci 50:1235–1236
Xu L, Li SF, Deng QY, Zhou LN, Gong JC, Xu R (2007) Polymorphism of 15 STR loci in Mao Nan minority of Guangxi. J Guangxi Med Univ 24:180–183 (in Chinese)
Ossmani HE, Talbi J, Bouchrif B, Chafik A (2009) Allele frequencies of 15 autosomal STR loci in the southern Morocco population with phylogenetic structure among worldwide populations. Leg Med (Tokyo) 11:155–158
Seah LH, Jeevan NH, Othman MI, Jaya P, Ooi YS, Wong PC, Kee SS (2003) STR Data for the AmpFlSTR Identifiler loci in three ethnic groups (Malay, Chinese, Indian) of the Malaysian population. Forensic Sci Int 138:134–137
Bouabdellah M, Ouenzar F, Aboukhalid R, Elmzibri M, Squalli D, Amzazi S (2008) STR data for the 15 AmpFlSTR Identifiler loci in the Moroccan population. Forensic Science Int 1:306–308
Acknowledgments
All authors would like to thank Mr. Zhanhai Wang for his hard work on collecting the blood samples. This project was supported by National Natural Science Foundation of China (NSFC, No. 30700470) and the Item of Science Technology Foundation of Shaanxi Province, People’s Republic of China (No. 2008K08-02).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Deng, Yj., Zhu, Bf., Shen, Cm. et al. Genetic polymorphism analysis of 15 STR loci in Chinese Hui ethnic group residing in Qinghai province of China. Mol Biol Rep 38, 2315–2322 (2011). https://doi.org/10.1007/s11033-010-0364-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0364-z