Abstract
Recently, differentiated somatic cells had been reprogrammed to pluripotential state in vitro, and various tissue cells had been elicited from those cells. Epigenetic modifications allow differentiated cells to perpetuate the molecular memory needed for the cells to retain their identity. DNA methylation and histone deacetylation are important patterns involved in epigenetic modification, which take critical roles in regulating DNA expression. In this study, we dedifferentiated NIH/3T3 fibroblasts by 5-aza-2-deoxycytidine (5-aza-dC) and Trichstatin A (TSA) combination, and detected gene expression pattern, DNA methylation level, and differentiation potential of reprogrammed cells. As the results, embryonic marker Sox2, klf4, c-Myc and Oct4 were expressed in reprogrammed NIH/3T3 fibroblasts. Total DNA methylation level was significant decreased after the treatment. Moreover, exposure of the reprogrammed cells to all trans-retinoic acid (RA) medium elicited the generation of neuronal class IIIβ-tubulin-positive, neuron-specific enolase (NSE)-positive, nestin-positive, and neurofilament light chain (NF-L)-positive neural-like cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Because of the ability to grow indefinitely and differentiate into cells of all three germ layers, human embryonic stem (ES) cells might be used to treat a host of diseases, such as Parkinson’s disease, spinal cord injury and diabetes etc. [1, 2]. However, there are ethical difficulties regarding the use of human embryos, as well as the problem of tissue rejection following transplantation in patients. One way to circumvent these issues is to generate pluripotential cells directly from the patients’ own cells.
Nuclear transfers, cellular fusion, the use of cell-free extracts and culture-induced reprogramming have been used to converse a differentiated cell into a pluripotent state. However, these processes are complex and difficult to repeat [3–5].
Recently, exalting progress has been achieved in induced pluripotent stem cells (iPS). Four transcription factors, Oct 4, Sox 2, Klf 4, and c-Myc, which were transfected into mouse or human fibroblast cells producing embryonic pluripotential state cells, and sharing almost all of the ES characteristics [6–8]. Furthermore, a humanized sickle cell anemia mouse model has been rescued after transplantation with hematopoietic progenitors obtained in vitro from autologous iPS cells [9]. However, the use of retroviral vectors for gene delivery which carry the risk of insertional mutagenesis, and using of oncogenes for reprogramming may lead to teratomas formation. These problems need to be resolved before iPS cells can be considered for clinical application.
To alter the phenotype of cells in a rational way, molecular parameters that distinguish these different cell types must be modified. Epigenetic regulation of gene expression is recognized as a key mechanism governing cell determination, commitment, differentiation and maintenance of those states as well [10, 11]. DNA methylation and histone deacetylation are the main components of an epigenetic program. Disturbances of any of these components may shift the balance between an active and silent chromatin conformation, resulting in an altered transcriptional state [12–14]. Pharmacological agents that interfere with this system active expression of many genes, including those required for maintenance of pluripotent or multipotent state of the cells [15–18].
In this study, we induced expression of embryonic marker, Sox2, klf4, c-Myc, and Oct4 in reprogrammed NIH/3T3 fibroblasts through using pharmacological epigenetic modifiers. By cultured in neural environment with RA supplement, we were able to generate the neuronal class IIIβ-tubulin-positive, NSE-positive, nestin-positive, and NF-L-positive neural-like cells from these reprogrammed fibroblasts.
Materials and methods
Reagents, cell culture
All reagents were purchased from Sigma Chemical Co. unless stated otherwise. NIH/3T3 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Hyclone) supplemented with 10% Newborn Calf Serum (Invitrogen).
Exposing NIH/3T3 Cells to epigenetic modifiers such as 5-aza-2′-deoxycytidine(5-aza-dc) and Trichostatin A(TSA)-Prior to treatment with 5-aza-dc (DNA methylation inhibitor), NIH/3T3 cells were pre-cultured for 24 h and then cultured for 3 days in embryonic stem cell maintenance medium (ESCmm, 15% fetal bovine serum(Invitrogen), 1 mM sodium pyruvate, 100 μg ml−1/100 μg ml−1 Penicilline G/Streptomycin, 20 ng/ml leukemia inhibitory factor (LIF), high glucose DMEM (Hyclone), 2 mM glutamine, 0.1 mM non-essential amino acids),which containing 0, 0.01, or 5 μM 5-aza-dc. For treatment with TSA (inhibitor of histone deacetylasel), NIH/3T3 cells were incubated for 24 h in ESCmm with 5-aza-dc and then exposed to 0.08 or 0.4 μM TSA with or without 0.01 or 5 μm 5-aza-dc for 72 h.
Analysis of Oct4, Sox2, c-Myc, and Klf4 expression by real-time RT-PCR
Total RNA was prepared with TRIzol reagent (Invitrogen). The mixture of total RNA was converted into first-strand cDNA synthesis using an RT-PCR kit (AMV, TaKaRa). Real-time PCR was performed with Power SYBR® Green PCR master mix (ABI) according to manufacturer’s instructions. Signals were detected with an ABI 7500 Real-time PCR system (Applied Biosystems). Primer sequences are listed in Table 1.
Quantification of total levels of cellular DNA methylation by flow cytometry
The levels of total cellular DNA methylation were quantified by flow cytometry through measuring the fluorescent levels of cells after incubation in anti-5-methyl cytosine (Abcam) primary antibodies (whose specificity toward the methyl group on carbon 5 of the pyrimidine ring) and fluorescent-conjugated secondary antibodies. The procedures for flow cytometry were followed as described in references [18, 19].
Neural induction and immunocytochemistry
NIH/3T3 cells grown in neural cell medium (NCm) (15% fetal bovine serum (Invitrogen), 100 μg/ml penicillin G, 100 μg/ml streptomycin, 10 ng/ml leukemia inhibitory factor (LIF), low-glucose DMEM/F12 (Hyclone), 2% B27, 1% N2, 20 ng/ml bFGF, 20 ng/ml EGF) in the presence of epigenetic modifiers (5 μM 5-aza-dc, 0.4 μM TSA) were replated on PO/L-coated glass coverslips and exposed to 5 μM RA in the same NCm. After 3 days, the samples were fixed for immunocytochemical studies.
For immunocytochemistry, cells were permeabilized (30 min with 0.1% Triton X-100 in PBS and 20 min with 2% BSA) and incubated for 1 h with one of the following primary antibodies monoclonal anti-nestin (SantaCruz), monoclonal anti- neuronal class IIIβ-tubulin (SantaCruz), monocloned anti-neurofilament light chain (NF-L) and a polyclonal anti-NSE antibody (Santa Cruz). Immunoreactive cells were visualized using fluorescence-conjugated (FITC) goat anti-rabbit/mouse IgG.
Results
Ectopic expression of Oct4, Sox2, KIf4, and c-Myc after 5-aza-dc and TSA treatment
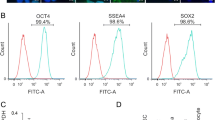
These four transcription factors were not detected in NIH/3T3 cells, but expression of Sox2 and c-Myc was effectively induced by treatment with 5-aza-dc in the presence or absence of TSA. Klf4 expression was induced by treatment with relatively high concentration (5 μM) of 5-aza-dc in the presence or absence of TSA. Oct4 expression was induced strictly with a combination of 5-aza-dc (0.01 or 5 μM) and TSA (0.08 or 0.4 μM; Fig. 1).
Real-time RT-PCR analysis (n = 3 independent PCR reactions; error bars indicate s.d.) of Oct4, Sox2, C-Myc and Klf4 with untreated NIH/3T3 cells (1#), with NIH/3T3 cells treated with 5 μM 5-aza-dc (2#), with NIH/3T3 cells epigenetically modified by 5 μM 5-aza-dc + 0.4 μM TSA (3#), with 0.01 μM 5-aza-dc-treated NIH/3T3 cells (4#) and with NIH/3T3 cells treated with a mixture of 0.01 μM 5-aza-dc and 0.08 μM TSA (5#).Transcript levels were normalized to Gapdh expression, with expression levels in NIH/3T3 fibroblasts set as 1. Inhibition of DNA methylation by 5-aza-dc with or without histone deacetylation by TSA elicited c-Myc expression in NIH/3T3 cells. Klf4 expression were induced by treatment with relatively high concentration (5 μM) of 5-aza-dc with or without TSA. Combined treatment of cells with 5-aza-dc (0.01 or 5 μM) and TSA (0.08 or 0.4 μM) could reactivate the expression of Oct4 and Sox2 mRNA
Total levels of cellular DNA methylation
In epigenetic modified NIH/3T3 cells, DNA is globally hypomethylated compared to that of untreated cells. The DNA methylation level was decreased by treatment with 5-aza-dc alone, and the lower methylation lever was presented when NIH/3T3 cells were treated with a combination of 5-aza-dc (0.01 or 5 μM) and TSA (0.08 or 0.4 μM; Fig. 2).
Examples of wave plots obtained with untreated NIH/3T3 cells (a), with NIH/3T3 cells treated with 0.01 μM 5-aza-dc (b), with NIH/3T3 cells epigenetic modified by 0.01 μM 5-aza-dc + 0.08 μM TSA (c), with 5 μM 5-aza-dctreated NIH/3T3 cells (d) and with a mixture of 5 μM 5-aza-dc and 0.4 μM TSA treated NIH/3T3 cells (e). All with anti-5-methyl cytosine antibodies. NIH/3T3 cells untreated, treated, and labeled as describedunder Materials and methods. R2, fluorescent-positive cells
Morphology and immunoreactivity of neural-like cells derived from induced NIH/3T3 fibroblasts
NIH/3T3 cells grown in 6-well plastic dishes in ESCmm containing 5 μM 5-aza-dc and 0.4 μM TSA for 3 days were cultured in ESCmm containing 5 μM RA. Three days later, neuronal class IIIβ-tubulin, nestin, NF-L and NSE-immunopositive clusters of cells with a circular or ovoid shape were observed. NSE, neuronal class IIIβ-tubulin, nestin and NF-L immunostaining was visualized with a green colour, and nuclei are shown in blue. Untreated NIH/3T3 cells, and NIH/3T3 cells only treated with 5-aza-dc/TSA or RA were NSE, neuronal class IIIβ-tubulin, nestin and NF-L negative, with fibroblastic morphology (Fig. 3).
Immunostaining demonstrated the expression of neuronal class IIIβ-tubulin (b, g, l, q) NSE (c, h, m, r), nestin (d, i, n, s) and NF-L (e, j, o, t) in NIH/3T3 cells treated with (a, b, c, d, e) or without (f, g, h, i, j) 5 μM 5-aza-dc/0.4 μM TSA/5 μM RA/NCm, and cells treated with 5-aza-dc and TSA but not with RA (k, l, m, n, o), and cells treated with RA but not with 5-aza-dc or TSA (p, q, r, s, t). Scale bar = 50 μm
Discussion
In the previous reports, Sox2, klf4, c-Myc and Oct4 are restricted to totipotent and pluripotent cells [7, 8]. Cell differentiation always associates with the demethylation and methylation of the genomic DNA to form the required cell or tissue type [20, 21]. Inhibitor of DNA methylation (5-aza-dc) and histone deacetylation (TSA) allowed NIH/3T3 cells to express the Oct4 gene de novo, and the clear link between Oct4 expression and epigenetic control by DNA methylation and chromatin modification has been verified [22]. Our findings suggested the Sox2, klf4, and c-Myc may be other candidates for regulation via DNA methylation. The link between these three factors’ expression and epigenetic modifiers requires further study.
It has been demonstrated that relatively high concentration (1.25 or 5 μM) of TSA could induce global histone hyperacetylation in G0/G1-and G2/M-stage cells but had no effect on DNA methylation [18]. However, in our experiments, TSA combining with DNA methylation inhibitor, 5-aza-dc, promoted the decrease of DNA methylation. It might indicate that chromatin structure modification by histone acetylation is also involved in the epigenetic regulation of DNA methylation.
Neuron-specific enolase is one of three enolase isoenzymes found in mammals. This isoenzyme, a homodimer, has a molecular weight of 78,000 Da. It is found in mature neurons and cells of neuronal origin. A switch from alpha enolase to gamma enolase occurs in rat and primate neural tissue during development. Nestin, neuronal class IIIβ-tubulin and NF-L are intermediate filament protein expressed in the developing nervous system and are markers of neural stem/progenitor and stem cell populations [23–25].Characteristic perinuclear patterns of NSE, neuronal class IIIβ-tubulin, nestin and NF-L expression were observed extensively in cells cultured in neural culture medium (NCm) containing 5 μM RA. This inducing approach led to the derivation of neural-like cells from NIH/3T3 fibroblasts.
Our data indicated that inhibitor of DNA methylation (5-aza-dc) and histone deacetylation (TSA) could modify the epigenetic state of somatic cells. This method avoids the risk of converting genetic elements which may rise in retrovirus transgenic iPS. Even though neural-like cells were induced from our reprogrammed fibroblast, more researches, such as other differentiation form those cells and mechanism of chemical induced reprogramming are still needed.
References
Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A (2000) Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol 18:399–404. doi:10.1038/74447
Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS et al (1998) Embryonic stem cell lines derived from human blastocysts. Science 282:1145–1147. doi:10.1126/science.282.5391.1145
Hochedlinger K, Jaenisch R (2006) Nuclear reprogramming and pluripotency. Nature 441:1061–1067. doi:10.1038/nature04955
Collas P, Taranger CK (2006) Epigenetic reprogramming of nuclei using cell extracts. Stem Cell Rev 2:309–317. doi:10.1007/BF02698058
Alberio R, Campbell KH, Johnson AD (2006) Reprogramming somatic cells into stem cells. Reproduction 132:709–720. doi:10.1530/rep.1.01077
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K et al (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872. doi:10.1016/j.cell.2007.11.019
Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K et al (2007) In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448:318–324. doi:10.1038/nature05944
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676. doi:10.1016/j.cell.2006.07.024
Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, Jaenisch R (2007) Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 318:1920–1923. doi:10.1126/science.1152092
Li E (2002) Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet 3:662–673. doi:10.1038/nrg887
Jaenisch R, Bird A (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33:245–254. doi:10.1038/ng1089
Alexanian AR (2007) Epigenetic modifiers promote efficient generation of neural-like cells from bone marrow-derived mesenchymal cells grown in neural environment. J Cell Biochem 100:362–371. doi:10.1002/jcb.21029
Kass SU, Pruss D, Wolffe AP (1997) How does DNA methylation repress transcription? Trends Genet 13:444–449. doi:10.1016/S0168-9525(97)01268-7
Benjamin D, Jost JP (2001) Reversal of methylationmediated repression with short-chain fatty acids: Evidence for an additional mechanism to histone deacetylation. Nucleic Acids Res 29:3603–3610. doi:10.1093/nar/29.17.3603
Kohyama J, Abe H, Shimazaki T, Koizumi A, Nakashima K, Gojo S et al (2001) Brain from bone: efficient “meta-differentiation” of marrow stroma-derived mature osteoblasts to neurons with Noggin or a demethylating agent. Differentiation 68:235–244. doi:10.1046/j.1432-0436.2001.680411.x
Reik W, Dean W, Walter J (2001) Epigenetic reprogramming in mammalian development. Science 293:1089–1093. doi:10.1126/science.1063443
Milhem M, Mahmud N, Lavelle D, Araki H, DeSimone J, Saunthararajah Y et al (2004) Modification of hematopoietic stem cell fate by 5-aza-2′-deoxycytidine and trichostatin A. Blood 103:4102–4110. doi:10.1182/blood-2003-07-2431
Enright BP, Kubota C, Yang X, Tian XC (2003) Epigenetic characteristics and development of embryos cloned from donor cells treated by trichostatin A or 5-aza-2′-deoxycytidine. Biol Reprod 69:896–901. doi:10.1095/biolreprod.103.017954
Habib M, Fares F, Bourgeois CA, Bella C, Bernardino J et al (1999) DNA global hypomethylation in EBV- transformed interphase nuclei. Exp Cell Res 249:46–53. doi:10.1006/excr.1999.4434
Imamura T, Ohgane J, Ito S, Ogawa T, Hattori N, Tanaka S et al (2004) CpG island of rat sphingosine kinase-1 gene: tissue-dependent DNA methylation status and multiple alternative first exons. J Biol Chem 279:17063–17069. doi:10.1074/jbc.M309002200
Ohgane J, Aikawa J, Ogura A, Hattori N, Ogawa T, Shiota K (1998) Analysis of CpG islands of trophoblast giant cells by restriction landmark genomic scanning. Dev Genet 22:132–140. doi:10.1002/(SICI)1520-6408(1998)22:2<132::AID-DVG3>3.0.CO;2-7
Hattori N, Nishino K, Ko YG, Hattori N, Ohgane J, Tanaka S et al (2004) Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J Biol Chem 279:17063–17069. doi:10.1074/jbc.M309002200
Lendahl U, Zimmerman LB, McKay RD (1999) CNS stem cells express a new class of intermediate filament protein. Cell 60:585–595. doi:10.1016/0092-8674(90)90662-X
Cattaneo E, McKay R (1990) Proliferation and differentiation of neuronal stem cells regulated by nerve growth factor. Nature 347:762–765. doi:10.1038/347762a0
Li M, Pevny L, Lovell-Badge R, Smith A (1998) Generation of purified neural precursors from embryonic stem cells by lineage selection. Curr Biol 8:971–974. doi:10.1016/S0960-9822(98)70399-9
Acknowledgments
This work was supported by National Natural Science Foundation of China (Grant No. 30671025), the Project of Abroad Researcher Foundation of Heilongjiang (Grant No. 1151hz031), the Project of Science Researcher Foundation of Heilongjiang (Grant No.11531145) and a grant from the Innovative Fund of Harbin Medical University graduate student (HCXB2007004).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Zhang, XM., Li, QM., Su, DJ. et al. RA induces the neural-like cells generated from epigenetic modified NIH/3T3 cells. Mol Biol Rep 37, 1197–1202 (2010). https://doi.org/10.1007/s11033-009-9489-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-009-9489-3