Abstract
Rock carp, Procypris rabaudi (Tchang), is an endemic fish species in China. We sequenced the complete mitochondrial genome of it by high-fidelity polymerase chain reaction with conserved primers and primer walking sequencing method. The complete mitochondrial genome of rock carp is 16595 bp in length and contains 13 protein-coding genes, two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes and one control region, with an identical order to that of most other vertebrates. The origin of L-strand replication (OL) in rock carp mitochondrion is located in a cluster of five tRNA genes (WANCY region) with 35 nucleotides in length. The control region is located between the tRNA-Pro and tRNA-Phe genes and is 943 bp in length. Three conserved sequence blocks (CSB), an extended termination associated sequence (ETAS), an AT-repeat microsatellite sequence and a putative promoter sequence for H-strand transcription (HSP) were identified within this region. The microsatellite sequence has a very low variation, with only one repeat alteration in 50 checked individuals (from 12 to 13 repeats). The phylogenetic analysis for rock carp was performed with Bayesian and Maximum likelihood (ML) methods based on the concatenated nucleotide sequence of 12 protein-coding genes on the heavy strand. The result suggested that traditional taxonomic barbines possibly originated more early than cyprininaes; rock carp was placed at the position between barbines and cyprininaes, while has a closer relationship with cyprininaes than barbines.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rock carp, Procypris rabaudi (Tchang), is an endemic species to China and mainly distributed in upper reaches of the Yangtze River drainage [1]. The fish prefers to inhabit slow flowing deep water with plenty of rocks at bottom, and lives through winter in the holes of rocks at bottom of deep pool or bed of river. As omnivorous fish, it mainly feeds on zoobenthos, such as aquatic insect, Limnoperna, aquatic Oligochaeta, and etc., and secondly on oddment of plant and phytoplankton [1, 2]. It is an important commercial fish due to its good taste, rich nutrition, and potential in aquaculture. Because of heavy fishing, dam construction and water pollution in the Yangtze River drainage, the wild populations of this species have rapidly declined in recent years. Now this species is listed as vulnerable in China [2]. In order to prevent the population decline from anthropogenic impacts, rock carp has been recommended as a second-class state protected animal in China (Wenxuan Cao, personal communication).
Rock carp belongs to the family Cyprinidae, and was further classified to Cyprininae based on its mainly morphological characters [3]. In phylogenetic relationships, Cyprininae is closest to Barbinae within Cyprinidae. Cyprininae is distinguished from Barbinae based on one mainly morphological character, the presence of strong last single ray in anal fin [3]. Some species of Puntioplites and Procypris in Cyprininae, including rock carp, were considered having closer relative to Barbinae based on their other morphological characters, such as pharyngeal tooth [3, 4] and the fist vertebra [5]. Phylogenetic analysis based on RAPD also suggested that Procypris rabaudi is closer to Barbinae than Cyprininae [6]. However, to date, the deeper phylogenetic relationship of rock carp remains ambiguous, and there is no more information available about phylogenetic knowledge for rock carp.
The typical vertebrate mitochondrial genome is circular, ranging in size from approximately 15 to 18 kb and generally containing 37 genes (22 tRNAs, two rRNAs, and 13 proteins) and a control region (D-loop) [7–10]. The gene order of mitochondrial genome is high conserved in fish with a few exception reported so far [8, 10–15]. Because of its maternal inheritance, relative lack of recombination, fast evolutionary rate compared to nuclear DNA, and the ability to provide an abundance of genotype, the mitochondrial DNA is a useful molecular marker for population genetic and phylogenetic studies [16–18]. In fish, numerous phylogenetic analyses have been conducted based on cytochrome b, rRNA, and control region of mitochondrion [19–27]. However, short sequence data may give rise to some misleading conclusions for resolution of deeper evolutionary branches [28–30]. Recent studies have shown that longer sequence, especially the 12 concatenated protein-coding genes, have great potential in phylogenetic inferences of deeper branches [8, 18, 31–36]. There are more than 600 complete mitochondrial DNA sequences of teleosts been deposited in GenBank. However, it is quite insufficient for complete mitochondrial data available to insight into the phylogenetic relationships in the family Cyprinidae. In this paper, we reported the complete sequence of the mitochondrial genome of rock carp and determined the mitochondrial genomic structure, gene order, codon usage and base composition. Based on rock carp’s complete mitochondrial DNA sequence and along with the sequences of 40 other cyprinids, we recovered the phylogenetic relationships to further clarify the relative phylogenetic position of P. rabaudi in Cyprinidae. We hope that the knowledge of the mitochondrial genome sequence of this species could contribute to the phylogeny clarification of cyprinids.
Materials and methods
Fish sample, mitochondrial DNA extraction, PCR amplification and sequencing
The rock carp was collected from Mudong in Chongqing, which was one of the main regions for distribution of this species in the Yangtze River. Total mitochondrial DNA (mtDNA) was extracted from the muscle tissue using the method described by Tapper et al. [37].
We used eight sets of primers to amplify contiguous, overlapping segments of the complete mitochondrial genome in rock carp. The primer sequences were shown in Table 1. The primers were designed from the mitochondrial conservative region based on a result of multiple sequence alignment of complete mitochondrion of Cyprinus carpio, Carassius carassius, Barbus barbus, Puntius ticto, Cyprinella spiloptera, Danio rerio, Ischikauia steenackeri, and Labeo batesii (GenBank accession nos. were shown in Table 2). PCR amplification was conducted on iCycler PCR System (Bio-Rad, USA) in a 25 μl reaction volume containing about 30 ng mt DNA, 1× La PCR buffer II (TaKaRa, China), 1.5 mM MgCl2, 2 μM of each primer, 0.5 mM dNTP and 1.0 U La Taq DNA polymerase (TaKaRa, China). PCR condition was 94°C for 4 min, 30 cycles consisting of 94°C for 30 s, 58°C for 50 s, 68°C extension 2–4 min, with a final extension at 72°C for 10 min. PCR products were electrophoresed on 1.0% agarose gel and sized relative to molecular weight marker D2000 (TIANGEN, China).
PCR products were purified using QUIEXII Kit (OMEGA, USA), and then directly sequenced using the primer walking method on ABI 3730 Genetic Analyzer (Applied Biosystems). The self-designed primers used in PCR and BigDye Termination v3.1 Cycle Sequencing Kit (Applied Biosystems) were used for sequencing.
Sequence analysis
DNA sequences were analyzed using the software DNAMAN version 3.0 (Lynnon Biosoft, Quebec, Canada). The locations of protein-coding and rRNA genes were determined by comparison with the corresponding known sequences of other three cyprinid fishes, Cyprinus carpio [38], Carassius carassius [39] and Barbodes gonionotus [34]. The tRNA genes were identified using the program tRNAscan-SE 1.21 [40]. Some tRNA genes, which could not be found by the tRNAscan-SE were identified by their secondary structure [41] and specific anti-codons.
Phylogenetic analysis
In order to acquire some implications about phylogenetic position of Procypris rabaudi within cyprinid, the nucleotide sequence data of 12 heavy-strand protein-coding genes were used for phylogenetic analysis. The ND6 was excluded from the phylogenetic analysis, because it is encoded by the opposite strand with considerably different base composition and codon bias. ND6 might possess a different evolutionary pattern from 12 protein-coding genes on the heavy-strand [30]. After removal of the gaps, all ambiguous sites around the gaps, overlapping region and stop codons, a 10865 nucleotide sequence set was obtained. Twelve concatenated protein-coding gene sequence of mitochondrion from rock carp and 40 other cyprinid fishes (Table 2) were used in the phylogenetic analysis. Leptobotia mantschurica, Vaillantella maassi, Gyrinocheilus aymonieri and Hypentelium nigricans (Table 2) were used as outgroups. Multiple alignments of the 12 concatenated protein-coding gene sequences were conducted using Clustal X [42] with the default settings. Two different methods, Maximum-likelihood (ML) and Bayesian, were used to construct phylogenetic relationship. For ML analysis, the best fitting models of sequence evolution were determined with Modeltest 3.06 [43]; heuristic searches were executed in 100 replicates with all characters unordered and equally weighted, and using tree bisection reconnection (TBR) branch swapping in the program PHYML [44]; bootstrapping proportions with 100 replicates were used for nodal evaluation. The Bayesian analysis was conducted using MrBayes 3.1.2 [45]. The Bayesian posterior probabilities were estimated with 2 million generations, sampling trees every 100 generations. About 20% of sampling trees were discarded (the burnin) after estimating with a conservative approach. Then a consensus tree was calculated using the remaining 16000 trees (whose log-likelihoods converged to stable values). Two separate runs with four Markov chains were performed. The genetic distances among P. rabaudi and other species in Cyprininae and Barbinae were calculated based on the concatenated nucleotide data of 12 heavy-strand protein-coding gene sequences using MEGA 3.1 [46] with the Kimura 2-parameter model.
Results and discussion
Genome organization
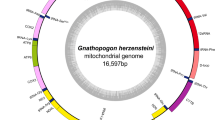
The complete mitochondrial genome sequence of rock carp was determined to be 16595 bases in length and was deposited in GenBank (Accession no. EU082030). As shown in Fig. 1, the organization of mitochondrial genome of rock carp is similar to that of typical vertebrate mitochondrial genome, consisting of 13 protein-coding genes, two rRNA genes and 22 tRNA genes. These genes are arranged in line in rock carp mitochondrial genome (Table 3). Also as in other vertebrates, most rock carp mitochondrial genes are encoded on the H-strand, except ND6 and eight tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Pro, tRNA-Glu), which are encoded on the L-strand. The overall base composition of H-strand of rock carp mitochondrial genome is A: 32.27%, T: 25.20 %, C: 26.92% and G: 15.60%, with an A + T rich feature as that of other vertebrate mitochondrial genome.
Protein-coding genes
Among rock carp mitochondrial protein-coding genes, the open reading frames of two pairs of contiguous genes overlap occurred on the same strand: ATPase8-ATPase6 and ND4L-ND4, and they overlap by seven nucleotides, respectively. ND5 and ND6 overlap by four nucleotides as well, whereas they are encoded on the opposition strand. All 13 protein-coding genes in rock carp mitochondrial genome use ATG as the initiation codon except the COI gene, which uses GTG as initiation codon. All COI genes in reported fishes use GTG as initiation codon, thus, the feature that COI uses GTG as initiation codon seems to be prevalent among nontetrapod vertebrates [30]. However, termination codons vary among different fish species [11]. Six protein-coding genes in rock carp mitochondrial genome end with complete stop codons, TAA (ND1, COI, ND4L, ND5, ND6) and TAG (ATPase8), the rest seven genes end with incomplete stop codons, either TA (ATPase6, COIII) or T (ND2, COII, ND3, ND4, Cytb), which are presumably completed as TAA after transcriptions [47]. The codon usage in rock carp mitochondrial genome was given in Table 4. The frequency of CTA (Leu) is the highest (count: 290), and TGT (Cys) and AAG (Lys) are the lowest (count: 6, respectively) among codons used in rock carp mitochondrial genome. This codon usage bias might be associated with the available tRNA in organism.
Base composition of rock carp mitochondrial protein-coding genes is given in Table 5. Similar to other vertebrates, the base composition of 12 protein-coding genes on the H-strand is bias against G and strong bias against G at the third codon position. The most frequent nucleotide at the third codon position is A, which is consistent with most cyprinid fish, such as Cyprinus carpio, Carassius carassius, Barbus barbus, Danio rerio, but inconsistent with Opsariichthys bidens [36], where C is the most frequent nucleotide. The most frequent nucleotide at the second codon position is T (40%) and pyrimidine is over-represented (T + C=68%), owing to the hydrophobic character of the proteins [48]. However, the ND6 possesses markedly different base composition and codon bias, having more G than C or A both in total base composition and at the third codon position. To explore the codon evolution of 12 protein-coding genes in teleostean mitochondrion, we investigated the nucleotide frequency of codon using 20 teleostean data sets reported in GenBank. As shown in Table 6, the significant changes of nucleotide frequency at the third codon position indicate that there are different nucleotide preferences for the codon ends among teleosteans. Whereas, there are highly conservative nucleotide frequencies at the second codon position, with extremely minor nucleotide frequency alteration (T: 40–41%, C: 27–29%, A: 18–19%, G: 13–14%). At the second codon position, the frequency of pyrimidine seldom alternated. Most teleosteans have a pyrimidine frequency of 68%, and it ranges from 68% to 69% in 20 investigated teleosteans. These results suggested that the frequency of non-synonymous mutations is very low in 12 mitochondrial protein-coding genes of teleosteans. In order to compare the codon evolution pattern of teleosteans to that of other vertebrates, we further investigated the codon nucleotide frequency of 12 mitochondrial protein-coding genes in four elasmobranches, two amphibians, three reptiles, five birds and five mammals. As shown in Table 6, there is a similar nucleotide frequency variation as that in teleosteans, exhibiting a relatively conservative nucleotide frequency at the second codon position and an obviously various nucleotide frequency at the third codon position among different species. Compared with nucleotide frequency at the second codon position of teleosteans, there is a slightly higher variation among the investigated amniotes as follows: T: 39–42%, C: 27–31%, A: 18–20%, G: 11–12%. This might be associated with diverse habitats. Most of the amniotes have a pyrimidine frequency of 69% or 70%.
Ribosomal and transfer RNA genes
The 12S and 16S rRNA genes of rock carp mitochondrion are 958 and 1683 bp in length, respectively. As in other vertebrates, they are located between tRNA-Phe and tRNA-Leu (UUR) genes and separated by tRNA-Val gene (Fig. 1). The base composition of the two rRNA gene sequences is 35.4% A, 20.37% G, 19.88% T and 24.35% C. The content of A + T (55.28%) is higher than that of C + G (44.72%). Thus, the rock carp mitochondrial rRNA genes also exhibit A+T content-rich like as other bony fishes [49–53]. Rock carp mitochondrial genome contains 22 tRNA genes, which are interspersed between the rRNA and protein-coding genes and range from 67 bp (tRNA-Cys) to 77 bp (tRNA-Lys) in size (Table 3). Twenty-one tRNA genes, which could fold into the typical cloverleaf secondary structure, were identified by tRNAscan-SE v.1.21 [40]. Due to lacking of the complete dihydrouridine arms (D-arms), the tRNA-Ser (AGY) gene was determined by proposed secondary structures [41] and the anti-codon. The anti-codons of the 22 tRNA genes of rock carp mitochondrial genome have no unique characteristics compared to other vertebrates.
Control region
The major non-coding sequence (control region) of the rock carp mitochondrial genome is located between the tRNA-Pro and tRNA-Phe genes and is 943 bp in length. The conserved sequence blocks (CSB1-3), which were thought to be involved in positioning RNA polymerase both for transcription and priming replication [54, 55], were identified in the positions 615–641, 698–715 and 741–759 nt downstream of the 5′ end, respectively. The extended termination associated sequences (ETAS) was found in the position 25–61 nt downstream of the 5′ end, which can form a stable hairpin-loop structure. The sequence ACCAAAAACTTCCAAAAAATA, which is a putative promoter for H-strand transcription (HSP) [49], was found at 55 nt upstream of the 5′ end of tRNA-Phe gene. The HSP has a few nucleotide substitutions at the two underlined positions compared to that of Cyprinus carpio [49]. An AT-repeat microsatellite sequence was also identified at 45 nt upstream of the 5′ end of HSP. The microsatellite has only one repeat variation in 50 tested individuals of rock carp (from 12 to 13 repeats) (Jun Song et al. unpublished data). It also presents at other teleostean mitochondrion control region, but the repeats of AT might be different, for example, Cyprinus carpio: 9 repeats (AP009047), Barbus barbus: 14 repeats (AB238965). This microsatellite sequence might be useful in some interspecies identification. As like as most vertebrates, the origin of L-strand replication (OL) in rock carp mitochondrion is located in a cluster of five tRNA genes (WANCY region). The region is 35 bp in length, overlaps the tRNA-Cys gene by 3 bp and has the potential to fold into a stable stem–loop secondary structure consisting of 22 bp in the stem and 13 bp in the loop. The conserved motif 5′-GGCGGG-3′ also presents in the stem of tRNA-Cys gene. Compared with the OL of Cyprinus carpio, the rock carp OL exhibits five nucleotide substitutions and one nucleotide deletion in the loop, yet they are completely identical in the stem.
Phylogenetic analysis
To investigate the phylogenetic position of rock carp, the concatenated nucleotide sequence of the 12 heavy-strand protein-coding genes were used to construct the phylogenetic relationships by Bayesian and ML methods (Fig. 2). Hierarchical likelihood ratio tests indicate that the (GTR +I + Г) model of substitution and gamma distribution was the best for our data (GTR + I + Г, −lnL = 280184.91; Ts/Tv ratio = 5.66; distribution shape parameter = 0.6044). The yielded Bayesian tree had a nearly same topology as that of ML (Fig. 2). In the clade C of Fig. 2, Barbodes gonionotus (subfamily Barbinae) was placed on the basal position, sister to the clade comprising of P. rabaudi, C. carpio and C. carassius. In addition, as shown in clade B, three other barbines diverged prior to cyprininaes as well. This suggested that the traditional taxonomic barbines possibly originated more early than cyprininaes. Procypris rabaudi diverged after the emergence of B. gonionotus of Barbinae and prior to C. carpio and C. carassius of Cyprininae. Also, the genetic distances of P. rabaudi to C. carpio and C. carassius were 0.1260 and 0.1382, respectively, and that to B. gonionotus, B. barbus, B. trimaculatus and P. ticto were 0.1396, 0.1498, 0.2242 and 0.1872, respectively. Both the phylogenetic tree and genetic distances showed that P. rabaudi had a closer relationship to Cyprininae than Barbinae, which was incongruent with the opinion that P. rabaudi was closer to Barbinae than Cyprininae inferred from RAPD analysis [6]. Our result appeared to be consistent with the results from morphological characters of rock carp compared with that of the Cyprininae and Barbinae [3–5]. However, considering only four species of Barbinae, and two other species of Cyprininae have been incorporated into the phylogenetic analysis, it is needed more evidences rooted in the more species of Barbinae and Cyprininae to confirm the phylogenetic position of rock carp in Cyprinidae in the future.
Phylogenetic relationships of the family Cyprinidae by Bayesian and Maximum likelihood (ML) methods based on the concatenated nucleotide sequence of 12 protein-coding genes on the heavy strand. Numbers in the nodes: posterior probabilities for Bayesian analysis and bootstrap values for ML analysis. Less than 0.95 of Bayesian posterior probabilities and 50% of bootstrap values were omitted. If only less than 0.95 of Bayesian posterior probability, or 50% of bootstrap value in the same clade, both were kept to avoid the confusion between them
In the present study, the fishes of Gobinoninaes formed an independent monophyletic group (clade A in Fig. 2), which agreed with the traditional Gobinoninae grouping [1, 3, 4]. In clade B, the Labeo batesii was placed at the basal-most position, sister to the clade consisting of barbines, cyprininaes and a schizothoracine. Several traditional taxonomic subfamilies, such as Cultrinae, Danioninae and Leuciscinae, represented polyphyletic in the phylogenetic tree, which was accordant to the conclusions of Saitoh et al. [34]. This may be due to the result that some similar morphological characters were from either convergent evolution or retained ancestral characters shared across some taxa, which were used to group by the traditional subfamily classification within Cyprinidae [56]. However, the relationships of the genera within Labeoninae, Schizothoracinae, Xenocyprinae and Acheilognathinae were ambiguous because of lack of complete mitochondrial data of various genera in these subfamilies.
References
Ding R (1994) The fishes of Sichuan, China. Sichuan Publishing House of Science and Technology, Chengdu, pp 411–413
Yue P, Chen Y (1998) Pisces. In: Wang S (ed) China red data book of endangered animals. Science Press, Beijing, pp 170–172
Yue P (2000) Fauna sinica: osteichthyes, cypriniformes III. Science Press, Beijing, pp 391–433
Wu X (1964) Cyprinidae sinica III. Shanghai Science and Technique Press, Shanghai, pp 395–438
Chen X, Yue P, Lin R (1984) Major groups within the family Cyprinidae and their phylogenetic relationships. Acta Zootaxonomica Sin 9(4):424–438
Liu S, Sun Y, Yang F, Han S, Wang X, Xie R, He S (2004) Identification analysis of Procypris rabaudi by RAPD. J Wuhan Univ (Nat Sci Edn) 50(4):477–481
Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27:1767–1780
Miya M, Takeshima H, Endo H, Ishiguro N, Inoue JG, Mukai T, Satoh TP, Yamaguchi M, Kawaguchi A, Mabuchi K, Shirai SM, Nishida M (2003) Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol Phylogenet Evol 26:121–138
Wilson AC, Cann RL, Carr SM, George M, Gyllensten UB, Helm-Bychowski KM, Higuchi RG, Palumbi SR, Prager EM, Sage RD, Stoneking M (1985) Mitochondrial DNA and two perspective on evolutionary genetics. Biol J Linn Soc 26:375–400
Miya M, Tsukamoto K, Nishida M (2005) The phylogenetic position of toadfishes (order Batrachoidiformes) in the higher ray-finned fish as inferred from partitioned Bayesian analysis of 102 whole mitochondrial genome sequences. Biol J Linn Soc 85:289–306
Miya M, Nishida M (1999) Organization of the mitochondrial genome of a deep-sea fish, Gonostoma gracile (Teleostei: Stomiiformes): first example of transfer RNA gene rearrangements in bony fish. Mar Biotechnol 1:416–426
Inoue JG, Miya M, Tsukamoto K, Nishida M (2003) Evolution of the deep-sea gulper eel mitochondrial genomes: large-scale gene rearrangements originated within the eels. Mol Biol Evol 20:1917–1924
Miya M, Kawaguchi A, Nishida M (2001) Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol 18:1993–2009
Mabuchi K, Miya M, Satoh TP, Westneat MW, Nishida M (2004) Gene rearrangements and evolution of tRNA pseudogenes in the mitochondrial genome of the parrotfish (Teleostei: Perci-formes: Scaridae). J Mol Evol 59:287–297
Satoh TP, Miya M, Endo H, Nishida M (2006) Round and pointed-head grenadier fishes (Actinopterygii: Gadiformes) represent a single sister group: evidence from the complete mitochondrial genome sequences. Mol Phylogenet Evol 40:129–138
Moore WS (1995) Inferring phylogenies from mtDNA variation: mitochondrial gene trees versus nuclear-gene trees. Evolution 49:718–726
Broughton RE, Reneau PC (2006) Spatial covariation of mutation and nonsynonymous substitution rates in vertebrate mitochondrial genomes. Mol Biol Evol 23:1516–1524
Peng Z, Wang J, He S (2006) The complete mitochondrial genome of the helmet catfish Cranoglanis bouderius (Siluriformes: Cranoglanididae) and the phylogeny of otophysan fishes. Gene 376:290–297
Bargelloni L, Ritchie PA, Patarnello T, Battaglla B, Lambert DM, Meyer A (1994) Molecular evolution at subzero temperatures: mitochondrial and nuclear phylogenies of fishes from Antarctica (suborder Nothotenoidei), and the evolution of antifreeze glycopeptides. Mol Biol Evol 11:854–863
Lee WJ, Conroy J, Howell WH, Kocher TD (1995) Structure and evolution of teleost mitochondrial control regions. J Mol Evol 41:54–66
Lydeard C, Roe KJ (1997) The phylogenetic utility of the mitochondrial cytochrome b gene for inferring relationships among actinopterygian fish. In: Kocher TD, Stepien CA (eds) Molecular systematics of fishes. Academic Press, San Diego, pp 285–303
Stepien CA, Dillon AK, Brooks MJ, Chase KL, Hubers AN (1997) The evolution of blennioid fishes based on an analysis of mitochondrial 12S rDNA. In: Kocher TD, Stepien CA (eds) Molecular systematics of fishes. Academic Press, San Diego, pp 245–270
Simons AM, Mayden RL (1998) Phylogenetic relationships of the Western North American phoxinins (Actinopterygii: Cyprinidae) as inferred from mitochondrial 12S and 16S ribosomal RNA sequences. Mol Phylogenet Evol 9:308–329
Doadrio I, Carmona JA, Machordom A (2002) Haplotype diversity and phylogenetic relationships among the Iberian barbells (Barbus, Cyprinidae) reveal two evolutionary lineages. J Hered 93:140–147
Perdices A, Cunha C, Coelho MM (2004) Phylogenetic structure of Zacco platypus (Teleostei, Cyprinidae) populations on the upper and middle Chang Jiang (=Yangtze) drainage inferred from cytochrome b sequences. Mol Phylogenet Evo 31:192–203
Mihara M, Sakai T, Nakao K, Martins LO, Hosoya K, Miyazaki J (2005) Phylogeography of loaches of the genus Lefua (Balitoridae, Cypriniformes) inferred from mitochondrial DNA sequences. Zool Sci 22:157–168
Rvan JR, Esa YB (2006) Phylogenetic analysis of hampala fishes (subfamily Cyprininae) in Malaysia inferred from partial mitochondrial cytochrome b DNA sequences. Zool Sci 23:893–901
Cummings MP, Otto SP, Wakeley J (1995) Sampling properties of DNA sequence data in phylogenetic analysis. Mol Biol Evol 12:814–822
Zardoya R, Meyer A (1996) Phylogenetic performance of mitochondrial protein-coding genes in resolving relationships among vertebrates. Mol Biol Evol 13:933–942
Saitoh K, Hayashizaki K, Yokoyama Y, Asahida T, Toyohara H, Yamashita Y (2000) Complete nucleotide sequence of Japanese flounder (Paralichthys olivaceus) mitochondrial genome: structural properties relationships. J Hered 91:271–278
Koepfli KP, Wayne RK (2003) Type ISTS markers are more informative than cytochrome b in phylogenetic reconstruction of the Mustelidae (Mammalia: Carnivora). Syst Biol 52:571–593
Jondeung A, Sangthong P, Zardoya R (2006) The complete mitochondrial DNA sequence of the Mekong giant catfish (Pangasianodon gigas), and the phylogenetic relationships among Siluriformes. Gene 387:49–57
Kim BC, Kang TW, Kim MS, Kim CB (2006) The complete mitogenome of Rhodeus uyekii (Cypriniformes, Cyprinidae). DNA Seq 17:181–186
Saitoh K, Sado T, Mayden RL, Hanzawa N, Nakamura K, Nishida M, Miya M (2006) Mitogenomic evolution and interrelationships of the Cypriniformes (Actinopterygii: Ostariophysi): the first evidence toward resolution of higher-level relationships of the world’s largest freshwater fish clade based on 59 whole mitogenome sequences. J Mol Evol 63:826–841
Yue GH, Liew WC, Orban L (2006) The complete mitochondrial genome of a basal teleost, the Asian arowana (Scleropages formosus, Osteoglossidae). BMC Genomics 7:242
Wang X, Wang J, He S, Mayden RL (2007) The complete mitochondrial genome of the Chinese hook snout carp Opsariichthys bidens (Actinopterygii: Cypriniformes) and an alternative pattern of mitogenomic evolution in vertebrate. Gene 399:11–19
Tapper DP, van Etten RA, Clayton DA (1983) Isolation of mammalian mitochondrial DNA and RNA and cloning of the mitochondrial genome. Methods Enzymol 97:426–434
Mabuchi K, Miya M, Senou H, Suzuki T, Nishida M (2006) Complete mitochondrial DNA sequence of the Lake Biwa wild strain of common carp (Cyprinus carpio L.): further evidence for an ancient origin. Aquaculture 257:68–77
Guo X, Liu S, Liu Y (2007) Evidence for maternal inheritance of mitochondrial DNA in allotetraploid. DNA Seq 18:247–256
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Kumazawa Y, Nishida M (1993) Sequence evolution of mitochondrial tRNA genes and deep-branch animal phylogenetics. J Mol Evol 37:380–398
Tompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DJ (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Guindon S, Gascuel O (2003) PhyML––a simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Anderson S, Bankier AT, Barell BG, de Bruijin MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreir PH, Smith AJH, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465
Naylor GJP, Collins TM, Brown WM (1995) Hydrophobicity and phylogeny. Nature 373:565–566
Chang Y-S, Huang F-L, Lo T-B (1994) The complete nucleotide sequence and gene organization of carp (Cyprinus carpio) mitochondrial genome. J Mol Evol 38:138–155
Zardoya R, Meyer A (1997) The complete DNA sequence of the mitochondrial genome of a ‘living fossil’, the coelacanth (Latimeria chalumnae). Genetics 146:995–1010
Kim IC, Lee JS (2004) The complete mitochondrial genome of the rockfish Sebastes schlegeli (Scorpaeniformes, Scorpaenidae). Mol Cells 17:322–328
Kim I-C, Kweon H-S, Kim YJ, Kim C-B, Gye MC, Lee W-O, Lee Y-S, Lee J-S (2004) The complete mitochondrial genome of the javeline goby Acanthogobius hasta (Perciformes, Gobiidae) and phylogenetic considerations. Gene 336:147–153
Oh DJ, Kim JY, Lee JA, Yoon WJ, Park SY, Jung YH (2007) Complete mitochondrial genome of the rock bream Oplegnathus fasciatus (Perciformes, Oplegnathidae) with phylogenetic considerations. Gene 392:174–180
Clayton DA (1991) Nuclear gadgets in mitochondrial DNA replication and transcription. Trends Biochem Sci 16:107–111
Shadel GS, Clayton DA (1997) Mitochondrial DNA maintenance in vertebrates. Ann Rev Biochem 66:409–435
He S, Mayden RL, Wang X, Wang W, Tang KL, Chen W-J, Chen Y (2008) Molecular phylogenetics of the family Cyprinidae (Actinopterygii: Cypriniformes) as evidenced by sequence variation in the first intron of S7 ribosomal protein-coding gene: Further evidence from a nuclear gene of the systematic chaos in the family. Mol Phylogenet Evol 46(3):818–829
Acknowledgements
We would like to thank Prof. Xuefu He for his help in sample collecting. This work was funded by the National Natural Science Foundation of China (No. 30670290) and 973 Project (No. 2007CB411605).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, X., Yue, B., Jiang, W. et al. The complete mitochondrial genome of rock carp Procypris rabaudi (Cypriniformes: Cyprinidae) and phylogenetic implications. Mol Biol Rep 36, 981–991 (2009). https://doi.org/10.1007/s11033-008-9271-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-008-9271-y