Abstract
Rapeseed (Brassica napus) is an important oil crop that supplies a considerable amount of global vegetable oil production. Genetic transformation system is important to gene functional analysis and molecular breeding. Here, an efficient Agrobacterium-mediated transformation protocol using hypocotyl of rapeseed as explants is described. To develop this protocol, we compared several essential factors that would affect the transformation efficiency, such as Agrobacterium strains, selection marker genes, and genotypes of rapeseed. Comparison of different Agrobacterium strains showed that the GV3101 had higher transformation efficiency than that of C58C1 and EHA105. HPTII, NPTII, and RePAT were used as selection marker genes in tissue culture. The results showed that the transformation efficiency was 3.7–4.8%, 2.2–22.5%, and 1.6–5.9% when the hypocotyl of Westar was infected by GV3101 and screened under hygromycin, kanamycin, and basta, respectively. The transformation efficiency of Westar was the highest and ZS11 was the lowest when five different genotypes of rapeseed (Westar, ZS9, ZS11, GY284, and WH3417) were infected by GV3101. Using this protocol, it will take 8–10 weeks to obtain transgenic plants. This protocol has been used to study gene function in several genotypes of rapeseed in our laboratory. These results indicate that it is efficient to obtain transgenic plant of rapeseed using this protocol.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rapeseed (Brassica napus, AACC, 2n = 38) is one of the most important oil crops and it produces approximately 15% of edible oil globally (Gracka et al. 2016; Yu et al. 2016). It originates from a spontaneous hybridization between Brassica rapa (AA, 2n = 20) and Brassica oleracea (CC, 2n = 18) (Chalhoub et al. 2014; Rahman et al. 2017). Conventional breeding of rapeseed is labor resource intensive and time consuming, which takes eight to ten generations to develop a new variety (Mason and Snowdon 2016). Genetic transformation technology is a key tool to study gene function and provides new genetic resource for molecular breeding, such as herbicide and disease resistance that is hardly achieved by conventional breeding (Bhalla and Singh 2008). Moreover, most traits introduced by gene-transfer method are dominant. Thus, an efficient gene-transfer system suitable for different genotypes of rapeseed would shorten the length of downstream breeding process, and develop a commercial product when using agronomic inferiority model varieties. Additionally, this would help reveal gene function in rapeseed.
Many DNA-transfer methods have been explored for plant transformation, including PEG-mediated DNA uptake, electroporation, microinjection, and particle bombardment (Poulsen 1996). PEG-mediated or electroporation-mediated protoplasts transformations have been attempted in Arabidopsis, B. oleracea, and B. napus (Bergman and Glimelius 1993; Eimert and Siegemund 1992; Mukhopadhyay et al. 1991; Radchuk et al. 2002; Yoo et al. 2007). However, regeneration of plant from the protoplast of Brassica is genotype dependent (Hansen et al. 1999). In addition, the use of protoplast requires a longer tissue culture period, which will lead to a rising risk of contamination and undesirable somaclonal variations (Bhalla and Singh 2008; Davey et al. 2005). Thus, application of PEG-mediated or electroporation-mediated protoplasts transformation method for regenerating transgenic plants is limited. Exogenous DNA or constructs also can be delivered into explant generated from microspores of rapeseed by microinjection and particle bombardment technology. The particle bombardment method involves propelling DNA-coated gold particles into intact plant tissues or cells followed by regeneration of transgenic plants (Kikkert et al. 2005). However, it leads to fragmentation of DNA during bombardment, insertion of backbone vector DNA, and insertion of multiple gene copies (Bhalla and Singh 2008). Multiple copies of the transgene can lead to gene silencing, and integration of vector DNA is undesirable, especially in the present regulatory and consumer environment (Bhalla and Singh 2008).
Agrobacterium-mediated transformation is widely used in many crop species (Tzfira and Citovsky 2006), and it has become the most common method for Brassica transformation (Bhalla and Singh 2008). The floral-dip is a classic Agrobacterium-mediated transformation method which has been successfully used in Arabidopsis and Camelina (Sitther et al. 2018; Clough and Bent 1998), which is also suitable for rapeseed transformation (Verma et al. 2008). However, the floral-dip method is difficult to expand in rapeseed because of low transformation efficiency and limited space for plant growth. The ability to regenerate transgenic plants from Agrobacterium-mediated transformed cells is vital for successful transformation. Hypocotyl and cotyledon explants have been used to obtain transgenic B. napus and B. oleracea (Bhalla and Singh 2008). However, most of the transformation protocols reported are relatively specific to a model cultivar such as Westar, which is not agronomically desirable (Rani et al. 2013).
Here, we described an efficient Agrobacterium-mediated transformation method for rapeseed using hypocotyl as explants. Following this protocol, thirteen constructs carrying hygromycin or kanamycin or basta selection marker gene were transformed into different genotypes of rapeseed independently. The results indicate that this transformation method has acceptable transformation efficiency for different genotypes of rapeseed, and it is suitable for different constructs containing different selection markers. It will benefit gene functional study and molecular breeding of rapeseed.
Materials and methods
Preparation of stock reagent
2,4-D (1.0 mg/mL)
Dissolve 50 mg 2,4-D (Sangon Biotech, Cat. No. A600166-0100) in 1 mL ethanol. Add millipore water to 50 mL, then filter the solution using a 0.22-μm syringe filter. Stock at − 20 °C.
Kinetin (0.3 mg/mL)
Dissolve 15 mg kinetin (Sigma, Cat. No. A600745-0025) in 1 mL 1 M HCl. Add millipore water to 50 mL, then filter the solution using a 0.22-μm syringe filter. Stock at − 20 °C.
Zeatin (1 mg/mL)
Dissolve 100 mg zeatin (Sangon Biotech, Cat. No. A600748) in 1 mL 1 M NaOH. Add millipore water to 100 mL, then filter the solution using a 0.22-μm syringe filter. Stock at − 20 °C.
Indole-3-acetic acid (IAA, 0.5 mg/mL)
Dissolve 25 mg IAA (Sangon Biotech, Cat. No. A600723-0025) in 1 mL 95% ethanol. Add millipore water to 50 mL, then filter the solution using a 0.2-μm syringe filter. Stock at − 20 °C.
Acetosyringone (AS, 100 mM)
Dissolve 0.92 g acetosyringone (Sangon Biotech, Cat. No. A601111) in 50 mL dimethyl sulfoxide (DMSO). Stock at − 20 °C.
Timentin (300 mg/mL)
Dissolve 15 g timentin (Goldbio, Cat. No. T-104-100) in 40 mL millipore water, then add millipore water to 50 mL and filter the solution using a 0.22-μm syringe filter. Stock at − 20 °C.
Silver thiosulfate (STS, 15 mM)
Slowly pour AgNO3 solution (Sinopharm, Cat. No. 10018461) (dissolve 509.61 mg in 30 mL millipore water) into the prepared Na2S2O3 solution (Sinopharm, Cat. No. 10021218) (dissolve 1.74 g in 70 mL millipore water) with stirring. Filter the mixed solution using a 0.22-μm syringe filter after the reaction finished. Stock at − 20 °C.
Indole-3-butyric acid (IBA, 0.5 mg/mL)
Dissolve 25 mg IBA (Sangon Biotech, Cat. No. A600725-0025) in 1 mL 95% ethanol. Add millipore water to 50 mL. Stock at room temperature.
Kanamycin (50 mg/mL)
Dissolve 2.5 g kanamycin (BBI, Cat. No. A600286) in 50 mL millipore water. Filter the solution using a 0.22-μm syringe filter. Stock at − 20 °C.
Basta (5 mg/mL)
Dissolve 250 mg glufosinate (Phyto technology laboratories, Cat. No. P679) in 50 mL millipore water. Filter the solution using a 0.22-μm syringe filter. Stocked at − 20 °C.
Preparation of work reagent
LB media (liquid)
Dissolve 10 g tryptone (OXOID, Cat. No. LP0042), 5 g yeast extract (OXOID, Cat. No. LP0021), and 10 g NaCl (Sinopharm, Cat. No. 10019318) in millipore water to make up a total volume of 1 L, pH 7.0, autoclave (103.4 kPa, 121.0 °C 20 min). Recommended concentration of antibiotic is added to screen the Agrobacterium before the media using.
LB media (solid)
Dissolve 10 g tryptone, 5 g yeast extract, and 10 g NaCl in millipore water to make up a total volume of 1 L, pH 7.0. Addition of 10 g agar powder (Biofroxx, Cat. No. 8211GR500) into liquid LB media before it autoclaves (103.4 kPa, 121.0 °C 20 min). Recommended concentration of antibiotic is added to screen the Agrobacterium before the media using.
Seed germination media (M0)
Dissolve 2.2 g MS powder (4.4 g/L, Duchefa Biochemie, Cat. No. P16438.01) in millipore water to make up a total volume of 1 L. Adjust the pH to 6.0 before addition of 10 g agar powder. Autoclave (103.4 kPa, 121.0 °C 15 min).
Infection and Agrobacterium suspension media (DM)
Mix 4.4 g MS powder and 30 g sucrose in millipore water to make up a total volume of 1 L. Adjust pH to 5.8 and autoclave (103.4 kPa, 121.0 °C 15 min). Addition of 100 μM AS into the media before it uses.
Cocultivation media (M1)
Mix 4.4 g MS powder, 30 g sucrose, 18 g mannitol (Biofroxx, Cat. No. 69-65-8), and 6 g agarose (Tsingke, Cat. No. TSJ001) in millipore water to make up a total volume of 1 L. Adjust pH to 5.8 before addition of agarose, and then autoclave (103.4 kPa, 121.0 °C 15 min). Addition of 1 mg/L 2,4-D, 0.3 mg/L kinetin, and 100 μM AS into the media before it solidifies.
Callus induction media (M2)
Mix 4.4 g MS powder, 30 g sucrose, 18 g mannitol, and 6 g agarose in millipore water to make up a total volume of 1 L. Adjust pH to 5.8 before addition of agarose, and then autoclave (103.4 kPa, 121.0 °C 15 min). Addition of 1 mg/L 2,4-D, 0.3 mg/L kinetin, 30 μM STS, 300 mg/L timentin, and 25 mg/L kanamycin (or 5 mg/L basta, or 20 mg/L hygromycin for different antibiotic screening) into the media before it solidifies.
Shoot initiation media (M3)
Mix 4.4 g MS powder, 10 g glucose (Sinopharm, Cat. No. 10010518), 0.25 g xylose (Sinopharm, Cat. No. 63012037), 0.6 g MES (Sangon Biotech, Cat. No. A610341-0100), and 6 g agarose in millipore water to make up a total volume of 1 L. Adjust pH to 5.8 before addition of agarose, and then autoclave (103.4 kPa, 121.0 °C 15 min). Addition of 2 mg/L zeatin, 0.1 mg/L IAA, 300 mg/L timentin, and 25 mg/L kanamycin (or 5 mg/L basta, or 20 mg/L hygromycin for different antibiotic screening) into the media before it solidifies.
Root initiation media (M4)
Mix 4.4 g MS powder, 10 g sucrose, 0.5 mg/L IBA, and 10 g agar powder in millipore water to make up a total volume of 1 L. Adjust pH to 5.8 before addition of agar powder, and then autoclave (103.4 kPa, 121.0 °C 15 min). Addition of 25 mg/L kanamycin (or 5 mg/L basta, or 20 mg/L hygromycin for different antibiotic screening) into the media before it solidifies.
Plant materials and growth conditions
Different genotypes of rapeseed including inbred spring-type Westar and semi-winter type GY284, WH3417, ZS9 (Zhongshuang 9), and ZS11 (Zhongshuang 11) were used in this study. Seeds were collected from self-crossed plant which were bagged during it flowering every generation in the field. The rooted plantlets from tissue culture were transplanted into the field, or the pot (12 cm × 15 cm) in growth room under the condition of 16 h of light and 8 h of dark at 25 °C.
Statistics and calculation
Each construct was transformed 2–8 times independently. The green seedling acquisition rate is calculated as number of final green seedlings on M4 / number of initial explants. The transformation efficiency is calculated as number of PCR positive seedlings / number of initial explants. The genome editing efficiency is calculated as number of edited plants / number of PCR positive plants.
Agrobacterium and vector
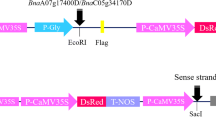
Three Agrobacterium strains—C58C1, EHA105, and GV3101—were used for infection. Five kinds of binary vectors with different resistance screening marker such as kanamycin (pKYLX71 (Hong et al. 2008), p35S-FAST (Lu et al. 2019), pKSE401 (Xing et al. 2014), and pCAMBIA1300 (Lu et al. 2013)) and hygromycin (pMDC83 (Curtis and Grossniklaus 2003)) carrying various genes were transformed into the competent cells of Agrobacterium by electric shock. For pCAMBIA1300S construction, the sequence of hygromycin phosphotransferase II (HPTII) was cloned and linked to pCAMBIA1300 to replace the neomycin phosphotransferase II (NPTII) gene. The RePAT gene was amplified from the vector of pU130 (Cui et al. 2016). The product was linked to pCAMBIA1300S, and then the HPTII gene was deleted. Finally, the vector of pCAMBIA1300S-1 was constructed. The pKSE401-GENE-sgRNA vectors were constructed as previously described (Yang et al. 2017). The modified vectors carrying various genes were transformed into the competent cells of Agrobacterium by electric shock. The primers used for testing the vectors were listed (Table S1). The confirmed Agrobacterium stain was propagated and stored at − 80 °C.
Procedure of transformation
Overview of the steps of Agrobacterium-mediated transformation of rapeseed is shown in Fig. 1. The detail transformation procedure is as follows.
Seed germination and preparing of hypocotyl explant
The rapeseed (Westar, GY284, WH3417, ZS9, and ZS11) seeds are sterilized by 75% ethanol for 1 min, and washed by sterile water for 3 times. Then, the seeds are followed sterilizing by 0.15% HgCl2 for 15 min, and washed by sterile water for 3 times. The sterilized seeds are transferred onto M0 solid media in a Petri dish (diameter = 6 cm). The Petri dish without cover is moved into a sterilized transparent box and kept in a dark room for 7 days under 25 °C. The hypocotyl is picked out (remove the cotyledons and roots carefully) and cut into pieces of 0.6–0.8 cm length in a Petri dish (diameter = 9 cm) with 20 mL DM media. Usually, there are 150–200 pieces of explants in one Petri dish.
Preparation of Agrobacterium
Agrobacterium stain introduced with vector is kept on a solid LB plate with suitable antibiotic. Single colony is picked and put into a glass bottle with 5 mL liquid LB and suitable antibiotic after the seed germination of 5th day. The bottle is fixed in a constant temperature shaker with 200 rpm at 28 °C. Usually, the OD value of the culture solution will reach 0.6–0.8 in 36–48 h.
Infection
Cultured Agrobacterium is transferred into a 10-mL sterilized tube and centrifuged at 6000 rpm for 10 min. Discard the supernatant, resuspend, and wash the pellet twice by adding 5 mL DM media. Addition of 2 mL suspension solution above into the Petri dish (diameter = 9 cm) which contains prepared explants and 20 mL DM solution. The explants are infected for 30 min with shaking one time every 10 min.
Cocultivation
Discard the DM solution after finishing the infection step above. Absorb the residual liquid using sterilized filter paper. The explants are transferred onto M1 media for 2–3 days in a dark room at 25 °C.
Callus induction
The explants are transferred onto M2 media and kept in the tissue culture room for 21 days, at the condition of 16 h day/8 h dark, 26/22 °C.
Shoot differentiation
The explants are transferred onto M3 media and kept in the tissue culture room for 14 days, at the condition of 16 h day/8 h dark, 26/22 °C. Subsequently, the explants are transferred onto a fresh M3 media every 14 days until the shoots come out.
Root initiation and seedling transplantation
Green shoot is cut from the junction of the explant, removed the extra dead tissue, and transplanted into the sterilized transparent box containing M4 media. The box is put in the tissue culture room at 26/22 °C under 16 h day/8 h dark. Usually, the root will grow well in half a month.
DNA extraction and identification of transgenic plants by PCR
Cut a small piece of fully expanded leaf and extract the DNA using shorty buffer (Lu et al. 2016). Transgenic plants are identified by PCR using gene-specific primers. The PCR-confirmed transgenic plants are transplanted into the pots with soil in the growth room, or the field. Further, RNA and protein can be extracted for evaluation of target gene expression by semi-quantitative PCR, real-time PCR, or immunoblotting at this stage if necessary (Lu et al. 2013).
Screening of transgenic plants using antibiotics or basta
The seeds generated from T0 plants can be screened using antibiotics or basta. For kanamycin resistance screening, the seeds are germinated on wetted filter paper for 2–3 days until the embryonic axis exposed from seed coat. Then, the germinated seeds are transferred onto filter paper wetted with 1.2 mg/mL kanamycin solution and cultured at 26/22 °C 16 h day/8 h dark for 7 days. The cotyledon color is green if the seedling is kanamycin resistance, while the cotyledon of negative transgenic plants is yellow or purple. For hygromycin resistance screening, the seeds are directly germinated on filter paper wetted with 150 μg/mL hygromycin solution and cultured at 26/22 °C 16 h day/8 h dark for 7 days. The color of cotyledon is green if the seedling has hygromycin resistance. For basta resistance screening, the seeds are sown in the field, and commercial basta (Bayer, www.premeo.de) under recommended concentration is sprayed on leaf during 5–6 leaf period. The plant grows well and the color of leaf is green if it has basta resistance.
Results
GV3101 Agrobacterium strain is more efficient for rapeseed transformation
We compared the transformation efficiency of three Agrobacterium strains, C58C1, EHA105, and GV3101. Two different vectors, pKYLX71-AtPLDα3 and p35S-FAST-AtPLDε with NPTII selection marker gene, were transformed into these three Agrobacterium strains, respectively. Then, the explants from Westar were infected by each Agrobacterium strain. As shown in Table 1, it was hardly to obtain green seedling after the explants were infected by C58C1. About 0–17 green seedlings were obtained from 161 to 678 explants after transfected by EHA105. The green seedling acquisition rate was 0.5–3.7% and 0–2.3% of pKYLX71-AtPLDα3 and p35S-FAST-AtPLDε, respectively (Table 1). Previous study reported that the green seedlings in M4 media were not all real positive transgenic plants (Zhang et al. 2020). Thus, the green seedlings were further confirmed by PCR using gene-specific primers. The results showed that the transformation efficiency was 0–0.15% and 0–0.16% of pKYLX71-AtPLDα3 and p35S-FAST-AtPLDε, respectively (Table 1). Meanwhile, the green seedling acquisition rate was 16.2–31.3% and 7.2–28.6%, and the transformation efficiency was 2.7–15.1% and 2.2–7.0% when explants were infected by GV3101 carrying the same constructs (Table 1). These results indicate that GV3101 is more efficient for rapeseed transformation than the other two Agrobacterium strains.
Hygromycin, kanamycin, and basta resistance are efficient selection markers for rapeseed transformation
The HPTII and NPTII genes are two major selection markers in plant transgenic research (Berg et al. 1975; Gritz and Davies 1983). The HPTII and NPTII genes confer transgenic plant resistance to hygromycin (Hyg) and kanamycin (Kan), respectively. In plant, Hyg and Kan normally inhibit protein synthesis by binding to ribosomes (Davey et al. 2010). Glufosinate ammonium (basta) is a chemical reagent that can inhibit the activity of glutamine synthetase. Two glufosinate-resistant genes of BAR and PAT, encoding a phosphinothricin N-acetyltransferase, can detoxify glufosinate ammonium by acetylation of the amino group, which are also widely used as selection markers in transgenic plant research (Davey et al. 2010; Donn and Köcher 2002; Cui et al. 2016). Recently, expression of a novel PAT coding gene (RePAT) in rice was reported to improve plant resistance to basta (Cui et al. 2016). Thus, the NPTII gene in pCAMBIA1300 was replaced by HPTII and RePAT genes, and renamed as pCAMBIA1300S and pCAMBIA1300S-1, respectively (Supplemental Figure 1).
The constructs of pCAMBIA1300S-BnKCS (Hyg), pMDC83-BnlncRNA (Hyg), p35S-FAST-BnPLDα1 (Kan), p35S-FAST-BnPLDδ (Kan), pCAMBIA1300-AtPLDα1 (Kan), and pCAMBIA1300S-1-RePAT (basta) were introduced into Agrobacterium GV3101, which were used to infect Westar hypocotyls. Finally, 25–236 of green seedlings were obtained from 286 to 599 explants by Kan selection. The green seedling acquisition rate was 9.6–60.1%, and the transformation efficiency was 2.1–22.5% (Table 1). For Hyg screening, 38–60 of green seedlings were obtained from 732 to 1530 explants. The green seedling acquisition rate was 3.9–5.5%, and the transformation efficiency was 3.7–4.8% (Table 1). Furthermore, 25–83 of green seedlings were obtained from 321 to 768 explants after basta screening. The green seedling acquisition rate and the transformation efficiency was 7.8–16.7% and 1.6–5.9%, respectively (Table 1). Although the highest transformation efficiency was 22.5% using NPTII as selection marker, the transformation efficiency of HPTII and RePAT was acceptable. These results suggest that HptII, NptII, and RePAT are suitable as the selection marker genes for rapeseed transformation (Fig. 2).
Varied transformation efficiency of different genotypes of rapeseed
The spring-type Westar is commonly used for rapeseed transformation because of the high transformation efficiency (Bhalla and Singh 2008; Zhang et al. 2005). To test this transformation procedure which is also applied to other genotypes of rapeseed, hypocotyls from four semi-winter genotypes of GY284, WH3417, ZS9, and ZS11 were infected by GV3101 containing the vector of pKSE401 or pCAMBIA1300. Two sgRNAs target gene-specific region of fatty acid elongase 1 (FAE1), fatty acid desaturase 2 (FAD2), and ketoacyl-CoA synthase (KCS) and were amplified and ligated to pKSE401 before it introduced into GV3101, respectively (Tables 1 and 2). The results showed that the green seedling acquisition rate of GY284, WH3417, ZS9, and ZS11 was 3.9–5.8%, 1.1–3.9%, 0–3.8%, and 0, respectively (Tables 1 and 2). The transformation efficiency was 3.7–5.6%, 1.1–3.5%, 0–0.87%, and 0 for GY284, WH3417, ZS9, and ZS11 (Tables 1 and 2). pKSE401 has been used as a genome editing toolbox in plants (Tang et al. 2018; Yang et al. 2017). Further, we tested the genome editing efficiency of pKSE401-BnFAE1, pKSE401-BnFAD2, and pKSE401-BnKCS by PCR products sequencing. The results indicated that the genome editing efficiency of pKSE401-BnFAE1, pKSE401-BnFAD2, and pKSE401-BnKCS was 23.3–24.0%, 9.5–22.2%, and 13.2–52.6% (Table 2), which was consistent with previous reports (Tang et al. 2018; Yang et al. 2017). These results suggest that the transformation efficiency of rapeseed is genotype dependent, which is similar to previous research (Bhalla and Singh 2008).
Confirmation of the positive transgenic plants
To investigate the transgenic-positive efficiency of regenerated plantlets, genomic DNA was extracted from these putative transgenic plantlets. PCR was performed to detect target genes by using gene-specific primers (Table S1). As expected, most of the putative transgenic plants presented gene-specific bands (Fig. 3a). Western blot was also performed to check protein expression of target gene (e.g., BnPLDα1) in the seedlings (Fig. 3b). Furthermore, the T1 seeds from transgenic lines were screened by antibiotics or basta. The results displayed that the seedling growth from negative transgenic plants was inhibited and the color of its cotyledon observed yellow under Kan (Fig. 4a) and Hyg screening (Fig. 4b). The non-transgenic plants were killed by basta in the field (Fig. 4c). In brief, some regenerated green plantlets are false transgenic plants, and molecular or chemical methods need to be done to confirm their reliability.
The positive transgenic plants were confirmed by PCR and western blot. a The image shows the transgenic plants confirmed by PCR using vector-specific primers. b The image shows the transgenic plants confirmed by western blot using PLDα1-specific antibody. Ten micrograms of total protein of each line was loaded and separated by 8% SDS-PAGE, and then transferred to a polyvinylidene difluoride membrane (PVDF membrane). PLDα1 was detected by PLDα1-specific antibody (Lu et al. 2013). The band in the lane of WT was the coloration of endogenous PLDa1 protein
Discussion
Due to its important position in agronomy, rapeseed has become a hot spot in scientific research, and rapeseed biotechnology has also made considerable progress. Previously, our transformation method had been used to study gene function and generate rapeseed with improved agronomic characteristics (Lu et al. 2013; Lu et al. 2016; Tang et al. 2018; Yang et al. 2017). However, the detail information of the transformation procedure is not described. In addition, most of researches use the model genotype of Westar as transformation receptor. The factors that influence the transformation efficiency such as type of Agrobacterium, selection marker, and genotype of rapeseed have not been compared (Lu et al. 2013; Lu et al. 2016; Tang et al. 2018; Yang et al. 2017). Here, we comprehensively described and compared multiple factors that affect the transformation efficiency by transforming different constructs into five genotypes of rapeseed via three Agrobacterium strains. The results showed that transgenic plants could be easily obtained in four genotypes of rapeseed, suggesting that this protocol is efficient for rapeseed transformation.
Agrobacterium-mediated transformation is the most successful method for producing of genetically modified plants (Tzfira and Citovsky 2006). It has been used for transformation of many plant species (Herrera-Estrella et al. 2004). The transformation has been tested in a variety of explant types such as bud, cotyledon, hypocotyl, embryo, and callus (Bhalla and Singh 2008; Ishida et al. 2007; Zhang et al. 2005; Clough and Bent 1998; Raineri et al. 1990). Transgenic plant of rapeseed can be obtained from hypocotyl and cotyledon which are infected by Agrobacterium (Bhalla and Singh 2008; Rani et al. 2013). These studies reported that the transformation efficiency was 7.7–68.1%, and 5.3–50.1% after Agrobacterium infected cotyledon and hypocotyl from five different rapeseeds, respectively (Bhalla and Singh 2008; Zhang et al. 2005). However, the transformation efficiency was calculated by the number of shoots on selection media divided by total explant number (Bhalla and Singh 2008; Zhang et al. 2005). This statistical method cannot accurately reflect the real transformation efficiency, because not all the green shoots will generate green seedlings (Zhang et al. 2020), or survive on the rooted media (M4 media). Additionally, the false-positive transgenic plants cannot be eliminated by antibiotic screening (Fig. 3a). We presented two values to evaluate the transformation efficiency, including the green seedling acquisition rate (number of final green seedlings on M4 divided by number of initial explants) and transformation efficiency (PCR positive seedlings divided by number of initial explants). In fact, the transformation efficiency reported by previous studies is equivalent to our green seedling acquisition rate. Strictly speaking, our statistic of transformation efficiency is more rigorous. Previous studies showed that the highest transformation efficiency was 21.9% and 33.1% after the hypocotyl and cotyledon of Westar infected by LAB4404 containing NPTII gene, respectively (Zhang et al. 2005). In this study, the highest ratio of green seedling acquisition was 60.1%. The highest ratio of green seedling acquisition is 2–3 times than that of previous study, indicating that our transformation method is more efficient to obtain transgenic plants.
The transformation method using hypocotyl as explant can greatly reduce the number of seed consuming compared with that using cotyledon. One seedling only provides two cotyledon explants, but can provide more than ten hypocotyl explants (0.6–0.8 cm) at least. In addition, the operation of cotyledon transformation requires the incision of each explant dipping into Agrobacterium liquid for 10–30 s, and then keeping the incision inserting into the solid media vertically (Bhalla and Singh 2008; Zhang et al. 2005). These operations are complex and time consuming. Moreover, it takes 10–14 weeks to obtain transgenic plants (Bhalla and Singh 2008), which is longer than our transformation procedure (8–10 weeks). Together, hypocotyl transformation is time-saving and easy to operate compared with previous cotyledon transformation (Bhalla and Singh 2008; Zhang et al. 2005).
Regeneration of rapeseed in transformation is highly genotype dependent. A study reported that the variation range of regeneration rate was from 0 to 91% in 100 cultivars of rapeseed (Ono et al. 1994). We found that the regeneration rate of Westar was the highest among five genotypes of rapeseed. Westar is a disease-susceptible cultivar, and ZS11 is a highly disease-resistant cultivar. Higher resistance of ZS11 might repress the Agrobacterium infection and reduce the transformation efficiency, suggesting that the regeneration of callus is affected by plant resistance. In addition, the Agrobacterium strain is found to be a critical factor for the successful transformation (Rani et al. 2013). We compared the transformation efficiency among three Agrobacterium stains including C58C1, EHA105, and GV3101. The efficiency of obtaining green seedlings and transformation efficiency of GV3101 was 7.2–31.3% and 2.2–15.1%, which were much higher than that of C58C1 and EHA105. It is more efficient than that of using LBA4404 stain infected hypocotyl of Westar in previous study (Zhang et al. 2005). Taken together, GV3101 is more suitable for the transformation of rapeseed.
Screening gene of antibiotic resistance is an essential factor for improving transformation efficiency (Rani et al. 2013). Efficient selection depends on kinds of antibiotics employed and their concentration used. The green seedling acquisition rate of using NPTII and RePAT as selection gene was higher than that of using HPTII (Table 1). However, the ratio of positive transgenic plants in green seedlings confirmed by PCR was 92.1–100.0% when using HPTII as selection gene. It was higher than that of using NPTII and RePAT. Usually, the transformation efficiency was not much different among these three selection markers, suggesting that HPTII, NPTII, and RePAT are suitable as selection genes for rapeseed transformation. Although the transformation efficiency was acceptable when HPTII, NPTII, and RePAT were used as selection genes, the variation of transformation efficiency was still greatly among different experiments, even the recipient material and construct were the same (Table 1 and Table 2). The reason might be due to this system project that was done by different researchers in different time, which would influence the transformation efficiency, such as the different operation of different person and the different situation of explants and Agrobacterium strains of different experiments.
In summary, we describe an efficient Agrobacterium-mediated transformation method using hypocotyl as explants, which is applicable to various genotypes of rapeseed. It takes approximately 8–10 weeks to complete the procedure from seed germination to obtaining of rooted plantlets. This method is highly efficient and time-saving. In addition, compared with the transformation method using cotyledon as explants, the explants are easier to obtain and the operation is simpler in our method. Together, this method will benefit gene functional study, especially in high-throughput molecular biology research in rapeseed.
References
Berg DE, Davies J, Allet B, Rochaix JD (1975) Transposition of R factor genes to bacteriophage lambda. Proc Natl Acad Sci U S A 72(9):3628–3632. https://doi.org/10.1073/pnas.72.9.3628
Bergman P, Glimelius K (1993) Electroporation of rapeseed protoplasts – transient and stable transformation. 88(4):604–611. https://doi.org/10.1111/j.1399-3054.1993.tb01378.x
Bhalla PL, Singh MB (2008) Agrobacterium-mediated transformation of Brassica napus and Brassica oleracea. Nat Protoc 3(2):181–189. https://doi.org/10.1038/nprot.2007.527
Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, Correa M, Da Silva C, Just J, Falentin C, Koh CS, Le Clainche I, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao M, Edger PP, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Le Paslier MC, Fan G, Renault V, Bayer PE, Golicz AA, Manoli S, Lee TH, Thi VH, Chalabi S, Hu Q, Fan C, Tollenaere R, Lu Y, Battail C, Shen J, Sidebottom CH, Wang X, Canaguier A, Chauveau A, Berard A, Deniot G, Guan M, Liu Z, Sun F, Lim YP, Lyons E, Town CD, Bancroft I, Wang X, Meng J, Ma J, Pires JC, King GJ, Brunel D, Delourme R, Renard M, Aury JM, Adams KL, Batley J, Snowdon RJ, Tost J, Edwards D, Zhou Y, Hua W, Sharpe AG, Paterson AH, Guan C, Wincker P (2014) Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345(6199):950–953. https://doi.org/10.1126/science.1253435
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743. https://doi.org/10.1046/j.1365-313x.1998.00343.x
Cui Y, Liu Z, Li Y, Zhou F, Chen H, Lin Y (2016) Application of a novel phosphinothricin N-acetyltransferase (RePAT) gene in developing glufosinate-resistant rice. Sci Rep 6:21259–21259. https://doi.org/10.1038/srep21259
Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133(2):462–469. https://doi.org/10.1104/pp.103.027979
Davey MR, Anthony P, Power JB, Lowe KC (2005) Plant protoplasts: status and biotechnological perspectives. Biotechnol Adv 23(2):131–171. https://doi.org/10.1016/j.biotechadv.2004.09.008
Davey MR, Soneji JR, Rao MN, Kourmpetli S, Bhattacharya A, Kole C (2010) Generation and deployment of transgenic crop plants: an overview. In: Kole C, Michler CH, Abbott AG, Hall TC (eds) Transgenic crop plants: principles and development. Springer, Berlin Heidelberg, pp 1–29. https://doi.org/10.1007/978-3-642-04809-8_1
Donn G, Köcher H (2002) Inhibitors of glutamine synthetase. In pp 87–101. https://doi.org/10.1007/978-3-642-59416-8_4
Eimert K, Siegemund F (1992) Transformation of cauliflower (Brassica oleracea L. var. botrytis)--an experimental survey. Plant Mol Biol 19(3):485–490. https://doi.org/10.1007/BF00023396
Gracka A, Jelen HH, Majcher M, Siger A, Kaczmarek A (2016) Flavoromics approach in monitoring changes in volatile compounds of virgin rapeseed oil caused by seed roasting. J Chromatogr A 1428:292–304. https://doi.org/10.1016/j.chroma.2015.10.088
Gritz L, Davies J (1983) Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene 25(2–3):179–188. https://doi.org/10.1016/0378-1119(83)90223-8
Hansen LN, Ortiz R, Andersen SB (1999) Genetic analysis of protoplast regeneration ability in Brassica oleracea. Plant Cell Tissue Organ Cult 58(2):127–132. https://doi.org/10.1023/A:1006359804328
Herrera-Estrella L, Simpson J, Martínez-Trujillo M (2004) Transgenic plants: an historical perspective. In: Peña L (ed) Transgenic plants: methods and protocols. Humana Press, Totowa, pp 3–31. https://doi.org/10.1385/1-59259-827-7:003
Hong Y, Pan X, Welti R, Wang X (2008) Phospholipase Dalpha3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 20(3):803–816. https://doi.org/10.1105/tpc.107.056390
Ishida Y, Hiei Y, Komari T (2007) Agrobacterium-mediated transformation of maize. Nat Protoc 2(7):1614–1621. https://doi.org/10.1038/nprot.2007.241
Kikkert JR, Vidal JR, Reisch BI (2005) Stable transformation of plant cells by particle bombardment/biolistics. Methods Mol Biol 286:61–78. https://doi.org/10.1385/1-59259-827-7:061
Lu S, Bahn SC, Qu G, Qin H, Hong Y, Xu Q, Zhou Y, Hong Y, Wang X (2013) Increased expression of phospholipase Dalpha1 in guard cells decreases water loss with improved seed production under drought in Brassica napus. Plant Biotechnol J 11(3):380–389. https://doi.org/10.1111/pbi.12028
Lu S, Fadlalla T, Tang S, Li L, Ali U, Li Q, Guo L (2019) Genome-wide analysis of phospholipase D gene family and profiling of phospholipids under abiotic stresses in Brassica napus. Plant Cell Physiol 60:1556–1566. https://doi.org/10.1093/pcp/pcz071
Lu S, Yao S, Wang G, Guo L, Zhou Y, Hong Y, Wang X (2016) Phospholipase Depsilon enhances Braasca napus growth and seed production in response to nitrogen availability. Plant Biotechnol J 14(3):926–937. https://doi.org/10.1111/pbi.12446
Mason AS, Snowdon RJ (2016) Oilseed rape: learning about ancient and recent polyploid evolution from a recent crop species. Plant Biol (Stuttg) 18(6):883–892. https://doi.org/10.1111/plb.12462
Mukhopadhyay A, Topfer R, Pradhan AK, Sodhi YS, Steinbiss HH, Schell J, Pental D (1991) Efficient regeneration of Brassica oleracea hypocotyl protoplasts and high frequency genetic transformation by direct DNA uptake. Plant Cell Rep 10(8):375–379. https://doi.org/10.1007/BF00232604
Ono Y, Takahata Y, Kaizuma N (1994) Effect of genotype on shoot regeneration from cotyledonary explants of rapeseed (Brassica napus L.). Plant Cell Rep 14(1):13–17. https://doi.org/10.1007/BF00233290
Poulsen GB (1996) Genetic transformation of Brassica. Plant Breed 115(4):209–225. https://doi.org/10.1111/j.1439-0523.1996.tb00907.x
Radchuk VV, Ryschka U, Schumann G, Klocke E (2002) Genetic transformation of cauliflower (Brassica oleracea var. botrytis) by direct DNA uptake into mesophyll protoplasts. Physiol Plant 114(3):429–438. https://doi.org/10.1034/j.1399-3054.2002.1140313.x
Rahman H, Bennett RA, Kebede B (2017) Mapping of days to flower and seed yield in spring oilseed Brassica napus carrying genome content introgressed from Brassica oleracea. Mol Breed 37(1). https://doi.org/10.1007/s11032-016-0608-2
Raineri DM, Bottino P, Gordon MP, Nester EW (1990) Agrobacterium–mediated transformation of rice (Oryza sativa L.). Nat Biotechnol 8(1):33–38. https://doi.org/10.1038/nbt0190-33
Rani T, Yadav R, Yadav N, Rani A, Singh D (2013) Genetic transformation in oilseed brassicas -a review. Indian J Agric Sci 83(4):367–373
Sitther V, Tabatabai B, Enitan O, Dhekney S (2018) Agrobacterium-mediated transformation of Camelina sativa for production of transgenic plants. J Biol Methods 5(1):e83. https://doi.org/10.14440/jbm.2018.208
Tang T, Yu X, Yang H, Gao Q, Ji H, Wang Y, Yan G, Peng Y, Luo H, Liu K, Li X, Ma C, Kang C, Dai C (2018) Development and validation of an effective CRISPR/Cas9 vector for efficiently isolating positive transformants and transgene-free mutants in a wide range of plant species. Front Plant Sci 9:1533. https://doi.org/10.3389/fpls.2018.01533
Tzfira T, Citovsky V (2006) Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr Opin Biotechnol 17(2):147–154. https://doi.org/10.1016/j.copbio.2006.01.009
Verma S, Chinnusamy V, Kc B (2008) A simplified floral dip method for transformation of Brassica napus and B. carinata. J Plant Biochem Biotechnol 17:197–200. https://doi.org/10.1007/BF03263286
Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14:327. https://doi.org/10.1186/s12870-014-0327-y
Yang H, Wu JJ, Tang T, Liu KD, Dai C (2017) CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci Rep 7(1):7489. https://doi.org/10.1038/s41598-017-07871-9
Yoo S-D, Cho Y-H, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2(7):1565–1572. https://doi.org/10.1038/nprot.2007.199
Yu B, Boyle K, Zhang W, Robinson SJ, Higgins E, Ehman L, Relf-Eckstein J-A, Rakow G, Parkin IAP, Sharpe AG, Fobert PR (2016) Multi-trait and multi-environment QTL analysis reveals the impact of seed colour on seed composition traits in Brassica napus. Mol Breed 36(8). https://doi.org/10.1007/s11032-016-0521-8
Zhang K, He J, Liu L, Xie R, Qiu L, Li X, Yuan W, Chen K, Yin Y, Kyaw MMM, San AA, Li S, Tang X, Fu C, Li M (2020) A convenient, rapid and efficient method for establishing transgenic lines of Brassica napus. Plant Methods 16:43. https://doi.org/10.1186/s13007-020-00585-6
Zhang Y, Singh MB, Swoboda I, Bhalla PL (2005) Agrobacterium-mediated transformation and generation of male sterile lines of Australian canola. Aust J Agric Res 56(4):353. https://doi.org/10.1071/ar04175
Acknowledgments
We are grateful to Prof. Qijun Chen at China Agricultural University for kindly providing pKSE401 and pCBC-DT1T2 vectors for constructing CRISPR/Cas9 system.
Funding
This study was supported by the National Program of Transgenic Variety Development of China (2018ZX08020-001), the National Natural Science Foundation of China (31701458), the Fundamental Research Funds for the Central Universities (2662018QD064), and the China Postdoctoral Science Foundation (2015M580651, 2016T90704).
Author information
Authors and Affiliations
Contributions
SP.L., C.D., and L.G. designed the research. YQ. L, L.L, ZL.D., SL. L., X.T., SJ. L., B.Y., W.Y., and J.W. performed the experiments. SP.L., and C.D. analyzed the data. C.D. and SP.L. wrote the manuscript. L.G. revised the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Dai, C., Li, Y., Li, L. et al. An efficient Agrobacterium-mediated transformation method using hypocotyl as explants for Brassica napus. Mol Breeding 40, 96 (2020). https://doi.org/10.1007/s11032-020-01174-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-020-01174-0