Abstract
Soybean [Glycine max (L.) Merr.] is one of the major legume crops for human and livestock, and the double cropping system with soybean have been widely applied to increase arable land utilization rate. To employ the soybean for multiple cropping system, cultivars, which are photoperiod insensitive and early maturing, are required. The region-specific crop adaptation could be achieved by the successful flowering followed by progeny production, and it is important to understand the mechanisms underlying the transition from the vegetative to reproductive stage. In soybean, 10 genes/QTLs conferring flowering time were identified, and four genes, E1, E2, E3, and E4, are mainly involved in geographic adaptation of soybean. Here, to develop the photoperiod-insensitive early-maturing soybean varieties, QTL analysis was conducted using RILs from the crosses between photoperiod-insensitive early-maturing soybean cultivar, Keunol, and late maturity soybean cultivar, Sinpaldal. Furthermore, to identify the topological distribution within Korea peninsula, SNPs in exon region of 40 soybean varieties were investigated by sequencing of the exon region in E2 and comparing to their origin information. The results showed that early flowering is mainly controlled by the E2 in five different environments, and there are three nonsynonymous SNPs in E2. Two of them, SNPs in exon 2 and 10, produce the premature stop codon. The first SNP in exon 2 was strongly linked to topological distribution, which is mainly restricted to southern area of Korea peninsula. This result could provide the useful information to develop photo-insensitive early-maturing soybean cultivar, which could be employed in various cropping system.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean [Glycine max (L.) Merr.] is one of the major legume crops to feed livestock and provide protein and oil sources for human (Masuda and Goldsmith 2009). To improve yield through increasing arable land utilization rate, double cropping systems with soybean have been widely applied. For double cropping system, early-maturing soybean cultivars were preferred due to their short cultivation period. However, photoperiod-sensitive short-season soybean cultivars showed significant yield losses caused by insufficient vegetative stage. In late sowing, these cultivars could not complete canopy establishment prior to reproductive development (Ball et al. 2000). To overcome this bottleneck, photoperiod-insensitive soybean varieties, which flowering date is not affected by the season change, could be a useful genetic resource for soybean-breeding program.
From an agricultural perspective, it is important to understand the mechanisms underlying the transition from the vegetative to the reproductive stage of crops, which play a key role in the control of reproductive success as flowering enables the completion of seed development under favorable environment conditions (Andres and Coupland 2012). Furthermore, the region-specific crop adaptation could be available, when successful flowering followed by progeny production. Many physiological studies have revealed that the timing of flowering is regulated by a complex network of genetic pathways, enabling plants to response to several external factors including photoperiod, temperature, and abiotic stresses, and internal factors including gibberellin level, and age. In the model plant Arabidopsis thaliana, it appears that there are four genetic pathways, which regulate flowering time including vernalization, autonomous, hormonal, and photoperiod pathways (Sharma et al. 2016). And there are numerous genes identified that are associated with the regulation of flowering time, such as CONSTANS (CO), FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC), FCA, FVE, and FLOWERING LOCUS C (Kobayashi et al. 1999; Michaels and Amasino 1999; Turck et al. 2008). Among the four pathways, photoperiod is considered as one of the most important environmental signals for decision to flower, where the circadian clock is known to be associated with measuring photoperiod and day length is used for perception of the season (Davis 2002; Wang et al. 2010). For example, short-day plants, such as rice, maize, sorghum, soybean, tobacco, and sugarcane, require long night for flowering; in contrast, long-day plants will flower when exposed to long days. Long-day plants include wheat, barley, ryegrass, peas, spinach, and lettuce (Nakamichi 2015). In A. thaliana, the GIGANTEA (GI), CONSTANS (CO), and FLOWERING LOCUS T (FT) genes are involved in a circadian clock-controlled photoperiodic flowering pathway, and this GI-CO-FT module is highly conserved in many plants including rice (Oryza sativa L.) (Hayama et al. 2003). The unique plant-specific nuclear protein, GI, is a key regulator of photoperiodic flowering which promotes CO transcription, the central component involved in measuring the day length (Fowler et al. 1999; Park et al. 1999).
Soybean is a typical short-day plant, which requires the day length decrease below some critical value to trigger the flowering. Under the long-day condition, floral induction and flowering are suppressed. The domestication period based on genetic studies of G. max from its wild relative, Glycine soja, is generally dating back to 6000~9000 years ago (Carter et al. 2004). Since then, soybeans have gone through the process of domestication, which resulted in a variety of soybean landraces that have adapted to wide range of environmental conditions. As a consequence, the domestication syndrome has occurred resulting in a common suite of traits that distinguishes wild progenitors from domesticated crops, and those traits include a loss of seed dormancy, photoperiod sensitivity, and synchronized flowering (Doebley et al. 2006). Generally, several genes in various crops control these distinctive morphological and physiological traits in domesticated crops. For example, the pseudoresponse regulator gene Ppd1, TaGI1 and the vernalization genes VERNALIZATION 1 (VRN1) and VRN2 in wheat, and Ppd-H1, HvGI, HvVRN1 and HvVRN2, HvCO1, EARLY MATURITY 8, and EARLY FLOWERING 3 in barley (Peng et al. 2015). Even though a number of genes are known to be associated with domestication in soybean based on the information from whole genome sequencing, only a few genes have been functionally identified as domestication-related genes (Sedivy et al. 2017). GmTfl1 alleles for controlling determinate growth habit, GMSHAT1-5 for mediating pod shattering, and GmHs1-1 for controlling hard-seededness (Dong et al. 2014; Sun et al. 2015; Tian et al. 2010).
In soybean, currently 10 genes/QTLs, E1–9, and J have been identified to associate with flowering time and maturity (Bernard 1971; Bonato and Vello 1999; Buzzell 1971; Buzzell and Voldeng 1980; Cober et al. 2010; Cober and Voldeng 2001; Kong et al. 2014; McBlain and Bernard 1987; Ray et al. 1995). Among these, four genes, E1, E2, E3, and E4, are involved in geographic adaptation of soybean, and they have cloned through either map-based cloning or candidate gene approach (Jiang et al. 2014). The E2 gene is an ortholog of Arabidopsis GIGANTEA, GmGIa (E2, Glyma.10g221500) (Watanabe et al. 2011). In addition, E3 and E4 are homologs of phytochrome A (PHYA), which controls the response to light quality and photoperiod length, and encode the photoreceptors GmPHYA3 (E3, Glyma.19g224200) and GmPHYA2 (E4, Glyma.20g090000), respectively (Liu et al. 2008; Watanabe et al. 2009). Interestingly, three GIGANTEA homologs, GmGI1a, GmGI1, and GmGI2, are present in soybean genome, and their molecular functions are found to be different (Wang et al. 2016), although GmGI1a has multiple functions in the circadian clock and flowering (Watanabe et al. 2011). A recent study has revealed that one of the GmGIa haplotypes, H1, is the most successful for early flowering and may have facilitated the radiation of domesticated soybeans (Wang et al. 2016).
The objectives of this research were to investigate the photoperiod-insensitive soybean adaptation in Korea based on the flowering time at different latitudes, using natural regional genotype variation in the short-period maturity soybean landraces using molecular approach. First, F8-derived F12 recombinant inbred lines (RILs) developed from a cross between photoperiod-insensitive early flowering variety, Keunol, and late flowering variety, Sinpaldal were used to construct a genome-wide molecular genetic linkage map to identify the soybean flowering time responsible QTL using the recently developed high-density 180K Axiom® SoyaSNP assay. Furthermore, we investigated sequence variations of identified soybean flowering gene using 60 Korean accessions collected from different latitudes to develop SNP markers that could be valuable in breeding soybean cultivars for photoperiod-insensitive early flowering, which could be adapted to multiple-cropping system in Korea.
Materials and methods
Plant materials
A total of 115 F8-derived F12 RILs were developed through single-seed descent from the cross between two parents, which showed significant difference in flowering time (Kim et al. 2006). Early flowering variety, Keunol showed the characteristic of photoperiod insensitivity, in which long-day condition does not suppress the flowering, and Sinpaldal was late flowering variety. Flowering date (DF), the number of days from sowing to the day when the first flower had opened between 40 to 50% of the plants in each plot, was recorded at three different locations and year, Suwon (37° 15′ N, 126° 58′ E), Yeoncheon (38° 10′ N, 127° 06′ E), and Cheonan, Korea (36° 83′ N, 127° 17′ E), in 2005, 2006, and 2015, respectively.
Total 60 soybean varieties, which is consisting of landraces or breeding lines developed by Korean breeders that flowered within 33 days, were selected from about 5000 accessions conserved in the Rural Development Administration (RDA) in Korea (Supplementary Table 1). A genotype variation of a DF-associated candidate gene region was investigated to show the soybean adaptability according to the region and latitude in Korea (Supplementary Table 1). They were sown in pots of 12 cm diameter filled with horticulture compost soil and sand (3:1) and grown in a growth chamber under a long-day condition of 18/6 h (light/dark) photoperiod at day/night and short-day condition of 12/12 h at temperature of 28/23 °C.
Genetic map construction and QTL analysis
Young three trifoliate leaves from three different plants per RIL line were harvested for genomic DNA extraction. Genomic DNA was extracted using the cetyltrimethylammonium bromide (CTAB) method with minor modification (Doyle 1987). To construct a genome-wide molecular genetic linkage map, two parents and 115 F8-derived F12 RILs were genotyped with 180,961 SNP markers developed by Lee et al. (2015) using the 180K Axiom SoyaSNP assay (Affymetrix, Santa Clara, CA, USA) and scanned with a GeneTitan® Scanner (Affymetrix). Of 180,961 SNPs, 169,028 high-quality SNPs were used to construct a genetic linkage map and QTL analysis, because 1185 SNPs and 10 SNPs were located in scaffolds and chloroplast, respectively.
A genetic linkage map was constructed using JoinMap software v.4.1 (Van Ooijen 2006) with a ML mapping algorithm for calculation of genetic distances with default parameters. The recombination fractions were converted into genetic map distances (cM) between markers using the Kosambi map function. Markers that detected to be scorable were 169,028, of which a total of 26,054 SNP markers showed polymorphism. Segregation distortion of SNP markers was calculated using the chi-square at the P < 0.05 level of significance. With segregation distortion and redundant markers being removed, 8691 SNP markers were used for the construction of genetic linkage map. Candidate QTL regions associated with DF were identified with composite interval mapping (CIM) based on MapQTL 6 software (Van Ooijen 2009). To determine QTL significance thresholds, a permutation test with 1000 replications with a walk speed of 1 cM was performed with a logarithm of odds (LOD) value greater 3.0 at a significance level of 0.05. The manufacturer’s instructions based on the Williams 82 genome assembly were implemented to determine the physical locations of the two nearest markers (Schmutz et al. 2010). The annotated genes within the two nearest markers were identified by SoyBase (Grant et al. 2010).
SNP variation for the candidate gene
Genomic DNAs from all 60 soybean varieties were extracted using previously described method. The polymerase chain reaction (PCR) was conducted in a final volume of 100 μl reaction system contained 50 μl of AmpONE™ Taq (4U, GeneAll), 30 μl of distilled water, 5 μl of each forward and reverse primers, and 20 ng of genomic DNA. The primers were designed according to the E2 gene sequences from Williams 82 using the PrimerQuest software. Ten pairs of forward and reverse primers were designed and used for amplification (Supplementary Table 2). The PCR reaction was conducted by an initial denaturation step at 95 °C for 5 min, followed by 35 cycles at 95 °C for 30 s, annealing at 52 °C for 30 s, and extension at 72 °C for 30 s to 1 min. PCR products were purified using the Favorprep™ GEL/PCR Purification Kit (Favorgen), and sequencing was performed using ABI 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA).
Results
Flowering date evaluation in five different environments
The phenotypic variation attributed to DF was evaluated in the RIL mapping population and their parents, early flowering variety, Keunol, and late flowering variety, Sinpaldal (Fig. 1). The early flowering parent, Keunol, exhibited shorter DF values, while the late flowering parent, Sinpaldal, showed longer DF values in Suwon (40 vs. 45 for Keunol in 2005 and 36 vs. 56 for Sinpaldal in 2006). Although DF of the parents varied among three different sites, the DF was similarly observed from Yeoncheon and Cheonan. In Yeoncheon, Keunol flowered at 35 and 31 days, while Sinpaldal flowered at 38 and 40, respectively. In 2015, DF was also measured at Cheonan and showed that DF for Keunol was at 43 days, while 54 days for Sinpaldal. In the RIL population, the phenotypic frequency distributions for DF observed from three different places had a relatively normal distribution, ranging from 31, 26, and 21 days to 60, 55, and 65 days in Suwon, Yeoncheon, and Cheonan, respectively. Moreover, extreme flowering times were between 41 and 45 in Suwon and Cheonan, and 31–35 for 2005, and 41–45 for 2006 in Yeoncheon. All RILs showed a continuous, unimodal phenotype distribution, indicating quantitative regulation of flowering time (Fig. 1).
Composite interval mapping of QTLs associated with flowering date
The high-density genetic linkage map for 115 RILs was constructed using the recently developed genotyping platform, the 180K Axiom® SoyaSNP assay. Of the 8691 SNP markers that were available for the construction of genetic linkage map, a total of 7321 SNP markers were mapped into 20 linkage groups corresponding to the 20 soybean chromosome pairs (Supplementary Table 3). The map spanned a total of 3302 cM with an average interval between adjacent markers of 0.5 cM. Compared to the previously constructed genetic linkage map using Illumina Infinium SoySNP6K BeadChip (Akond et al. 2015), the number of markers mapped and the total length of the genetic linkage map increased by 4.9 and 2.3 times, respectively. Moreover, our map was much denser than the previous report where intervals between markers decreased by 53%.
To identify the candidate QTL regions, composite interval mapping was used and detected 2 QTLs conferring flowering date in the F8:12 RILs from the cross between Keunol and Sinpaldal. One QTL in the marker interval of AX-90386027 and AX-90459836 on chromosome 10 was detected to be associated with DF from all five environments, Suwon, Yeoncheon, and Cheonan, which was positioned between 45,283,870 and 45,329,450-bp genomic region on chromosome according to Wm82.a2.v1 soybean genome assembly (Table 1). Another QTL controlling flowering date was also identified on chromosome 16 between two flanking SNP markers, AX-90422463–AX-90460180 only from Yeoncheon 2005. The QTLs conferring flowering date on chromosome 10 had the LOD score ranged from 5.12 to 11.92, while the QTL on chromosome 16 had the LOD score of 4.96. The additive effects of the QTL on chromosome 10 from all five environments were negative, while the QTL on chromosome 16 was positive. Within QTL interval on chromosome 16, there are several genes conferring soybean flowering time, including GmFT2a. However, this QTL was not identified in other environments, including Yeoncheon 2006. This result may be caused by the different sowing date and growth climatic condition followed by the different length of vegetative stage between 2005 and 2006 in Yeoncheon. Also, flowering date frequency distribution showed significant difference (Fig. 1). To elucidate the SNP variations underlying flowering time, the potential candidate genes underlying the major QTL on Chr 10 were identified based on the Williams 82 reference genome according to the USDA-ARS soybean genetic database (Grant et al. 2010). Based on the search, there were three candidate genes between 45,323,870 and 45,329,450 bp, which were Glyma.10g221500, Glyma.10g221600, and Glyma.10g221700. A previous report identified that Glyma.10g221500 was associated with GIGANTEA gene associated with maturity and flowering, which is a photoperiod response gene, E2, in soybean (Watanabe et al. 2011). Therefore, Glyma.10g221500 was used to detect the SNP variations in the early flowering variety, Keunol, and the late flowering variety, Sinpaldal (Fig. 2).
SNP variation for the candidate gene
Using the identified candidate gene, Glyma.10g221500, sequence variability was screened between Keunol and Sinpaldal for mutations that alter nucleotides. The sequence analysis revealed four nucleotide substitutions in exon regions between Williams 82/Sinpaldal2 and Keunol. They were located within the 2nd exon (position 49; C to T), 6th exon (position 11; A to G), 7th exon (position 152; G to T), and 10th exon (position 137; G to T) (Fig. 3a). Furthermore, three nucleotide substitutions resulted in nonsynonymous amino acid substitutions. A C/T transition in the 2nd exon introduced a premature stop codon to truncated immature protein, while a A/G transition in the 6th exon and a G/T transition in the 10th exon led to nonsynonymous amino acid substitutions of isoleucine to valine and serine to isoleucine, respectively, leading to putative structural changes in the protein. There was a synonymous amino acid substitution in the 7th exon. Even though the position of SNP variation varied, previous studies exhibited that there was a premature stop codon in the 10th exon occurred in a mutant line of Japanese soybean varieties (Watanabe et al. 2011) and in Chinese soybean varieties (A/T transition) (Wang et al. 2016).
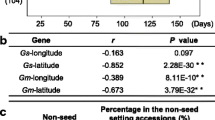
The DF difference between short-day condition and long-day condition for 60 accessions were evaluated to identify the photoperiod-insensitive early-maturing soybean varieties (Supplementary Table 4). These soybean varieties should show minimum difference of DF. The 40 varieties among total 60 soybean accessions showed the significantly small difference, less than 4 days, between long-day and short-day condition. To investigate the topological distribution of 40 photoperiod-insensitive early-maturing soybean, its origin information is obtained from the National Agrobiodiversity Center Database (http://genebank.rda.go.kr), and marked on the map (Fig. 3). Interestingly, most of soybean varieties with T allele in 2nd exon were obtained from southern area of Korea peninsula, and they are also different from Japan origin varieties. These SNPs could significantly contribute to regional adaptation and cultivation of current soybean varieties.
Three SNPs in E2 exon region to 43 Korean photoperiod-insensitive early-maturing soybean varieties and their topological origin. (A) Gene model of Glyma.10g221500. Single nucleotide polymorphism in exon region is marked with red asterisk. (B) Origin information of 43 Korean photoperiod-insensitive early-maturing soybean varieties and their SNP information. When its specific origin is not available, it is marked as NA. (C) Topological origin distribution in Korea peninsula. T allele in exon 2 is marked with red reverse triangle; the C allele is marked with blue. All North Korea and Japan origin varieties are marked in one representative reverse tringle
Discussion
Plant breeding is broadly described as the science, art, and business of the improvement of plants for human benefit (Bernardo 2002). Breeding is often performed by phenotype-based without knowledge composition of genes that control the trait being selected, and artificial selection accelerated phenotypic variations as early as domestication period. Since G. max was domesticated from G. soja in China about 6000~9000 years ago, cultivated G. max has gone through domestication processes involving artificial/natural selections and plant adaption to different environment conditions that gave rise to a loss of genetic diversity, and ultimately led to the domestication bottleneck (Carter et al. 2004). As a consequence, distinctive morphology and physiology changes, also known as domestication-related traits (DRTs), appear to have evolved in numerous crop species, enabling farmers to grow crops with desirable traits (Doebley et al. 2006). These DRTs include those enhancing adaptation to cultivation practices and desirability for human consumption and use (Gepts 2010). Knowing that understanding the genetic basis underlying DRTs is important for efficient breeding programs, extensive efforts have been made to identify the genomic regions responsible to DRTs in soybean such as flowering time, determinate habit, plant height, number of nodes, maximum internode length, twinning habit, pod dehiscence, seed weight, and hard seededness.
Among those DRTs, flowering time is an important adaptive trait in plant. A recent study has discovered the geographic radiation of the GIGANTEAa (GIa) that reflects soybean domestication processes. Using the wild soybean accessions originated from Korea, China, and Japan, the H1 haplotype, one of the GIa haplotypes, was only detected in soybeans restricted to the Yellow River region in China, whereas H1 was not detected in Korean and Japanese wild soybean germplasm (Wang et al. 2016). This study has hypothesized that H1 haplotype was originated from the Yellow River region of China and subsequently spread to Japan, Korea, and other parts of the world during domestication. Moreover, H1 showed a premature stop codon in the 10th exon, suggesting that H1 was probably more efficient involved in promotion of flowering compared to its closest GIa haplotype. In our study, among 41 soybean landraces collected from different parts of Korea, 12 varieties, as well as the early flowering variety, Keunol, showed a C/T transition in the 2nd exon resulted in a premature stop codon in Glyma10g221500 which is known to be associated with GIGANTE. All of these soybean accessions were surprisingly originated from the southern part of Korea, showing an interesting geographical pattern (Fig. 3). The Keunol variety was collected in Gyeongsangbuk-do, and the collection origin of other 12 soybeans were near the Gyeongsangbuk-do areas including Samcheok, Gangwon-do, Gyeongsangnam-do, Jeollanam-do, and Jeollabuk-do. This geographical pattern suggests that a premature stop codon in Glyma.10g221500 has undergone artificial selection by farmers in the southern part of Korea, yet it has not been introduced in other parts of Korea.
In conclusion, flowering time is an essential trait for reproductive and adaptation to a wide range of environmental conditions; therefore, identification of QTLs underlying DF will provide opportunities for breeders to develop new cultivars in changing environmental conditions. In addition, sequence analysis in other candidate genes controlling DF using a large set of soybean samples from other regions of Korea may provide the geographic radiation of soybeans during domestication in Korea, and ultimately create SNP markers for different flowering time that could be useful for soybean cultivation.
Abbreviations
- QTL:
-
Quantitative trait loci
- SNP:
-
Single nucleotide polymorphism
- RILs:
-
Recombinant inbred lines
References
Akond M, Liu S, Kantartzi SK, Meksem K, Bellaloui N, Lightfoot DA, Yuan J, Wang D, Anderson J, Kassem MA (2015) A SNP genetic linkage map based on the ‘Hamilton’ by ‘Spencer’ recombinant inbred line population identified QTL for seed isoflavone contents in soybean. Plant Breed 134:580–588
Andres F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13:627–639
Ball RA, Purcell LC, Vories ED (2000) Optimizing soybean plant population for a short-season production system in the southern USA This paper is published with the approval of the director of the Arkansas Agricultural Experimental Station (manuscript number 99094). Crop Sci 40:757–764
Bernard RL (1971) Two major genes for time of flowering and maturity in soybeans. Crop Sci 11:242–244
Bernardo R (2002) Breeding for quantitative traits. Stemma Press, Minneapolis
Bonato ER, Vello NA (1999) E6, a dominant gene conditioning early flowering and maturity in soybeans. Genet Mol Biol 22:229–232
Buzzell RI (1971) Inheritance of a soybean flowering response to fluorescent-daylength conditions. Can J Genet Cytol 13:703–707
Buzzell RI, Voldeng HD (1980) Inheritance of insensitivity to long daylength. Soybean Genet Newsl 7:26–29
Carter TE, Nelson RL, Sneller CH, Cui Z (2004) Genetic diversity in soybean. In: Boerma HR, Specht JE (eds) Soybeans: improvement, production, and uses. American Society of Agronomy, Crop Science Society of America, and Soil Science Society of America, Madison, pp 303–416
Cober ER, Voldeng HD (2001) A new soybean maturity and photoperiod-sensitivity locus linked to E1 and T ECORC contribution no. 991424. Crop Sci 41:698–701
Cober ER, Molnar SJ, Charette M, Voldeng HD (2010) A new locus for early maturity in soybean All rights reserved. No part of this periodical may be reproduced or transmitted in any form or by any means, electronic or mechanical, including photocopying, recording, or any information storage and retrieval system, without permission in writing from the publisher. Permission for printing and for reprinting the material contained herein has been obtained by the publisher. Crop Sci 50:524–527
Davis SJ (2002) Photoperiodism: the coincidental perception of the season. Curr Biol 12:R841–R843
Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127:1309–1321
Dong Y, Yang X, Liu J, Wang BH, Liu BL, Wang YZ (2014) Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat Commun 5:3352
Doyle JJ (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18:4679–4688
Gepts P (2010) Crop domestication as a long-term selection experiment. Plant Breeding Reviews. Wiley, pp 1–44
Grant D, Nelson RT, Cannon SB, Shoemaker RC (2010) SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res 38:D843–D846
Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422:719–722
Jiang B, Nan H, Gao Y, Tang L, Yue Y, Lu S, Ma L, Cao D, Sun S, Wang J, Wu C, Yuan X, Hou W, Kong F, Han T, Liu B (2014) Allelic combinations of soybean maturity loci E1, E2, E3 and E4 result in diversity of maturity and adaptation to different latitudes. PLoS One 9:e106042
Kim HK, Kang ST, Oh KW (2006) Mapping of putative quantitative trait loci controlling the total oligosaccharide and sucrose content of Glycine max seeds. J Plant Res 119:533–538
Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286:1960–1962
Kong F, Nan H, Cao D, Li Y, Wu F, Wang J, Lu S, Yuan X, Cober ER, Abe J, Liu B (2014) A new dominant gene E9 conditions early flowering and maturity in soybean. Crop Sci 54:2529–2535
Lee YG, Jeong N, Kim JH, Lee K, Kim KH, Pirani A, Ha BK, Kang ST, Park BS, Moon JK, Kim N, Jeong SC (2015) Development, validation and genetic analysis of a large soybean SNP genotyping array. Plant J 81:625–636
Liu B, Kanazawa A, Matsumura H, Takahashi R, Harada K, Abe J (2008) Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 180:995–1007
Masuda T, Goldsmith P (2009) World soybean production: area harvested, yield, and long-term projections. International Food and Agribusiness Management Review 12
McBlain BA, Bernard RL (1987) A new gene affecting the time of flowering and maturity in soybeans. J Hered 78:160–162
Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11:949–956
Nakamichi N (2015) Adaptation to the local environment by modifications of the photoperiod response in crops. Plant Cell Physiol 56:594–604
Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285:1579–1582
Peng FY, Hu Z, Yang RC (2015) Genome-wide comparative analysis of flowering-related genes in Arabidopsis, wheat, and barley. Int J Plant Genomics 2015:874361
Ray JD, Hinson K, Mankono JEB, Malo MF (1995) Genetic control of a long-juvenile trait in soybean. Crop Sci 35:1001–1006
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Sedivy EJ, Wu F, Hanzawa Y (2017) Soybean domestication: the origin, genetic architecture and molecular bases. New Phytol 214:539–553
Sharma N, Xin R, Kim DH, Sung S, Lange T, Huq E (2016) NO FLOWERING IN SHORT DAY (NFL) is a bHLH transcription factor that promotes flowering specifically under short-day conditions in Arabidopsis. Development 143:682–690
Sun L, Miao Z, Cai C, Zhang D, Zhao M, Wu Y, Zhang X, Swarm SA, Zhou L, Zhang ZJ, Nelson RL, Ma J (2015) GmHs1-1, encoding a calcineurin-like protein, controls hard-seededness in soybean. Nat Genet 47:939–943
Tian Z, Wang X, Lee R, Li Y, Specht JE, Nelson RL, McClean PE, Qiu L, Ma J (2010) Artificial selection for determinate growth habit in soybean. Proc Natl Acad Sci U S A 107:8563–8568
Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59:573–594
Van Ooijen JW (2006) JoinMap(r) 4, Software for the calculation of genetic linkage maps in experimental population. In: B.V. K (ed), Wageningen, Netherlands
Van Ooijen JW (2009) MapQTL(r) 6, Software for the mapping of quantitative trait loci in experimental populations of diploid species. In: B.V. K (ed), Wageningen, Netherlands
Wang C, Chen Y, Ku L, Wang T, Sun Z, Cheng F, Wu L (2010) Mapping QTL associated with photoperiod sensitivity and assessing the importance of QTLxenvironment interaction for flowering time in maize. PLoS One 5:e14068
Wang Y, Gu Y, Gao H, Qiu L, Chang R, Chen S, He C (2016) Molecular and geographic evolutionary support for the essential role of GIGANTEAa in soybean domestication of flowering time. BMC Evol Biol 16:79
Watanabe S, Hideshima R, Xia Z, Tsubokura Y, Sato S, Nakamoto Y, Yamanaka N, Takahashi R, Ishimoto M, Anai T, Tabata S, Harada K (2009) Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 182:1251–1262
Watanabe S, Xia Z, Hideshima R, Tsubokura Y, Sato S, Yamanaka N, Takahashi R, Anai T, Tabata S, Kitamura K, Harada K (2011) A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188:395–407
Funding
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through Agri-Bio industry Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (116141-01), and the Next-Generation BioGreen 21 Program (No. PJ0132132018), Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
Key Message
Three nonsynonymous SNPs on E2 could contribute topological distribution of photoperiod-insensitive early-maturing soybean varieties in Korea peninsula.
Electronic supplementary material
ESM 1
(DOCX 28 kb)
Rights and permissions
About this article
Cite this article
Kim, SK., Kim, ES., Kim, K.H. et al. Genetic variance for flowering time conferring E2 gene in photoperiod-insensitive early-maturing soybean accessions and topological distribution in Korea peninsula. Mol Breeding 38, 148 (2018). https://doi.org/10.1007/s11032-018-0887-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-018-0887-x