Abstract
Soybean [Glycine max (L.) Merr.] is an important oilseed crop which produces about 30 % of the world’s edible vegetable oil. The quality of soybean oil is determined by its fatty acid composition. Soybean oil high in oleic and low in linolenic fatty acids is desirable for human consumption and other uses. The objectives of this study were to identify quantitative trait loci (QTLs) for unsaturated fatty acids and to evaluate the genetic effects of single QTL and QTL combinations in soybean. A population of recombinant inbred lines derived from the cross of SD02-4-59 × A02-381100 was evaluated for fatty acid content in seven environments. In total, 516 polymorphic single nucleotide polymorphism markers, 477 polymorphic simple sequence repeat markers and three GmFAD3 genes were used to genotype the mapping population. By using the composite interval mapping and/or the interval mapping method, a total of 15 QTLs for the three unsaturated fatty acids were detected in more than two environments. Two QTLs for oleic acid on linkage groups G [chromosome (Chr) 18] (qOLE-G) and J (Chr 16) (qOLE-J), three QTLs for linoleic acid on linkage groups A1 (Chr 5) (qLLE-A1) and G (Chr 18) (qLLE-G-1 and qLLE-G-2), and five QTLs for linolenic acid on linkage groups C2 (Chr 6), D1a (Chr 1), D1b (Chr 2), F (Chr 13) and G (Chr 18) were consistently detected in at least three individual environments and the average data over all environments. Significant QTL × QTL interactions were not detected. However, significant QTL × environment interactions were detected for all the QTLs which were repeatedly detected. Some QTLs reported previously were confirmed, and seven new QTLs (two for oleic acid, two for linoleic acid and three for linolenic acid) were identified in this study. Comparisons of two-locus and three-locus combinations indicated that cumulative effects of QTLs were significant for all the three unsaturated fatty acids. QTL pyramiding by molecular marker-assisted breeding would be an appropriate strategy for the improvement of unsaturated fatty acids in soybean.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean [Glycine max (L.) Merr.] is the leading oilseed crop in the United States in terms of gross vegetable oil production and economic importance (Wilcox 2004). The quality and utilization of soybean oil is determined by its fatty acid composition. Different concentrations of a particular fatty acid may play a decisive role in the end use or application of soybean oil. Soybean oil consists predominantly of five fatty acids: palmitic (16:0), stearic (18:0), oleic (18:1), linoleic (18:2) and linolenic (18:3) (Wilson 2004). Palmitic and stearic acids are saturated fatty acids and stable to the oxidation process, but too much intake of them may cause problems like heart cerebrovascular disease and colon and prostate cancer (Hu et al. 1997; Henderson 1991). Oleic, linoleic and linolenic fatty acids are unsaturated fatty acids which may benefit human health. However, polyunsaturated linoleic and linolenic fatty acids are susceptible to the oxidation process, which negatively affects the stability and flavor of the oil (Crapiste et al. 1999). Hydrogenation of polyunsaturated fatty acids increases the oxidative stability and quality of soybean oil. However, hydrogenation may also form trans-fatty acids which are linked to heart disease (Mensink et al. 1994; Willett 1994). Decreasing the polyunsaturated fatty acids (linoleic and linolenic) and increasing the oxidatively stable mono-unsaturated oleic acid through breeding and genetic approaches is an economic and efficient solution to improving soybean oil quality and stability without the need for hydrogenation, thus making the oil more suitable for human consumption (Ha et al. 2010; Mounts et al. 1988; Oliva et al. 2006).
Oil content in soybean is quantitatively inherited (Burton et al. 1983; White et al. 1961), while for fatty acids, in addition to minor genes (Bachlava et al. 2009; Panthee et al. 2006; Wang et al. 2012), there may also be major genes regulating their inheritance (Bilyeu et al. 2006; Bachlava et al. 2009; Pham et al. 2010). In some cases, single major genes or mutant alleles, either transgenic or non-transgenic, can dramatically increase oleic acid or significantly decrease linolenic acid (Bilyeu et al. 2005; Pham et al. 2010). However, in many cases, there are many small or minor effect loci involved in the inheritance of fatty acids. Quantitative trait locus (QTL) analysis of oil content and fatty acids in soybean has been reported (Diers and Shoemaker 1992; Li et al. 2002; Fasoula et al. 2004; Hyten et al. 2004a, b). Hyten et al. (2004b) detected 12 QTLs on six linkage groups for the fatty acids in an F6-derived recombinant inbred line (RIL) population and identified a single marker interval on linkage group L [chromosome (Chr) 19] associated with palmitic, oleic, linoleic and linolenic acids. Panthee et al. (2006) mapped four QTLs on linkage groups E (Chr 15) and G (Chr 18) for oleic, linoleic and linolenic acids in an F6-derived RIL population. Monteros et al. (2008) identified six oleic QTLs on linkage groups A1 (Chr 5), D2 (Chr 17), G (Chr 18) and L (Chr 19) in the F2:3 population of G99-G725 × N00-3350 and confirmed all the QTLs in the F2:3 population of G99-G3438 × N00-3350. Ha et al. (2010) further developed single nucleotide polymorphism (SNP) assays associated with the six oleic QTLs in the F2:3 population of G99-G725 × N00-3350. Bachlava et al. (2009) used a population segregating for oleic acid and a cognate segregating population to confirm the QTLs. Six oleic QTLs were identified in each of the populations, but only two QTLs, one on linkage group F (Chr 13) and one on linkage group I (Chr 20), were consistently detected in both populations. Their study also detected several QTLs for linoleic and linolenic acids. Bilyeu and colleagues also developed DNA markers for the mutant alleles of GmFAD2 and GmFAD3, the microsomal ω–6 and ω–3 fatty acid desaturase genes, respectively, for high oleic and low linolenic acids (Bilyeu et al. 2003, 2005, 2006; Pham et al. 2010). These results are useful for the development of marker-assisted selection or backcrossing to alter fatty acids in soybean.
Although progress in the molecular analysis of unsaturated fatty acids has been made previously, the underlying genetic knowledge of the alteration of unsaturated fatty acids remains incomplete. The QTLs that can be consistently detected across multiple environments and different genetic backgrounds as well as the understanding of the effects of single QTL and QTL combinations is still limited. It is necessary to map and validate QTLs associated with unsaturated fatty acids in different environments and different genetic backgrounds to obtain a better understanding of the genetic basis of differences in fatty acids and to effectively use fatty acid germplasm resources. We have previously reported the QTLs associated with saturated fatty acids in a recombinant inbred population (Wang et al. 2012). The QTLs for unsaturated fatty acids in the same population were also analyzed (data unpublished). To obtain additional phenotypic data and further verify the results/conclusions, the same population was planted again in two locations in 2011. Meanwhile, 340 additional simple sequence repeat (SSR) markers were screened for polymorphism and 179 additional polymorphic markers were genotyped in the population to narrow the putative intervals which had shown associations with unsaturated fatty acids. In total, 516 polymorphic single nucleotide polymorphism (SNP) markers, 477 polymorphic SSR markers and three GmFAD3 genes were used to genotype the mapping population. The objectives of this study were: (1) to identify QTLs for unsaturated fatty acids in this population and confirm previously identified QTLs, (2) to compare the effects of the QTLs for individual loci and locus combinations, and (3) to provide suggestions for QTL selection and pyramiding in practical soybean breeding programs.
Materials and methods
Plant materials and fatty acid analysis
The same population derived from the cross SD02-4-59 × A02-381100 for QTL analysis of saturated fatty acids (Wang et al. 2012) was used for the analysis of unsaturated fatty acids. As discussed previously, the population was small, consisting of 87 F5-derived RILs, and the parents were not included due to lack or loss of seeds. To diminish the impact of this defect and enhance the reliability of the results, the population was planted in multiple years and locations. In addition to the previous five tests (Wang et al. 2012), the field experiments were also conducted again in two locations in 2011, and thus the phenotyping was performed in seven environments in total. Except for the experiments with single-row plots and one replication at Aurora, SD, USA in 2007 and 2008 (designated as E07 and E08, respectively), all the RILs were planted in two-row plots with two replications in a randomized complete block design at Aurora, SD in 2009 and 2010 (E09AU and E10AU), Beresford, SD in 2009 and 2011 (E09BF and E11BF), and Volga, SD in 2011 (E11VG). Plots were planted in rows 4.4 m long with 0.8 m row spacing, and the seeding rate was 26 seeds m−1.
Five-seed samples were taken at random from the combined seeds for each plot. The seed fatty acid content was determined using gas chromatography. The analysis was performed in the USDA/ARS laboratory in Peoria, IL, USA for experiments E07 and E08; and in the Iowa State University DNA Facility in Ames, IA, USA for E09AU, E09BF, E10AU, E11VG and E11BF.
Molecular markers and linkage map construction
Based on the map we constructed (Wang et al. 2012) and the preliminary results of QTL analysis of unsaturated fatty acids, 340 additional SSR markers were ordered and screened for polymorphism to narrow the putative intervals which showed associations with unsaturated fatty acids. In total, 516 polymorphic SNP markers and 477 polymorphic SSR markers were used to genotype the population. In order to detect the presence and effects of the alleles of three GmFAD3 genes in the population which might be derived from the parental line A02-381100 developed by Dr. Walter R. Fehr in Iowa State University, the DNA samples of all the 87 RILs and the parent A02-381100 were sent to Dr. Kristin Bilyeu’s Laboratory in USDA-ARS and genotyped there for the genes GmFAD3A, GmFAD3B and GmFAD3C (Bilyeu et al. 2006).
The genotypic data of all DNA markers and the three GmFAD3 genes were used to construct the genetic linkage map by using JoinMap version 3.0 (Van Ooijen and Voorrips 2001). Seventy-eight markers which exhibited significant segregation distortion (i.e. significant at P = 0.01) and 205 markers for which data were missing in more than 10 lines were excluded from the map construction (Wang et al. 2012). A logarithm of odds (LOD) score of 3.0 was set as the threshold value for linkage grouping. Finally, a total of 311 SNP and 399 SSR markers as well as the three GmFAD3 genes were mapped on 28 linkage groups. The new linkage map covered all the 20 chromosomes in soybean and spanned a total length of 2,099.9 cM with an average interval length of 3.2 cM. Due to lack of polymorphic markers in the related regions, four of the linkage groups, C1 (Chr 4), F (Chr 13), H (Chr 12) and K (Chr 9), were split into two unlinked sub-groups, and two linkage groups, J (Chr 16) and O (Chr 10), involved three unlinked sub-groups.
QTL mapping and statistical analysis
Analysis of variance (ANOVA) was performed to determine the significance of genotypic differences between the RILs with experiments/environments E09AU, E09BF, E10AU, E11VG and E11BF, except for E07 and E08 with only one replication. Heritability was estimated on the line mean basis as h 2 = σ 2g /[σ 2g + (σ 2ge /e) + (σ 2/re)], where h 2 represents the heritability, σ 2g is genotypic variance, σ 2ge is genotype × environment interaction variance, σ 2 is error variance, r is number of replications and e is number of environments (Fehr 1987).
QTL analysis was conducted in WinQTLCart version 2.5 (Wang et al. 2005) for each environment and the average data over seven environments. Single marker analysis (SMA), interval mapping (IM), and composite interval mapping (CIM) were performed. Multiple interval mapping (MIM) was performed to detect epistatic interactions between the QTLs detected. By treating the data from individual experiments as separate traits, multiple-trait IM or CIM analysis was performed to detect the QTL × environment interaction. According to the permutation tests (performed 1,000 times at α = 0.05 for experiment-wise Type I error) and referring to the empirical threshold values widely used for QTL mapping (Bachlava et al. 2009; Jiang et al. 2007; Panthee et al. 2006; Tucker et al. 2010; Winter et al. 2007; Yue et al. 2001), a LOD value of 2.5 was set as the threshold for significance of a QTL. In few cases, the QTLs with a LOD value above 2.0 (equivalently P = 0.002 and significant at P < 0.01 for ANOVA) that were detected in at least two environments and by at least two methods were also declared significant (Concibido et al. 1997; Cornelious et al. 2005; Li et al. 2002; Winter et al. 2007; Yesudas et al. 2013). However, the QTLs that could be detected in only one environment were not declared significant. Therefore, only the QTLs which were detected by CIM and/or IM as well as SMA in at least two individual environments and also confirmed by ANOVA are presented in this paper. To confirm or validate the results of QTL Cartographer (Wang et al. 2005), QTL analysis was also performed by inclusive interval mapping (IIM) and inclusive composite interval mapping (ICIM) in the software QTL IciMapping version 3.1 (Wang et al. 2011).
To verify the validation of QTLs and to provide information for marker-assisted selection (MAS), a comparison between two groups of RILs carrying different marker alleles from the parents was conducted based on ANOVA. Likewise, a comparison of different QTL/marker combinations for multiple loci was also computed based on the results of ANOVA (Jiang et al. 2007).
Results
ANOVA results showed that the differences between RILs were highly significant for all three unsaturated fatty acids (P < 0.01) (Table 1). The environmental differences and genotype × environment interaction effects were also highly significant (P < 0.01). The range of variation in the population was quite large for each of the traits, indicating that the population was appropriate for QTL analysis. Of the three unsaturated fatty acids, linolenic exhibited the highest level of heritability. A relatively high estimate of heritability was also obtained for oleic and linoleic acids.
QTL mapping and analysis
The results of QTL analysis for unsaturated fatty acids exhibited a high consistency between the software QTL Cartographer and QTL IciMapping in detecting the number of QTLs, in spite of slight differences in the magnitude of LOD, genetic effects and the phenotypic variance explained. Therefore, only the results from QTL Cartographer are presented here.
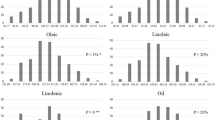
Six QTLs for oleic acid were identified on linkage groups E (Chr 15), G (Chr 18), I (Chr 20), J (Chr 16) and O (Chr 10) in two to four individual environments and/or the combined data (average over all seven environments) by CIM and/or IM analyses (Table 2). The linkage groups and locations of the detected QTLs are presented in Fig. 1. Two QTLs on linkage group E (Chr 15) (qOLE-E-1 and qOLE-E-2) explained 13.2–14.9 and 13.4–19.0 % of the phenotypic variation, respectively. The QTL on linkage group G (Chr 18) was consistently detected in three individual environments and the combined data, explaining 13.0–19.7 % of the phenotypic variation. The QTL on linkage group J (Chr 16) was repeatedly detected in four environments and the combined data, explaining 10.8–14.7 % of the total variation. The QTLs on linkage groups I (Chr 20) and O (Chr 10) were each detected in two single environments, but were not detected in the combined data. All the favorable alleles of the QTLs (increasing oleic acid) were inherited from the parent SD02-4-59, except qOLE-G with the favorable allele derived from the parent A02-381100.
For linoleic acid, four QTLs were detected in multiple environments and/or the combined average data by CIM and/or IM, and mapped on linkage groups A1 (Chr 5), G (Chr 18) and O (Chr 10) (Table 2; Fig. 1). On linkage group G (Chr 18), two QTLs (qLLE-G-1 and qLLE-G-2) with different sources of favorable alleles (decreasing linoleic acid) were consistently detected in four or five individual environments and the combined data. These two QTLs explained 10.2–22.6 and 8.5–29.1 % of the total variation, respectively. On linkage group A1 (Chr 5), a QTL (qLLE-A1) was consistently detected in five individual environments and the combined data, accounting for 7.6–17.6 % of the phenotypic variation. The QTL on linkage group O (Chr 10) (qLLE-O) was detected in three individual environments, explaining 26.1–38.6 % of the total variation.
For linolenic acid, five QTLs were detected by CIM analysis and/or by IM in all individual environments and in the combined data (Table 2). These QTLs were mapped on linkage groups C2 (Chr 6), D1a (Chr 1), D1b (Chr 2), F (Chr 13) and G (Chr 18) (Fig. 1). Two major QTLs (qLLN-D1b and qLLN-G) with the same source of favorable alleles (decreasing linolenic acid) were consistently detected not only in all the individual experiments and combined data but also by all the methods used, and they explained 34.5–43.4 and 5.6–18.7 % of the phenotypic variation, respectively. These two QTLs coincided with the GmFAD3B and GmFAD3C genes. The QTLs on linkage group F (Chr 13) (qLLN-F) and linkage group C2 (Chr 6) (qLLN-C2) were not detected by CIM, but were consistently detected by IM and IIM and confirmed by ANOVA with all the single-experiment data and combined data, accounting for 15.6–19.3 and 12.9–20.4 % of the variation, respectively. The QTL on linkage group D1a (Chr 1) (qLLN-D1a) was also detected by IM and IIM in all individual environments and the average data, accounting for 11.2–22.7 % of the total variation. The QTLs qLLN-C2, qLLN-D1a, qLLN-D1b and qLLN-G had the same source of favorable alleles for decreasing linolenic content, which were derived from the parental line A02-381100. However, the favorable allele of aLLN-F was derived from the SD02-4-59 parent.
QTL × QTL and QTL × environment interactions
Multiple interval mapping (MIM) and inclusive composite interval mapping (ICIM) analysis indicated that no significant QTL × QTL interactions or epistatic effects were detected between the QTLs for the unsaturated fatty acids as described above. However, significant QTL × environment interactions were detected by multiple trait CIM analysis (MT-CIM) for all the QTLs (Table 2), which were highly consistent with the results of ANOVA. For the unsaturated fatty acid QTLs, the additive effects or the proportions of phenotypic variation explained fluctuated over different environments, but they were still consistently identified in multiple environments.
Comparison of single QTL alleles
The results of ANOVA for group comparison of QTLs/markers over seven environments indicated that the differences between two alternative alleles were highly significant for all the unsaturated fatty acids investigated. The averages of alternative alleles of QTLs associated with unsaturated fatty acids and the differences between them are listed in Table 3. For oleic acid, the favorable alleles (increasing oleic content) of the QTLs on linkage groups E (Chr 15), I (Chr 20), J (Chr 16) and O (Chr 10) were inherited from the parent SD02-4-59, while the favorable allele of the QTL on linkage group G (Chr 18) was derived from the parent A02-381100. All the favorable alleles increased oleic acid content significantly (P < 0.001, except qOLE-O with P < 0.05). Over all the environments, the two major QTLs qOLE-E-1 and qOLE-G exhibited larger effects than the other QTLs (Table 3).
For linoleic acid, linkage group G (Chr 18) carried two QTLs with different sources of favorable alleles (decreasing linoleic content) and both QTLs showed larger effects than other QTLs. The favorable allele of the QTL on linkage group A1 (Chr 5) was inherited from A02-381100, while the favorable allele for qLLE-O was derived from SD02-4-59. All the favorable alleles significantly decreased linoleic acid content (P < 0.001).
For the QTLs associated with linolenic acid, the favorable allele (decreasing linolenic content) of qLLN-F was derived from the parent SD02-4-59, while the favorable alleles for other QTLs were inherited from the parent A02-381100. Among all the QTLs detected for linolenic acid, the QTL on linkage group D1b (Chr 2) (qLLN-D1b) showed the largest effect. The QTLs on linkage group F (Chr 13) and G (Chr 18) exhibited effects of similar magnitudes, but in reverse directions.
Cumulative effects of QTL combinations for multiple loci
The results of two-locus comparisons showed that the mean values of the RILs carrying favorable alleles at both loci were higher for oleic acid, or lower for linoleic and linolenic acids, than those of the reciprocal genotypes in all cases (P < 0.05), and in most cases exhibited more desired fatty acid levels than those of RILs in which only one locus carried favorable alleles (Table 4). Likewise, the results of three-locus comparisons indicated that, for the same three-locus combination, the RILs carrying favorable alleles at all three loci generally exhibited better performance for the traits (higher for oleic acid, and lower for linoleic and linolenic acids) than the RILs carrying favorable alleles at only two loci, though the differences were not significant in some cases (Table 4).
Among all the two-locus combinations, the RILs bearing the favorable alleles at the QTL combinations of qOLE-G + qOLE-E-1, qOLE-G + qOLE-E-2, qOLE-G + qOLE-I, qOLE-E-1 + qOLE-I and qOLE-E-2 + qOLE-I for oleic acid, qLLE-G-1 + qLLE-A1 and qLLE-G-1 + qLLE-G-2 for linoleic acid, and qLLN-D1b + qLLN-G, qLLN-D1b + qLLN-F, qLLN-D1b + qLLN-D1a and qLLN-D1b + qLLN-C2 for linolenic acid were more desirable, leading to better performances of the traits, i.e. higher oleic acid, lower linoleic acid and lower linolenic acid, respectively. Among all the three-locus combinations, the RILs bearing the favorable alleles at qOLE-G + qOLE-E-1 + qOLE-E-2, qOLE-G + qOLE-E-1 + qOLE-I and qOLE-E-1 + qOLE-J + qOLE-I for oleic acid, qLLE-G-1 + qLLE-G-2 + qLLE-A1and qLLE-G-1 + qLLE-G-2 + qLLE-O for linoleic acid, and qLLN-D1b + qLLN-G + qLLN-F, qLLN-D1b + qLLN-G + qLLN-D1a and qLLN-D1b + qLLN-G + qLLN-C2 for linolenic acid were the best, resulting in the best performances of the traits.
Discussion
Comparison of QTLs detected in different studies
In this study, six QTLs were identified that were associated with oleic acid in multiple environments. Four of these QTLs were consistent with or similar to those reported previously. Panthee et al. (2006) identified an oleic QTL close to the marker Satt263 on linkage group E (Chr 15) and Bachlava et al. (2009) detected a QTL in the interval of Sat_273–Satt651 on linkage group E (Chr 15) in the FAF population (N97-3363-3 × PI 423893). In our study, two QTLs on linkage group E (Chr 15) in the marker intervals Satt606–BARCSOYSSR_15_0673 (qOLE-E-1) and Satt452–BARCSOYSSR_15_1262 (qOLE-E-2) were detected in two individual environments and in the combined data. Two-locus combination analysis showed that there were significant cumulative effects for these two QTLs (Table 4), indicating that they were different from or independent of each other even though they were located in close proximity to each other (about 9 cM apart for the peak markers). Referring to the public genetic map (Song et al. 2010, supplementary Table 1), the position of marker Satt263 is at 75.7 cM, and the interval of Sat_273–Satt651 is at 33.3–48.1 cM, while the interval of qOLE-E-1 is at 70.5–71.0 cM, and the interval of qOLE-E-2 is at 76.9–79.0 cM. Thus, the interval of these two QTLs detected in our study is approximately 1 or 5 cM away from the QTL detected by Panthee et al. (2006), but far away (>20 cM) from the QTL reported by Bachlava et al. (2009). This suggests that qOLE-E-2 is similar to or the same as the QTL reported by Panthee et al. (2006), while qOLE-E-1 should be a new QTL for oleic acid. Monteros et al. (2008) mapped two QTLs on linkage group G (Chr 18), with peaks at markers Satt394 and Satt191 (64 cM apart). Bachlava et al. (2009) detected a QTL on this linkage group in the marker interval Satt288–Satt472. Reinprecht et al. (2006) also found the SSR markers Satt136 and Satt288 to be associated with oleic acid. The genetic map (Song et al. 2010) indicates that the interval for qOLE-G detected in our study covered the two adjacent markers Satt191 and Satt472. Therefore, we suggest that qOLE-G is similar to or the same as previously reported QTL associated with markers Satt191 and Satt472 (Monteros et al. 2008; Bachlava et al. 2009). Bachlava et al. (2009) also reported QTLs on linkage groups I (Chr 20) and O (Chr 10) for oleic acid, and confirmed the QTL associated with marker Satt153 on linkage group O (Chr 10) identified by Monteros et al. (2008). In our study, the QTL on linkage group I (Chr 20) shared the same marker, Satt354, and the marker interval Satt243–BARC-050013-09288 for qOLE-O was partly covered by the interval Sat_108–Satt 153. Therefore, our results further confirmed the QTLs on linkage group I (Chr 20) and O (Chr 10) reported previously (Bachlava et al. 2009; Monteros et al. 2008). Diers and Shoemaker (1992) used restriction fragment length polymorphism (RFLP) markers to identify QTLs for fatty acids in a population of A81-356022 × PI 468916. They mapped three QTLs associated with oleic acid on linkage groups B, A and J, respectively. Since no common markers were used, our QTL could not be directly compared with their results. Thus, it seems that the QTL on linkage group J (Chr 16) (qOLE-J) detected in this study might be a new one for oleic acid.
Linoleic content in soybean oil is easily affected by environment (Wolf et al. 1982). In this study, however, four QTLs for linoleic acid were repeatedly detected in at least three individual experiments and/or in the combined data. The QTL qLLE-G-2 on linkage group G (Chr 18) was located adjacent to the marker interval of the QTL detected with the population N98-4445A × PI 423893 (Bachlava et al. 2009). However, their genetic relationship or allelism cannot be determined as there are no common parental sources in the pedigrees between these two populations. The peak marker for another QTL on linkage group G (Chr 18) (qLLE-G-1) was GmFAD3C, which was the same as the peak marker for a QTL for linolenic acid, indicating that this QTL/gene might have pleiotropic effects. In this study, two QTLs for linoleic acid were detected on linkage groups A1 (Chr 5) (qLLE-A1) and O (Chr 10) (qLLE-O). Referring to the genetic map (Song et al. 2010), the markers/intervals for qLLE-A1 and qLLE-O were located far from, and as such these two QTLs were different from, the QTLs identified by Monteros et al. (2008) and Bachlava et al. (2009). Thus, it is likely that the QTLs qLLE-A1 and qLLE-O identified in this study are new ones for linoleic acid.
Linolenic was the most stable trait of the three unsaturated fatty acids in this population and it also exhibited the highest estimate of heritability. Five QTLs were mapped on linkage groups C2 (Chr 6), D1a (Chr 1), D1b (Chr 2), F (Chr 13) and G (Chr 18) in all seven environments and in the overall mean data. Previous studies indicated that there were three versions of GmFAD3 genes which together could reduce linolenic acid content to as low as 1 % (Bilyeu et al. 2003, 2005, 2006). In order to determine the presence and potential effects of the three GmFAD3 genes in the population, the genes GmFAD3A, GmFAD3B and GmFAD3C (Bilyeu et al. 2006) were also genotyped in all the RILs along with the low linolenic parental line A02-381100. The results of QTL analysis showed that the QTLs on linkage groups D1b (Chr 2) (qLLN-D1b) and G (Chr 18) (qLLN-G) coincided with GmFAD3B and GmFAD3C, respectively, indicating that these two QTLs should be the same or similar allelic versions of the GmFAD3B and GmFAD3C genes. However, no QTL associated with linolenic acid on linkage group B2 (Chr 14) was detected by any method and in any environment. ANOVA-based group comparison for GmFAD3A indicated that the differences between the two alternative alleles were significant at P = 0.05 but not significant at P = 0.01 in most cases, with a difference of 0.9 % averaged over seven environments. The results indicated that the GmFAD3A locus did not play as important a role as GmFAD3B or GmFAD3C in this population, although it has been reported to contribute large reductions of linolenic acid in other cases (Bilyeu et al. 2006). No other marker on linkage group B2 (Chr 14) was significantly associated with linolenic acid, though GmFAD3A was significant at P = 0.05 in all cases but exhibited small effects. In our study, the GmFAD3B locus, followed by the GmFAD3C locus, showed the largest effects among all the QTLs detected, indicating that the genetic basis of low linolenic content in this population was not completely the same as that in the lines A29 and IA3017 (Bilyeu et al. 2006), in which all three GmFAD3 loci impacted linolenic level with GmFAD3A exhibiting the largest effect. In addition, three QTLs on linkage groups C2 (Chr 6), D1a (Chr 1) and F (Chr 13) were consistently detected in all environments and in the combined data. The major QTL qLLN-F was not detected by CIM, but showed a consistent association with linolenic acid by IM and IIM across all the environments (R 2 = 15.6–19.3 %). The significant effects of this QTL were further confirmed by group comparison (Table 3). Hyten et al. (2004b) reported a QTL with R 2 = 8.1 % at the position of 11.4 cM with the closest marker Satt269 on linkage group F (Chr 13) on the integrated soybean genetic map (Song et al. 2004). In their study, the QTL resulted in only a small change in linolenic acid concentration (1.9 g/kg), showing the smallest effect among all three QTLs for linolenic acid. In this study, qLLN-F alone reduced linolenic acid by about 1.2 % (i.e. 11.9 g/kg) (Table 3), close to the effect of the major gene GmFAD3C. Referring to the integrated soybean genetic map version 2004 and version 2010 (Song et al. 2004, 2010), the closest marker to qLLN-F, Sat_039, is at 27.9 and 11.6 cM, respectively, while the marker Satt269 closely associated with the QTL reported by Hyten et al. (2004a, b) is at 11.4 and 27.5 cM, respectively. This indicates that these two QTLs not only exhibited significantly different effects but were also located about 16 cM apart. Therefore, we speculate that qLLN-F detected in this study might be a new QTL for linolenic acid. No QTL on linkage groups C2 (Chr 6) or D1a (Chr 1) has been reported, suggesting that qLLN-C2 and qLLN-D1a detected in this study are also two novel QTLs for linolenic acid.
In general, the magnitude of phenotypic variation explained by a QTL is affected by several factors, such as the number of QTLs detected, the degree of association of the genetic markers being assayed with the genes controlling the trait, the range of variation within the population, and the population size. It also varies with genetic background and the environment. For QTL mapping, large populations are more effective in detecting QTLs, especially for minor-effect QTLs. In practice, the sample sizes varied considerably, from 60 to 380 (Melchinger et al. 2000). If the results can be repeatedly verified in multiple environments, a small population with appropriate variation should be considered effective (Cornelious et al. 2005; Yesudas et al. 2013). In this study, the population was small, consisting of 87 F5-derived RILs, and might be less effective in detecting minor-effect QTLs than a larger population. However, phenotyping was conducted in seven independent environments, and most of the QTLs identified were repeatedly or consistently detected in three individual environments and in the combined data over all environments. Therefore, the results are reliable because confirmation from multiple environments increases the reliability. The more environments a QTL is confirmed in, the less the possibility of Type I error would be. Our study also confirmed some previously identified QTLs for unsaturated fatty acids. It is noted that the proportions of total variation explained by single QTLs were close to or above 10 % in many cases. It is possible that the genetic effects of these QTLs were overestimated due to the small population size (Beavis 1994). In other words, one should not expect that similar high effects would be exhibited when the QTLs are used in practical breeding, especially with larger populations. However, the QTLs which were detected in multiple environments and/or validated in different studies may be used in practical breeding for the improvement of fatty acids. The QTLs may also be used as alternatives to diversify the genetic basis of soybean improvement, although large changes in fatty acid composition can now be achieved through combinations of mutant GmFAD2 and GmFAD3 genes (Pham et al. 2012). For those QTLs which could only be detected in a few single environments, confirmation and validation are needed.
QTL × QTL (epistasis) and QTL × environment interactions
In genetic studies on quantitatively inherited traits, epistasis or QTL × QTL interaction has received increased attention. Significant epistatic interactions have been reported in soybean for plant height (Lark et al. 1995), seed yield (Orf et al. 1999) and isoflavone content (Primomo et al. 2005). However, there is very little knowledge of epistasis for fatty acids in soybean (Wang et al. 2012). In this study, no significant QTL × QTL interaction was detected for any of the unsaturated fatty acids. This might be attributed in part to limited population size and the limited number of individuals or lines in certain genotypic classes (Tanksley 1993). Therefore, the possibility of QTL × QTL interaction cannot be completely ruled out until further investigation has been done with larger populations, because large populations are more effective. On the other hand, it indicates that the unsaturated fatty acids in this population were predominantly inherited in an additive or additive-dominance mode.
Significant genotype × environment interactions were detected for all the unsaturated fatty acids by ANOVA and multiple-trait CIM or IM analysis. The genetic effects of the QTLs fluctuated between different environments. This is in agreement with the general understanding of inheritance of quantitative traits, i.e. the expression of genes/QTLs controlling quantitatively inherited characteristics is easily affected by environmental conditions, even for the traits with high heritability. It is necessary to evaluate quantitative traits in multiple environments for QTL mapping. In this study, the multiple-trait CIM or IM analysis, which was highly consistent with the ANOVA analysis, showed that linolenic acid levels were most stable across environments. This is a result similar to that reported by Oliva et al. (2006), who determined the stability across seed filling temperature environments of unsaturated fatty acid levels in a set of 17 soybean genotypes with varying unsaturated fatty acid genotypes. Six QTLs for oleic acid, four QTLs for linoleic acid and five QTLs for linolenic acid were consistently detected in at least two environments and in the average data, and were also confirmed by ANOVA, even though the genetic effects of these QTLs were significantly affected by environments. The QTL × environment interaction may have two forms of expression. One form occurs when a QTL has significant influence on the trait or the mean of a population in some environments, but not in others. For the QTLs with such QTL × environment interaction, a breeder might not predict when the QTL will be effective or not. Multiple-environment tests will be helpful because the expression of a gene or QTL also depends on favorable environmental conditions, which is why multiple-environment tests are necessary for quantitatively inherited traits like yield. The other form occurs when a QTL exhibits significant effects in all environments but the magnitudes of the effect are significantly different. For instance, the QTLs for linolenic acid identified in this study were significant in all environments while the genetic effects fluctuated obviously with the environments (Table 2). For this kind of QTL, it is relatively easy to use and predict the effects of selection in practical breeding. Therefore, we suggest that these QTLs could be used in marker-assisted breeding.
QTL pyramiding for improvement of unsaturated fatty acids in breeding practice
Pyramiding of multiple genes or QTLs is a prospective strategy for enhancing or improving a quantitatively inherited trait in plant breeding. The cumulative effects of QTL pyramiding have been proven previously in soybean (Li et al. 2010; Njiti et al. 2001, 2002) and in other crops (Huang et al. 1997; Jiang et al. 2007; Richardson et al. 2006). Li et al. (2010) analyzed Phytophthora root rot tolerance in a RIL population and evaluated the cumulative effects of the detected QTL. Their results indicated that the plant tolerance to Phytophthora root rot was correlated with the number of loci carrying resistance alleles. The cumulative effects of QTL pyramiding for saturated fatty acids in soybean were also detected in our previous work (Wang et al. 2012). In the present study, the cumulative effects of QTL were determined for all the unsaturated fatty acids. Two-locus QTL comparison results showed that in all cases the RILs carrying favorable alleles at both loci exhibited significantly better performance than those of the reciprocal genotypes, and in most cases their performance was superior to those of RILs with favorable alleles at only one locus. For three-locus QTL pyramiding, the RILs carrying favorable alleles at three loci generally exhibited better performance than the RILs with only two favorable alleles, although the differences were not significant in some cases.
Based on the results of QTL pyramiding analysis, the following QTL combinations could be considered in practical molecular marker-assisted breeding. The two-locus combinations of qOLE-G + qOLE-E-1, qOLE-G + qOLE-E-2, qOLE-G + qOLE-I, qOLE-E-1 + qOLE-I and qOLE-E-2 + qOLE-I and the three-locus combinations of qOLE-G + qOLE-E-1 + qOLE-E-2, qOLE-G + qOLE-E-1 + qOLE-I and qOLE-E-1 + qOLE-J + qOLE-I for oleic acid, the two-locus combinations of qLLE-G-1 + qLLE-A-1 and qLLE-G-1 + qLLE-G-2 and the three-locus combinations of qLLE-G-1 + qLLE-G-2 + qLLE-A1 and qLLE-G-1 + qLLE-G-2 + qLLE-O for linoleic acid, the two-locus combinations of qLLN-D1b + qLLN-G, qLLN-D1b + qLLN-F, qLLN-D1b + qLLN-D1a and qLLN-D1b + qLLN-C2 and the three-locus combinations of qLLN-D1b + qLLN-G + qLLN-F, qLLN-D1b + qLLN-G + qLLN-D1a and qLLN-D1b + qLLN-G + qLLN-C2 for linolenic acid should be the appropriate options for desirable fatty acid profiles in soybeans.
In practical MAS breeding, however, fewer QTLs rather than more are preferred for ease and efficiency in selection. If a breeder can get desirable genotypes by combining fewer QTLs, the selection will be easily performed and the efficiency will be enhanced compared with the use of more QTLs. On the other hand, the efficiency of selection is also dependent upon the degree of influence of QTLs on the trait. Therefore, fewer major QTLs are always desired. For the population in this study, two-QTL combinations might be better in efficiency than three-QTL combinations, because the differences between the desirable two- and three-QTL combinations as described above were small. For instance, the average of the desired two-QTL combinations for oleic acid and linoleic acid was 31.2–31.8 and 53.5–53.6 %, compared to the desired three-QTL combinations (31.8–32.1 and 51.8–53.0 %). The average of linolenic acid content for the desired two-QTL combinations was 1.7–1.9 %, compared with 1.3–1.7 % for the desired three-QTL combinations. This was because two major QTLs were first considered in the combinations and the third QTL had smaller effects. Nevertheless, additional QTLs may still be helpful to some extent for the effect of trait improvement though not for the efficiency. The best five lines for oleic acid (average = 34.7 % with a range of 33.5–36.4 % over seven environments) or for linoleic acid (average = 50.6 % with a range of 49.9–51.4 %) of the population in this study bear favorable alleles at all the six or four loci shown in Table 3, respectively. Likewise, the lowest five lines for linolenic acid (average = 1.3 % with a range of 1.3–1.4 %) bear favorable alleles at all the five linolenic acid loci. This indicates that these lines integrated favorable alleles derived from either of the parents at multiple loci. Thus, not only could they be regarded as supporting evidence for QTL/gene pyramiding, but could be also used as new parents in soybean breeding for improved fatty acids.
References
Bachlava E, Dewey RE, Burton JW, Cardinal AJ (2009) Mapping and comparison of quantitative trait loci for oleic acid seed content in two segregating soybean populations. Crop Sci 49:433–442
Beavis WD (1994) The power and deceit of QTL experiments: lessons from comparative QTL studies. In: 49th annual corn and sorghum industry research conference. American Seed Trade Association, Washington, DC
Bilyeu KD, Palavalli L, Sleper DA, Beuselinck PR (2003) Three microsomal omega-3 fatty-acid desaturase genes contribute to soybean linolenic acid levels. Crop Sci 43:1833–1838
Bilyeu K, Palavalli L, Sleper D, Beuselinck P (2005) Mutations in soybean microsomal omega-3 fatty acid desaturase genes reduce linolenic acid concentration in soybean seeds. Crop Sci 45:1830–1836
Bilyeu K, Palavalli L, Sleper D, Beuselinck P (2006) Molecular genetic resources for development of 1% linolenic acid soybeans. Crop Sci 46:1913–1918
Burton JW, Wilson RF, Brim CA (1983) Recurrent selection in soybeans: IV. Selection for increased oleic acid percentage in seed oil. Crop Sci 23:744–747
Concibido VC, Denny R, Lange DA, Orf JH, Young ND (1997) Genome mapping on soybean cyst nematode resistance genes in ‘Peking’, PI 90763 and PI 88788 using DNA markers. Crop Sci 37:258–264
Cornelious B, Chen P, Chen Y, de Leon N, Shannon JG, Wang D (2005) Identification of QTLs underlying water-logging tolerance in soybean. Mol Breed 16:103–112
Crapiste GH, Brevedan MI, Carelli AA (1999) Oxidation of sunflower oil during storage. J Am Oil Chem Soc 76:1437–1443
Diers BW, Shoemaker RC (1992) Restriction fragment length polymorphism analysis of soybean fatty acid content. J Am Oil Chem Soc 69:1242–1244
Fasoula VA, Harris DK, Boerma HR (2004) Validation and designation of quantitative trait loci for seed protein, seed oil, and seed weight from two soybean populations. Crop Sci 44:1218–1225
Fehr WR (1987) Principles of cultivar development—volume 1, theory and technique. Macmillan Publishing Company, New York
Ha B, Monteros MJ, Boerma HR (2010) Development of SNP assays associated with oleic acid QTL in N00-3350 soybean. Euphytica 176:403–415
Henderson MM (1991) Correlations between fatty acid intake and cancer incidence. In: Nelson GJ (ed) Health effects of dietary fatty acids. AOCS, Champaign, IL, p 136
Hu FB, Stampfer MJ, Manson JE, Rimm E, Colditz GA, Rosner BA, Hennekens CH, Willett WC (1997) Dietary fat intake and the risk of coronary heart disease in women. N Engl J Med 337:1491–1499
Huang N, Angeles ER, Domingo J, Magpantay G, Singh S, Zang G, Kumaravadivel N, Bennet J, Khush GS (1997) Pyramiding of bacterial blight resistance genes in rice: marker-assisted selection using RFLP and PCR. Theor Appl Genet 95:313–320
Hyten DL, Pantalone VR, Sams CE, Saxton AM, Landau-Ellis D, Stefaniak TR, Schmidt ME (2004a) Seed quality QTL in a prominent soybean population. Theor Appl Genet 109:552–561
Hyten DL, Pantalone VR, Saxton AM, Schmidt ME, Sams CE (2004b) Molecular mapping and identification of soybean fatty acid modifier quantitative trait loci. J Am Oil Chem Soc 81:1115–1118
Jiang G-L, Shi JR, Ward RW (2007) QTL analysis of resistance to Fusarium head blight in the novel wheat germplasm CJ 9306. I. Resistance to fungal spread. Theor Appl Genet 116:3–13
Lark KG, Chase K, Adler F, Mansur LM, Orf JH (1995) Interactions between quantitative trait loci in soybean in which trait variation at one locus is conditional upon a specific allele at another. Proc Natl Acad Sci USA 92:4656–4660
Li Z, Wilson RF, Rayford WE, Boerma HR (2002) Molecular mapping genes conditioning reduced palmitic acid content. Crop Sci 42:373–378
Li X, Han Y, Teng W, Zhang S, Yu K, Poysa V, Anderson T, Ding J, Li W (2010) Pyramided QTL underlying tolerance to Phytophthora root rot in mega-environment from soybean cultivars ‘conrad’ and ‘Hefeng 25’. Theor Appl Genet 121:651–658
Melchinger AE, Utz HF, Schon CC (2000) From Mendel to Fisher: the power and limits of QTL mapping for quantitative traits. Vortr Pflanzenzucht 48:132–142
Mensink RP, Temme EH, Hornstra G (1994) Dietary saturated and trans fatty acids and lipoprotein metabolism. Ann Med 26:461–464
Monteros MJ, Burton JW, Boerma HR (2008) Molecular mapping and confirmation of QTL associated with oleic content in N00-3350 soybean. Crop Sci 48:2223–2234
Mounts TL, Warner K, List GR, Kleiman R, Fehr WR, Hammond EG, Wilcox JR (1988) Effect of altered fatty acid composition on soybean oil stability. J Am Oil Chem Soc 65:624–628
Njiti VN, Johnson JE, Torto TA, Gray LE, Lightfoot DA (2001) Inoculum rate influences selection for field resistance to soybean sudden death syndrome in the greenhouse. Crop Sci 41:1726–1731
Njiti VN, Meksem K, Iqbal MJ, Johnson JE, Kassem MA, Zobrist KF, Kilo VY, Lightfoot DA (2002) Common loci underlie field resistance to soybean sudden death syndrome in Forrest, Pyramid, Essex, and Douglas. Theor Appl Genet 104:294–300
Oliva ML, Shannon JG, Sleper DA, Ellersieck MR, Cardinal AJ, Paris RL, Lee JD (2006) Stability of fatty acid profile in soybean genotypes with modified seed oil composition. Crop Sci 46:2069–2075
Orf JH, Chase K, Adler FR, Mansur LM, Lark KG (1999) Genetics of soybean agronomic traits: II. Interaction between yield quantitative trait loci in soybean. Crop Sci 39:1652–1657
Panthee DR, Pantalone VR, Saxton AM (2006) Modifier QTL for fatty acid composition in soybean oil. Euphytica 152:67–73
Pham A-T, Lee JD, Shannon JG, Bilyeu KD (2010) Mutant alleles of FAD2-1A and FAD2-1B combine to produce soybeans with the high oleic acids seed oil trait. BMC Plant Biol 10:195
Pham A-T, Shannon JG, Bilyeu KD (2012) Combinations of mutant FAD2 and FAD3 genes to produce high oleic acid and low linolenic acid soybean oil. Theor Appl Genet 125:503–515
Primomo VS, Poysa VW, Ablett GR, Jackson C, Gijzen M, Rajcan I (2005) Mapping QTL for individual and total isoflavone content in soybean seeds. Crop Sci 45:2454–2464
Reinprecht Y, Poysa VW, Yu K, Rajcan I, Ablett GR, Pauls KP (2006) Seed and agronomic QTL in low linolenic acid, lipoxygenase-free soybean (Glycine max (L.) Merrill) germplasm. Genome 49:1510–1527
Richardson KL, Vales MI, Kling JG, Mundt CC, Hayes PM (2006) Pyramiding and dissecting disease resistance QTL to barley stripe rust. Theor Appl Genet 113:485–495
Song Q, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB (2004) A new integrated genetic linkage map of the soybean. Theor Appl Genet 109:122–128
Song Q, Jia G, Zhu Y, Grant D, Nelson RT, Hwang E-Y, Hyten DL, Cregan PB (2010) Abundance of SSR motifs and development of candidate polymorphic markers (BARCSOYSSR_1.0) in soybean. Crop Sci 50:1950–1960
Tanksley SD (1993) Mapping polygenes. Annu Rev Genet 27:205–233
Tucker DM, Saghai-Maroof MA, Mideros S, Skoneczka JA, Nabati DA, Buss GR, Hoeschele I, Tyler BM, St. Martin SK, Dorrance AE (2010) Mapping quantitative trait loci for partial resistance to Phytophthora sojae in a soybean interspecific cross. Crop Sci 50:628–635
Van Ooijen JW, Voorrips RE (2001) JoinMap 3.0: software for the calculation of genetic linkage maps. Plant Research International, Wageningen, The Netherlands
Wang S, Basten CJ, Zeng ZB (2005) Windows QTL cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC
Wang J, Li H, Zhang L, Li C, Meng L (2011) QTL IciMapping V3.1. Institute of Crop Science and CIMMT China, Chinese Academy of Agriculture Sciences, Beijing, China
Wang X, Jiang G-L, Green M, Scott RA, Hyten DL, Cregan PB (2012) Quantitative trait locus analysis of saturated fatty acids in a population of recombinant inbred lines of soybean. Mol Breed 30:1163–1179
White HB, Quackenbush FW, Probsr AH (1961) Occurrence and inheritance of linolenic and linoleic acids in soybean seeds. J Am Oil Chem Soc 38:113–117
Wilcox JR (2004) World distribution and trade of soybean. In: Boerma RH, Specht JE (eds) Soybeans: improvement, production, and uses, 3rd edn. American Society of Agronomy, Madison, WI, pp 1–13
Willett WC (1994) Diet and health: what should we eat? Science 264:532–537
Wilson RF (2004) Seed composition. In: Boerma RH, Specht JE (eds) Soybeans: improvement, production, and uses, 3rd edn. American Society of Agronomy, Madison, WI, pp 621–669
Winter SMJ, Shelp BJ, Anderson TR, Welacky TW, Rajcan I (2007) QTL associated with horizontal resistance to soybean cyst nematode in Glycine soja PI464925B. Theor Appl Genet 114:461–472
Wolf RB, Cavins JF, Kleiman R, Black LT (1982) Effect of temperature on soybean seed constituents: oil, protein, moisture, fatty acids, amino acids and sugars. J Am Oil Chem Soc 59:230–232
Yesudas CR, Bashir R, Geisler MB, Lightfoot DA (2013) Identification of germplasm with stacked QTL underlying seed traits in an inbred soybean population from cultivars Essex and Forrest. Mol Breed 31:693–703
Yue P, Arelli PR, Sleper DA (2001) Molecular characterization of resistance to Heterodera glycines in soybean PI 438489B. Theor Appl Genet 102:921–928
Acknowledgments
This research was supported in part by USB Seed Composition Research project via USDA/ARS, and South Dakota Soybean Research and Promotion Council project. The determination of fatty acids was completed in the USDA/ARS Lab., in Peoria, IL and the Iowa State University DNA Facility in Ames, IA. Genotyping of three GmFAD3 genes was performed by Dr. Kristin Bilyeu’s Lab., USDA/ARS in Columbia, MO. The parental line A02-381100 was provided by Dr. Walter Fehr, Iowa State University. Dr. Kelly Whiting, the United Soybean Board, reviewed the manuscript and provided constructive suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, X., Jiang, GL., Green, M. et al. Quantitative trait locus analysis of unsaturated fatty acids in a recombinant inbred population of soybean. Mol Breeding 33, 281–296 (2014). https://doi.org/10.1007/s11032-013-9948-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-013-9948-3