Abstract
Using random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), simple sequence repeats (SSR), and morphological traits, the first genetic maps for Cucurbita pepo (2n=2x=40) were constructed and compared. The two mapping populations consisted of 92 F2 individuals each. One map was developed from a cross between an oil-seed pumpkin breeding line and a zucchini accession, into which genes for resistance to Zucchini Yellow Mosaic Virus (ZYMV) from a related species, C. moschata, had been introgressed. The other map was developed from a cross between an oil-seed pumpkin and a crookneck variety. A total of 332 and 323 markers were mapped in the two populations. Markers were distributed in each map over 21 linkage groups and covered an average of 2,200 cM of the C. pepo genome. The two maps had 62 loci in common, which enabled identification of 14 homologous linkage groups. Polyacrylamide gel analyses allowed detection of a high number of markers suitable for mapping, 10% of which were co-dominant RAPD loci. In the Pumpkin-Zucchini population, bulked segregant analysis (BSA) identified seven markers less than 7 cM distant from the locus n, affecting lignification of the seed coat. One of these markers, linked to the recessive hull-less allele (AW11-420), was also found in the Pumpkin-Crookneck population, 4 cM from n. In the Pumpkin-Zucchini population, 24 RAPD markers, previously introduced into C. pepo from C. moschata, were mapped in two linkage groups (13 and 11 markers in LGpz1 and LGpz2, respectively), together with two sequence characterized amplified region (SCAR) markers linked to genes for resistance to ZYMV.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Cucurbita includes some of the oldest domesticated plant species. Three of them, Cucurbita pepo L., Cucurbita maxima Duchesne, and Cucurbita moschata Duchesne, are economically important crop species worldwide (Robinson and Decker-Walters 1997), with C. pepo exhibiting the widest variation, especially with respect to fruit characteristics. Duchesne (1786) and Naudin (1856) concluded that C. pepo L. could be the most polymorphic species in the plant kingdom. Botanical classification based on allozyme variation recognized three subspecies: C. pepo subsp. pepo, C. pepo subsp. ovifera (L.) D.S. Decker [syn. C. pepo subsp. texana (Scheele) Filov], and C. pepo subsp. fraterna (Bailey) Lira, Andres and Nee (Andres 1987; Decker 1988; H. S. Paris personal communication). Using mainly fruit shape, Paris (1986) classified edible-fruited C. pepo into eight cultivar-groups: Pumpkin, Zucchini, Cocozelle, Vegetable Marrow, Acorn, Crookneck, Scallop, and Straightneck. The first four of these cultivar-groups named above belong to subsp. pepo and the latter four to subsp. ovifera (Paris 2001). Polymorphisms of DNA markers, including RFLP, RAPD, ISSR, AFLP, and SRAPD, have been used to study genetic relationships within C. pepo (Torres-Ruiz and Hemleben 1991; Katzir et al. 2000; Paris et al. 2003; Ferriol et al. 2003) and the results support these classifications. Also, the results obtained, using AFLP, ISSR, and SSR were very highly correlated (P < 0.000001).
Genetic maps have been constructed from inter-specific crosses in the genus Cucurbita (Lee et al. 1995; Brown and Myers 2002), and we recently reported the initiation of a map for C. pepo (Zraidi and Lelley 2004). Compared to other economically important Cucurbitaceae, genetic mapping of Cucurbita (2n = 2× = 40) is in its infancy. Mapping efforts in melon, Cucumis melo L., resulted in a large number of DNA-based maps (e.g., Baudracco-Arnas and Pitrat 1996; Wang et al. 1997; Périn et al. 1998; Oliver et al. 2001; Danin-Poleg et al. 2002). Similarly, in cucumber, Cucumis sativus L., several maps have been developed (e.g., Serquen et al. 1997; Park et al. 2000; Fazio et al. 2002). Efforts to anchor, align, and merge maps with SSR, RFLP or STS markers are underway for cucumber (Bradeen et al. 2001), and melon (Gonzalo et al. 2005).
In Austria, C. pepo has been cultivated for centuries primarily for seed oil production (Teppner 2000). Pumpkin seed-oil is a delicious salad oil (Murkovic et al. 1996). It is highly valued for its excellent nutritional quality and medicinal value, especially for preventing and curing benign prostate hyperplasia in its early stage (Blumenthal et al. 1998; Kreuter 2000; Schmidlin and Kreuter 2003), although the chemical agent responsible for this property of the seeds has not been identified yet (Murkovic et al. 2004). A special feature of Austrian oil-pumpkin varieties is their completely hull-less (non-lignified) seed coat (Teppner 2000; Zraidi et al. 2003), which is conferred largely by a single recessive gene, n (naked seed) (Paris and Brown 2005).
The principal objective of the present study was to construct a genetic map for C. pepo, using RAPD, AFLP, SSR, and morphological markers. Co-linear common markers segregating in two F2 populations that mapped to the same linkage group were considered to be locus specific, and were used to align and merge the linkage maps that were constructed. Another objective was the identification of markers tightly linked to the seed coat locus n, believed to be controlled by a single major gene (Grebenščikov 1954; Prym von Becherer 1955; Zraidi et al. 2003). For this bulked segregant analysis (BSA) (Michelmore et al.1991) was used.
Materials and methods
Plant material
Two C. pepo F2 populations, each derived from a single F1 fruit, were used for mapping. The first population, which consisted of 92 plants, is here designated as the Pumpkin–Zucchini population, following the horticultural classification of Paris (1986). This population was derived from an intra-subspecific (C. pepo subsp. pepo) cross between an oil-pumpkin breeding line (SZG1), developed by “Saatzucht Gleisdorf” (Gleisdorf, Austria), and a zucchini accession, “True French Resistant”, a near-isogenic line of the UK zucchini “True French” carrying three complementary dominant genes for resistance to Zucchini Yellow Mosaic Virus (ZYMV). These were introgressed from the Portuguese C. moschata “Menina” (Paris and Cohen 2000). Seeds of “True French Resistant” were obtained from H. S. Paris of the A. R. O. Newe Ya’ar Research Center, Ramat Yishay, Israel. The second population, which also consisted of 92 plants, is here designated as Pumpkin–Crookneck population. This population was derived from an inter-subspecific cross (C. pepo subsp. pepo × C. pepo subsp. ovifera) between the US oil-pumpkin variety “Lady Godiva” and the Italian crookneck variety “Bianco Friulano.”
Ten seeds from each of the 92 selfed F2 plants of the Pumpkin–Zucchini population were sown to produce a segregating F3 progeny. This was used to identify homozygous hull-less (n/n) and homozygous hulled (N/N) F2 genotypes to create contrasting DNA pools for BSA (Michelmore et al. 1991).
Morphological traits
Three qualitative gene loci, M (Mottled leaves), n (naked seeds), and Bu (Bush habit) (Paris and Brown 2005), showed clear segregation in the F2 populations and were scored. The leaf-mottle trait was scored as the presence (M/–) or absence (m/m) of silver patches in the axils of the leaf veins, without regard to the extent or intensity of silver. The seed coat was scored as hulled (N/–) or hull-less (n/n), irrespective of an occasional residual lignification observed in the hull-less seeds (Teppner 2000). The growth habit was scored as plants being either bush (Bu/–) (length of the vine maximum 1.5 m) or vining (bu/bu) (length of vine 2–6 m) types.
DNA extraction
DNA was isolated from fresh young leaves using the Wizard Genomic DNA Purification Kit (Promega Corp., Madison, WI, USA). DNA concentration was determined by the GenQuant RNA/DNA Calculator (Amersham Biosciences Europe, Freiburg, Germany) according to the manufacturer’s instructions.
RAPD markers
A total of 495 10-mer RAPD primers (Operon Technology Inc., Huntsville, USA) were screened for polymorphisms, using DNA-pools of ten plants from each parental genotype. PCR conditions, fragment separation and visualization were conducted as described by Stift et al. (2003), using 10% polyacrylamide gel. Primers producing clear bands, showing polymorphism in both populations, were selected for mapping. Co-dominant RAPD markers were identified by the clear 1:2:1 segregation of the two corresponding fragments and by the complete lack of a recessive allele (Fig. 1).
AFLP markers
The AFLP analysis (Vos et al. 1995) was done as described by Hartl et al. (1999) and Buerstmayr et al. (2002), using MseI and EcoRI enzymes. Selective amplification was performed using 28 EcoRI and MseI primer combinations with two or three randomly chosen selective nucleotides. RAPD and AFLP gels from the two populations were compared and bands, showing identical molecular weights in the two populations, were considered to be common markers.
SSR markers
A screen for potential SSR polymorphisms between the four parental genotypes used in the two crosses was done by J. E. Staub, University of Wisconsin, Madison, USA, using 102 Cucumis-SSR primers (Fazio et al. 2002). Eighteen C. pepo SSR primer pairs were developed by our laboratory, and were tested on the parents of the Pumpkin–Zucchini population.
Bulked segregant analysis (BSA)
Analysis of F3 progenies allowed the identification of N/N homozygotes from the F2 in the Pumpkin–Zucchini population. Two DNA pools were made, each containing a mixture of 12 DNA samples from homozygous dominant (hulled) and homozygous recessive (hull-less) plants for the n gene (Zraidi et al. 2003). A total of 1,068 RAPD markers were screened for polymorphism using the two DNA-bulks, and markers showing association with the n locus were tested on DNA from single plants of the two pools. Ultimately, markers associated with the trait, found through BSA, were used on the entire F2 population to calculate the degree of linkage.
Linkage analysis
Marker segregation was tested for fit to 3:1 (dominant loci) and 1:2:1 (SSR and co-dominant RAPD and AFLP markers) ratios at P = 0.05 probability. Linkage maps were constructed independently for each population using MAPMAKER/EXP 3.0 (Lander et al. 1987; Lincoln et al. 1992). Markers were associated with the group command with a likelihood of LOD ≥5 and with a maximum distance of 30. Marker-ordering within linkage groups was initially performed using the order command with a likelihood of LOD ≥3. Linkage groups containing more than six markers, which could not be ordered with this likelihood level, were ordered manually, using the compare command. The remaining markers were then integrated into these linkage groups, using the try command. Distances were calculated with the Kosambi function (Kosambi 1944). The order of the markers within linkage groups was double checked, using the ripple command. Linkage groups in the Pumpkin–Zucchini map are named LGpz, and those of the Pumpkin–Crookneck map LGpc.
Results
Polymorphism
Of the 495 tested RAPD primers, 227 (46%) and 296 (60%) showed polymorphism between the parents of the Pumpkin–Zucchini and the Pumpkin–Crookneck populations, respectively. Finally, 101 primers were selected for mapping because they showed polymorphism in both populations. These primers produced an average of 15 bands, each ranging in size from 150 to 1,900 bp. The average number of polymorphic loci per primer was 2.6 and 2.7 in the Pumpkin–Zucchini and Pumpkin–Crookneck populations, respectively.
AFLP primer combinations generated up to 60 bands, ranging in size from 50 to 700 bp. The 28 AFLP EcoRI/MseI primer combinations produced 118 polymorphic loci, with an average of 4.2 loci per primer combination, in the Pumpkin–Zucchini and 180 polymorphic loci, with an average of 6.4 loci per primer combination in the Pumpkin–Crookneck population.
None of the 102 Cucumis-SSR markers tested, were polymorphic between the parental genotypes. Of the 18 Cucurbita-SSR markers, five (27.7%) were polymorphic between the mapping parents of the Pumpkin–Zucchini population and three could be mapped in the Pumpkin–Zucchini map in linkage groups LGpz1, LGpz6, and LGpz11 (Fig. 2).
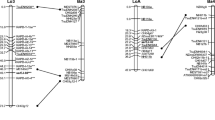
Linkage map of the Pumpkin–Zucchini population. The loci and their size are listed to the right of the bars. To the left are the distances between markers in centiMorgans. RAPD and AFLP are represented by their universal codes. Underlining indicates common markers. Bold font indicates co-dominant markers and italic font indicates that the markers originate from the source of resistance to ZYMV (C. moschata genotype)
Construction and characteristics of the maps
Only linkage groups (major linkage groups) containing more than four markers and including at least one common or one co-dominant marker (Fig. 1) were used in the construction of the two maps (Figs. 2, 3). A number of linkage groups containing four or less markers (minor groups) were at first excluded from the maps. These were eight doublets in the Pumpkin–Zucchini and ten doublets in the Pumpkin–Crookneck map, and two quadruplets and six triplets in each of the two maps. However, some of these linkage groups were later incorporated in the final maps based on the presence of common markers and after the alignment of the two maps. These were two doublets (LGpz14b and LGpz16b) in the Pumpkin–Zucchini map (Fig. 2) and one triplet in each of the two maps (LGpz16a in the Pumpkin–Zucchini map and LGpc13a in the Pumpkin–Crookneck map). Forty-one zucchini markers and 49 crookneck markers were unlinked.
Linkage map of the Pumpkin–Crookneck population. The loci and their size are listed to the right of the bars. To the left are the distances between markers in centiMorgans. RAPD and AFLP are represented by their universal codes. Underlining indicates common markers. Bold font indicates co-dominant markers
A total of 333 markers (247 RAPD, 82 AFLP, 3 SSR, and the n locus), 29 of which were co-dominant (21 RAPDs, 5 AFLPs, and 3 SSRs), were integrated in the Pumpkin–Zucchini map covering 2,140 cM of the genome and distributed over 21 major and 3 minor linkage groups (Fig. 2). The average size of the linkage groups was 90 cM, ranging from 10 cM in LGpz21 to 191 cM in LGpz4. The average number of markers per linkage group was 16 and the average distance between markers was 6.4 cM. Except linkage group LGpz21, all linkage groups contained at least one co-dominant marker, with a maximum of six in LGpz1, or one common marker, with a maximum of 11 in LGpz3 (Fig. 2).
The Pumpkin–Crookneck map contained 323 markers consisting of 196 RAPD markers, of which 20 were co-dominant, 125 AFLP with 12 co-dominant markers, and two gene loci, n on LGpc9 and Bu on LGpc12. This map covers 2,234 cM, distributed over 21 major and 3 minor linkage groups ranging in size from 38 cM in LGpc21 to 176 cM in LGpc3, with an average of 93 cM. Except LGpc20 which, however, contained 17 markers, all other linkage groups contained at least one co-dominant and/or one common marker (Fig. 3). The average number of markers was 15.5 per linkage group, and the distance between markers was 6.9 cM (Fig. 3). The distribution of RAPD and AFLP markers throughout the genome was markedly different (Figs. 2, 3). While the distribution of RAPD markers was relatively even in both maps, AFLP markers tended to cluster in most cases.
Segregation distortions
Chi-square tests (P = 0.05) revealed that the segregation of 20 (4.7%) loci in the Pumpkin–Zucchini and 26 (5.6%) loci in the Pumpkin–Crookneck map were distorted. Markers having distorted segregation were at first excluded from grouping and ordering analyses. They were mapped after creating linkage groups using only markers without segregation distortion. Ten distorted markers in the Pumpkin–Zucchini and 15 in the Pumpkin–Crookneck map were grouped in doublets or triplets and were not included in the final maps. Eleven distorted markers, distributed over five linkage groups, were included in the Pumpkin–Zucchini map, and ten, distributed over four linkage groups, in the Pumpkin–Crookneck map. Some of these markers were grouped together in the same linkage group in both maps, namely LGpz/LGpc5 and LGpz/LGpc8. The Pumpkin–Zucchini map contains 31 RAPD markers originating from the source of ZYMV resistance, C. moschata. None of these 31 markers showed segregation distortions.
Markers for ZYMV resistance and hull-less seed coat
About 24 of the 31 C. moschata specific markers were mapped onto two linkage groups in clusters of the Pumpkin–Zucchini map. LGpz1 contains 11 and LGpz2 contains 13 of these markers, two of which are closely linked to the ZYMV-resistance loci. These two markers have been converted to SCARs (unpublished results). The remaining seven markers were distributed over six linkage groups, namely two in LGpz7, and one in each of the following linkage groups: LGpz9, LGpz11, LGpz13, LGpz14b, and LGpz19 (see italic font in Fig. 2).
The BSA identified 148 of the 1,068 primers which were polymorphic between the two bulks, i.e., two pools of 12 DNA samples from homozygous plants for hulled and hull-less seeds, respectively. After testing these primers on the 12 individual DNA samples composing each of the two bulks, only seven showed association with the seed coat characteristic. Upon genotyping the 92 F2 plants of the Pumpkin–Zucchini population with these markers, five (AK11-340, AN10-340, AB14-235, H18-385, and AB07-590) were found, flanking the n locus in linkage group LGpz9 with a distance <1.5 cM (Fig. 2). Two markers (AO11-280 and AW11-420) were <6.5 cM away from the n locus. In five of the seven cases, the RAPD band was linked to the N allele, while two, AB14-235 and AW11-420, were linked to the n allele. While mapping the Pumpkin–Crookneck population, four markers (F09-670, AE07-850, AA12-340, and AB17-980) were found to be tightly linked to the N allele at a distance <7 cM. The fifth marker (AW11-420), linked to the n allele, only 3 cM distant, was present in both populations (Fig. 4).
Alignment of the two maps
Gels of both populations, containing either RAPD or AFLP fragments, were compared to identify common markers based on their identical fragment size. Fifty-two RAPD (19.4%) and 40 AFLP loci (33.8%), were detected. Of the 92 common markers scored, 62 were used in mapping to recognize identical linkage groups (Fig. 4). We initially identified ten linkage groups that contained at least three common markers. When using relaxed grouping, i.e., LOD ≥3 and recombination frequency of higher than 35, the two linkage groups LGpz8a and LGpz8b were joined in the Pumpkin–Zucchini map (Fig. 2). In the same way the two linkage groups LGpc6a and LGpc6b were joined in the Pumpkin–Crookneck map (Fig. 3). Based on the presence of common markers, LGpz14 and LGpz16 in the Pumpkin–Zucchini map and LGpc5 and LGpc13 in the Pumpkin–Crookneck map were connected (Fig. 4). Based on the presence of at least two common markers in each of the two maps, 14 linkage groups were aligned with an average of four common markers and a maximum of ten (LG3 in Fig. 4). LG2 contained three common markers in the Pumpkin–Zucchini map, yet only one of these markers could be mapped in the Pumpkin–Crookneck population. Therefore the alignment of this linkage group is based on only one marker. Although the order of common markers was, in most cases, conserved, markers in LG3 and LG4 had to be rearranged (Fig. 4). The average distances between common markers in the two populations were, in most cases, similar.
Discussion
By virtue of their marker coverage, density and consistency, the maps presented herein are suggested as reference maps for C. pepo. We have used mainly RAPD and AFLP markers as they are universal in their application and require no sequence information for their development. The combination of these markers allowed the construction of moderately saturated maps in both populations. Although they are, in general, dominant markers which limits their use in comparative mapping, they were successfully used here and in other studies for comparison of linkage groups and to align genetic maps (Plomion et al. 1995; Krutovskii et al. 1998; Laucou et al. 1998; Costa et al. 2000). Map alignment and map merging have generally been done on different populations, derived from the same two parental genotypes, or on two populations having one parental genotype in common. In our case, we used four different parental genotypes to create the two mapping populations, but two of the four parents are closely related, both being oil pumpkins. The number of common markers (92 in total, i.e., almost 30%) was relatively high. They allowed the establishment of a skeletal map for C. pepo, in which 62 mapped common loci were used to align 15 linkage groups. We consider these 15 linkage groups to represent different chromosomes, that is three quarters of the haploid number of chromosomes of C. pepo. The 62 common loci, represented by fragments of identical size in the two populations and therefore considered to be orthologous, were contributed in almost equal numbers by the four parents. This suggests an even distribution of alleles in the four parents. The order of these markers was conserved in most cases. Only in two cases rearrangements were observed, which could be due to the size of the mapping population, and/or to the algorithm used for arranging the markers (Périn et al. 2000). Other reasons could be genotyping errors, missing values and mapping distorted markers (Hackett and Broadfoot 2003).
Polymorphism
The significantly higher level of polymorphism, as revealed by both RAPD and AFLP markers, between the parents of the pumpkin/crookneck population than between the parents of the pumpkin/zucchini population clearly reflects the taxonomical position of the three cultivar-groups within the species. Pumpkin and zucchini belong to the same subspecies, C. pepo subsp. pepo. They were most probably domesticated in Mexico. The crookneck variety, on the other hand, belongs to C. pepo subsp. ovifera and was domesticated most likely in eastern US. The genetic relationship among these cultivar groups is solidly established by isozyme and DNA-markers (Decker 1988, Sanjur et al. 2002, Paris et al. 2003).
The level of polymorphism of RAPD in our mapping populations was higher than that found by Brown and Myers (2002), even though they used an inter-specific (C. pepo × C. moschata) mapping population. The greater amount of polymorphism detected in our populations may partly be due to the use of polyacrylamide instead of agarose for fragment separation (Stift et al. 2003).
Results of mapping using AFLP markers have not been previously published for Cucurbita and therefore comparison of the polymorphism level is not possible. Nevertheless, this polymorphism level in our study was similar to that found in C. melo (Wang et al. 1997), which is probably in part due to the similar genome size of the two species (Bennett and Leitch 2004). The AFLP technique produced twice as many polymorphic bands as the RAPD marker system (average polymorphism of the two populations: 5.4 vs. 2.7). Similar results have been reported in other species (Powell et al. 1996; Milbourne et al. 1997; Costa et al. 2000; Haussmann et al. 2002; Zhong et al. 2004).
SSRs are versatile, highly informative, co-dominant markers for many eukaryotic genomes (Wang et al. 1994). They are locus specific, abundant and evenly distributed throughout the genome, and were recommended as standard markers to be used in the construction of highly saturated genetic maps (Beckmann and Soller 1990). Unfortunately, they showed poor transferability among cucurbit genera, as predicted by Katzir et al. (1996). The four parents of the two maps were genotyped for more than 100 melon SSR markers. Only a few of the markers generated bands, and none of them were polymorphic between the parents used to create the mapping populations. This was not the case when using Cucurbita SSR markers. Five of the available 18 C. pepo specific SSR markers showed polymorphism, and three of them could be mapped.
Genome coverage
The genome coverage in the Pumpkin–Zucchini map (2,140 cM) and in the Pumpkin–Crookneck map (2,234 cM) was slightly higher than the genome coverage (1,954 cM) of the inter-specific map reported by Brown and Myers (2002). The average distance between markers they found (13 cM) was, however, twice as large as that of our study (∼6.6 cM), and is probably due to the low number of polymorphic markers they could use for mapping (148 vs. 330 markers). This is, however, understandable considering that Brown and Myers (2002) used a back-cross population for mapping, which effectively reduces the number of potential polymorphisms to half.
Marker distribution
The difference in distribution among RAPD and AFLP markers described here are in good agreement with several reports in the literature, especially on clustering of AFLP markers in other species (Qi et al. 1998; Bert et al. 1999; Boivin et al. 1999; Haanstra et al. 1999; Vuylsteke et al. 1999; Young et al. 1999; Haussmann et al. 2002). This can be attributed to the use of EcoRI that cuts in AT-rich domains mainly located in the centromeric and telomeric regions. In contrast, however, De la Rosa et al. (2003) reported even distribution of both marker types, AFLP (using EcoRI) and RAPD in olive.
Co-dominant markers
Polyacrylamide gel electrophoresis, due to its higher resolution, resulted in additional scorable RAPD fragments and identification of co-dominant RAPD markers (Fig. 1). In the two populations RAPD primers produced approximately 10% co-dominant loci, which was twice that found in the AFLP marker system. The number of RAPD-based co-dominant loci was considerably greater than that reported for other species (Grattapaglia and Sederoff 1994; Laucou et al. 1998; Krutovskii et al. 1998), likely due to the use of polyacrylamide instead of agarose gel for fragment separation. Two co-dominant RAPD markers, linked to ZYMV resistance, were isolated, sequenced, and successfully converted into SCAR markers. One allele pair differed in length by six nucleotides, the other pair differed in three base substitutions (SNPs). It is well known, that electrophoretic mobility of DNA in polyacrylamide is also affected by its base composition causing structural changes of the molecules. Thus, molecules of the same size can differ in mobility and can be distinguished using polyacrylamide gel electrophoresis.
Distorted segregation
In comparison to other crop species (Laucou et al. 1998; Haanstra et al. 1999; Bert et al. 1999; Cai et al. 2004), the number of distorted loci found in this study was relatively low (∼5%). It is worth mentioning the absence of any segregation distortion among the 31 markers originating from the source of resistance against ZYMV, C. moschata. Obviously, the integration of the genetic material from C. moschata into C. pepo, after backcrossing with the latter, was without any negative consequences. Segregation distortions have been reported in numerous genetic mapping studies, especially in populations derived from inter-specific crosses (e.g., Paterson et al. 1988; Keim et al. 1990; Heun et al. 1991; Echt et al. 1994). The mechanism of segregation distortion is not clearly understood, but it may have a number of different causes, such as chromosomal imbalance (aneuploidy), chromosomal rearrangements (translocations), competition among gametes, or genes affecting the viability of the embryo, as was suggested recently by Gonzalo et al. (2005). This can be the reason for the higher frequency (14%) of distorted markers in the inter-specific hybrid between C. pepo and C. moschata reported by Brown and Myers (2002).
Segregation distortion in C. pepo has been observed by Wilson and Payne (1994) and Paris (2000). In the first study, different pollen mixtures were applied in crosses between cultivated and free-living genotypes of C. pepo to study the competition of microgametophytes in fertilization. In the latter publication inter-subspecific crosses between two C. pepo genotypes were studied. Results of both experiments suggested an interaction between microgametophyte and gynoecium, which could, however, also be partly caused by the structure of gynoecium (Wilson and Payne 1994). Paris (2000) put forward the idea that interaction of microgametophyte with gynoecium and consequently gametophytic selection could partly be responsible for the continuous existence of groups of particular fruit shapes (cultivar-groups) throughout the centuries (Paris 2000). In our case, the average of 5% distorted markers in both populations was relatively low. Some of these markers were grouped in the same linkage groups in both maps, e.g., two in LGpz/LGpc5 and three in LGpz/LGpc8. The remaining were scattered individually in different linkage groups. No discernible difference between the intra- and inter-subspecific cross, with respect to segregation distortion, was observed.
Tagging the seed coat character
Of the seven markers, which were found linked to the n locus in the Pumpkin–Zucchini map, only two were in coupling phase with the hull-less allele. The marker AW11-420, which was 7 cM away from the hull-less allele in the Pumpkin–Zucchini map, was also found linked, at a distance of 3 cM, to the n/n (hull-less) allele in the Pumpkin–Crookneck map. This marker could play an important role in map-based cloning experiments, studying the genetics of the lignification of the seed-coat in C. pepo. The usefulness of this marker in selecting for hull-less genotypes has to be validated. Using BSA, Haley et al. (1994) identified RAPD markers in common bean, linked in both, coupling and repulsion, to a resistance allele against Bean Common Mosaic Virus (BCMV). In a case study they demonstrated that repulsion-phase linkages provided greater selection efficiency than coupling-phase linkage, even when the former had a greater linkage distance from the pest resistance allele. Repulsion-phase markers greatly improved selection efficiency for homozygous resistant genotypes. This we shall test too for the repulsion-phase markers in hull-less pumpkin.
Tagging ZYMV resistance
Three independent complementary genes confer resistance to ZYMV in C. pepo (Paris and Cohen 2000). About 24 of 31 bands introduced into C. pepo from the source of resistance, C. moschata, were mapped to LGpz1 and LGpz2. The remaining seven markers were distributed over six other linkage groups, and conceivably one of them is linked to the third putative resistance gene. Increasing map density may join some linkage groups into one, carrying the third independent resistance gene.
References
Andres TC (1987) Cucurbita fraterna, the closest wild relative and progenitor of C. pepo. Cucurbit Genet Coop Rep 10:69–71
Baudracco-Arnas S, Pitrat M (1996) A genetic map of melon (Cucumis melo L.) with RFLP, RAPD, isozyme, disease resistance and morphological markers. Theor Appl Genet 93:57–64
Beckmann JS, Soller M (1990) Towards a unified approach to genetic mapping of eukaryotes based on sequence tagged microsatellite site. Biotechnology 8:30–32
Bennett MD, Leitch IJ (2004) Plant DNA C-values database (release 3.0, Dec. 2004) http://www.rbgkew.org.uk/cval/homepage.html
Bert PF, Charmet G, Sourdille P, Hayward MD, Balfourier F (1999) A high-density molecular map for ryegrass (Lolium perenne) using AFLP markers. Theor Appl Genet 99:445–452
Blumenthal M, Busse WR, Goldberg A, Gruenwald J, Hall T, Riggins CW, Rister RS (eds), Klein S, Rister RS (trans) (1998) The complete German commission E monographs. S. 193. American Botanical Council; Boston; Integrative Medicine Communications, Austin
Boivin K, Deu M, Rami J-F, Trouche G, Hamon P (1999) Towards a saturated sorghum map using RFLP and AFLP markers. Theor Appl Genet 98:320–328
Bradeen JM, Staub JE, Wye C, Antonise R, Peleman J (2001) Towards an expanded and integrated linkage map of cucumber (Cucumis sativus L). Genome 44:111–119
Brown RN, Myers JR (2002) A genetic map of squash (Cucurbita ssp.) with randomly amplified polymorphic DNA markers and morphological markers. J Am Soc Hortic Sci 127:568–575
Buerstmayr H, Lemmens M, Hartl L, Doldi L, Steiner B, Stierschneider M, Ruckenbauer P (2002) Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. I. Resistance to fungal spread (type II resistance). Theor Appl Genet 104:84–91
Cai H, Inoue M, Yuyama N, Nakayama S (2004) An AFLP-based linkage map of Zoysiagrass (Zoysia japonica). Plant Breed 123:543–548
Costa P, Pot D, Dubos C, Frigerio J-M, Pionneau C, Bodénès C, Bertocchi E, Cervera M-T, Remington DL, Plomion C (2000) A genetic map of maritime pine based on AFLP, RAPD and Protein markers. Theor Appl Genet 100:39–48
Danin-Poleg Y, Tadmor Y, Tzuri G, Reis N, Hirschberg J, Katzir N (2002) Construction of a genetic map of melon with molecular markers and horticultural traits, and localization of genes associated with ZYMV resistance. Euphytica 125:373–384
Decker DS (1988) Origin(s), evolution, and systematics in Cucurbita pepo (Cucurbitaceae). Econ Bot 42:4–15
De la Rosa R, Angiolillo A, Guerriero C, Pellegrini M, Rallo L, Besnard G, Berville A, Martin A, Baldoni L (2003) A first linkage map of olive (Olea europaea L.) cultivars using RAPD, AFLP, RFLP and SSR markers. Theor Appl Genet 106:1273–1282
Duchesne AN (1786) Essai sur l’histoire naturelle des courges. Pankoucke CJ, Paris, p 46
Echt CS, Kidwell KK, Knapp SJ, Osborn TC, McCoy TJ (1994) Linkage mapping in diploid alfalfa (Medicago sativa). Genome 37:61–71
Fazio G, Staub JE, Chung SM (2002) Development and characterization of PCR markers in cucumber (Cucumis sativus L.) J Am Soc Hortic Sci 127:545–557
Ferriol M, Picó B, Nuez F (2003) Genetic diversity of a germplasm collection of Cucurbita pepo using SRAP and AFLP markers. Theor Appl Genet 107:271–282
Gonzalo MJ, Oliver M, Garcia-Mas J, Monfort A, Dolcet-Sanjuan R, Katzir N, Arus P, Monforte AJ (2005) Simple-sequence repeat markers used in merging linkage maps of melon (Cucumis melo L.). Theor Appl Genet 110: 802–811
Grattapaglia D, Sederoff R (1994) Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137:1121–1137
Grebenščikov I (1954) Zur Vererbung der Dünnschaligkeit bei Cucurbita pepo L. Züchter 24:162–166
Haanstra JP, Wye W, Verbakel H, Meijer-Dekens F, Van den Berg P, Odinot P, Van Heusden AW, Tanksley S, Lindhout P, Peleman J (1999) An integrated high-density RFLP-AFLP map of tomato based on two Lycopersicon esculentum x L. pennellii F2 populations. Theor Appl Genet 99:254–271
Hackett CA, Broadfoot LB (2003) Effects of genotyping errors, missing values and segregation distortion in molecular marker data on the construction of linkage maps. Heredity 90:33–38
Haley SD, Afanador L, Kelly JD (1994) Selection for monogenic pest resistance traits with coupling- and repulsion-phase RAPD markers. Crop Sci 34:1061–1066
Hartl L, Mohler V, Zeller FJ, Hsam SLK, Schweizer G (1999) Identification of AFLP markers closely linked to the powdery mildew resistance genes Pm1c and Pm4a in common wheat. Genome 42:322–329
Haussmann BIG, Hess DE, Seetharama N, Welz HG, Geiger HH (2002) Construction of a combined sorghum linkage map from two recombinant inbred populations using AFLP, SSR, RFLP, and RAPD markers, and comparison with other sorghum maps. Theor Appl Genet 105:629–637
Heun M, Kennedy AE, Anderson JA, Lapitan NLV, Sorrells ME, Tanksley SD (1991) Construction of a restriction fragment length polymorphism map for barley (Hordeum vulgare). Genome 34:437–447
Katzir N, Danin-Poleg Y, Tzuri G, Karchi Z, Lavi U, Cregan PB (1996) Length polymorphism and homologies of microsatellites in several Cucurbitaceae species. Theor Appl Genet 93:1282–1290
Katzir N, Tadmor Y, Tzuri G, Leshzeshen E, Mozes-Daube N, Danin-Poleg Y, Paris HS (2000) Further ISSR and preliminary SSR analysis of relationships among accessions of Cucurbita pepo. In: Katzir N, Paris HS (eds) Proceedings of the cucurbitaceae 2000. Acta Hortic 510:433–439
Keim P, Diers BW, Olson TC, Shoemaker RC (1990) RFLP mapping in soybean: association between marker loci and variation in quantitative traits. Genetics 126:735–742
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Kreuter MH (2000) Hormone disbalance and urinary bladder disease. Phyto Novum, Emil Flachsmann AG, Zürich Switzerland, pp 2–24
Krutovskii KV, Vollmer SS, Sorensen FC, Adams WT, Knapp SJ, Strauss SH (1998) RAPD genome maps of Douglas-fir. J Hered 89:197–205
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Laucou V, Haurogné K, Ellis N, Rameau C (1998) Genetic mapping in pea. 1. RAPD linkage map of Pisum sativum. Theor Appl Genet 97:905–915
Lee YH, Jeon HJ, Hong KH, Kim BD (1995) Use of random amplified polymorphic DNA for linkage group analysis in an interspecific cross hybrid F2 generation of Cucurbita. J Kor Soc Hortic Sci 36:323–330
Lincoln SE, Daly MJ, Lander ES (1992) Constructing genetic maps with MAPMAKER/EXP 3.0. Whitehead Inst Tech Rpt 3rd Edition
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease resistance gene by bulked segregant analysis: a rapid method to detect markers in specific genomic regions using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Milbourne D, Meyer R, Bradshaw JE, Baird E, Bonar N, Provan J, Powell W, Waugh R (1997) Comparison of PCR-based marker systems for the analysis of genetic relationship in cultivated potato. Mol Breed 3:127–136
Murkovic M, Hillebrand A, Winkler J, Leitner E, Pfannhauser W (1996) Variability of fatty acid content in pumkin seeds (Cucurbita pepo L.) Z. Lebensm Unters Forsch 203:216–219
Murkovic M, Piironen V, Lampi AM, Kraushofer T, Sontag G (2004) Changes in chemical composition of pumpkin seeds during the roasting process for production of pumpkin seed oil (Part 1: non-volatile compounds). Food Chem 84:359–365
Naudin C (1856) Nouvelles recherches sur les caractères spécifiques et les variétés des plantes du genre Cucurbita. Ann Sci Nat Bot IV 6:5–73, pl. 1–3
Oliver M, Garcia-Mas J, Cardús M, Pueyo N, López-Sesé AI, Arroyo M, Gómez-Paniagua H, Arús P, de Vicente MC (2001) Construction of a reference linkage map of melon. Genome 44:836–845
Paris HS (1986) A proposed subspecific classification for Cucurbita pepo. Phytologia 61:133–138
Paris HS (2000) Segregation distortion in Cucurbita pepo. In: Katzir N, Paris HS (eds) Proceedings of the cucurbitaceae 2000. Acta Hortic 510:199–202
Paris HS, Cohen S (2000) Oligogenic inheritance for resistance to zucchini yellow mosaic virus in Cucurbita pepo. Ann Appl Biol 136:209–214
Paris HS (2001) History of the cultivar-groups of Cucurbita pepo. Hortic Rev 25:71–170
Paris HS, Yonash N, Portnoy V, Mozes-Daube N, Tzuri G, Katzir N (2003) Assessment of genetic relationships in Cucurbita pepo (Cucurbitaceae) using DNA markers. Theor Appl Genet 106:971–978
Paris HS, Brown RN (2005) The genes of pumpkin and squash. HortScience 40:1620–1630
Park YH, Sensoy S, Wye C, Antonise R, Peleman J, Havey MJ (2000) A genetic map of cucumber composed of RAPDs, RFLPs, AFLPs, and loci conditioning resistance to papaya ringspot and zucchini yellow mosaic viruses. Genome 43:1003–1010
Paterson AH, Landers ES, Hewitt JD, Peterson S, Lincoln SE, Tanksley SD (1988) Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature 335:721–726
Périn C, Hagen L, Dogimont C, de Conto V, Pitrat M (1998) Construction of a genetic map of melon with molecular markers and horticultural traits. In: McCreight JD (ed) Cucurbitaceae 98: evaluation and enhancement of cucurbit germplasm. ASHS Press, Alexandria, VA, pp 370–376
Périn C, Hagen L, Dogimont C, de Conto V, Lecomte L, Pitrat M (2000) Construction of a reference genetic map of melon. In: Katzir N, Paris HS (eds) Proceedings of the cucurbitaceae 2000. Acta Hortic 510:367–374
Plomion C, O’Malley DM, Durel CE (1995) Genomic analysis in maritime pine (Pinus pinaster). Comparison of two RAPD maps using selfed and open-pollinated seeds of the same individual. Theor Appl Genet 90:1028–1034
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellites) markers for germplasm analysis. Mol Breed 2:225–238
Prym von Becherer L (1955) Untersuchungen zur Anatomie und zum Erbverhalten der Samenschalen von Cucurbita maxima DUCH. und Cucurbita pepo L. Züchter 25:1–14
Qi X, Stam P, Lindhout P (1998) Use of locus-specific AFLP markers to construct a high-density molecular map in barley. Theor Appl Genet 96:376–384
Robinson RW, Decker-Walters DS (1997) Cucurbits. CAB International, New York
Schmidlin CB, Kreuter MH (2003) Cucurbita pepo, mögliche Einfluss auf hormonelle Ungleichgewicht bei Inkontinenz. Phytotherapie 3:2–4
Serquen FC, Bacher J, Staub JE (1997) Mapping and QTL analysis of horticultural traits in a narrow cross in cucumber (Cucumis sativus L.) using random-amplified polymorphic DNA markers. Mol Breed 3:257–268
Sanjur OI, Piperno DR, Andres ThC, Wessel-Beaver L (2002) Phylogenetic relationships among domesticated and wild species of Cucurbita (Cucurbitaceae) inferred from mitochondrial gene: implications for crop plant evolution and areas of origin. Proc Natl Acad Sci USA 99:535–540
Stift G, Pachner M, Lelley T (2003) Comparison of RAPD fragment separation in agarose and polyacrylamide gel by studying Cucurbita species. Cucurbit Genet Coop Rep 26:62–65
Teppner H (2000) Cucurbita pepo (Cucurbitaceae)—history, seed coat types, thin coated seeds and their genetics. Phyton 40:1–42
Torres-Ruiz R, Hemleben V (1991) Use of ribosomal DNA spacer probes to distinguish cultivars of Cucurbita pepo L. and other Cucurbitaceae. Euphytica 53:11–17
Vos P, Hogers R, Bleeker M, Reijans M, van der Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Vuylsteke M, Mank R, Antonise R, Bastiaans E, Senior ML, Stuber CW, Melchinger AE, Lübberstedt T, Xia XC, Stam P, Zabeau M, Kuiper M (1999) Two high-density AFLP linkage maps of Zea mays L.: analysis of distribution of AFLP markers. Theor Appl Genet 99:921–935
Wang YH, Thomas CE, Dean RA (1997) A genetic map of melon (Cucumis melo L.) based on amplified fragment length polymorphism (AFLP) markers. Theor Appl Genet 95:791–798
Wang Z, Weber JL, Zhong G, Tanksley SD (1994) Survey of plant short tandem DNA repeates. Theor Appl Genet 88:1–6
Wilson HD, Payne JS (1994) Crop/weed microgametophyte competition in Cucurbita pepo (Cucurbitaceae). Am J Bot 81:1531–1537
Young WP, Schupp JM, Keim P (1999) DNA methylation and AFLP marker distribution in the soybean genome. Theor Appl Genet 99:785–790
Zhong D, Pai A, Yan G (2004) AFLP-based genetic linkage map for the red flour beetle (Tribolium castaneum). J Hered 95:53–61
Zraidi A, Obermayer R, Pachner M, Lelley T (2003) On the genetics and histology of the hull-less character of Styrian Oil-Pumpkin (Cucurbita pepo L.). Cucurbit Genet Coop Rep 26:57–61
Zraidi A, Lelley T (2004) Genetic map for pumpkin Cucurbita pepo using random amplified polymorphic DNA markers. In: Lebeda A, Paris HS (eds) Progress in cucurbit genetics and breeding research. Proceedings of the cucurbitaceae 2004, 8th EUCARPIA meeting on cucurbit genetics and breeding, Olomouc, pp 507–514
Acknowledgments
This research was financially supported by the Austrian Science Fund (FWF Project, No. P15773) and by the State of Lower Austria. Our thanks to J. E. Staub, USDA–ARS and University of Wisconsin, Madison, for testing the Cucumis-SSRs on the four parents used in our crosses. We are grateful to J. E. Staub and H. S. Paris for critically reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zraidi, A., Stift, G., Pachner, M. et al. A consensus map for Cucurbita pepo . Mol Breeding 20, 375–388 (2007). https://doi.org/10.1007/s11032-007-9098-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-007-9098-6