Abstract
The main aim of this study was to discover small molecule inhibitors against Cathepsin D (CatD) (EC.3.4.23.5), a clinically proven prognostic marker for breast cancer, and to explore the mechanisms by which CatD could be a useful therapeutic target for triple-positive and triple-negative breast cancers (TPBC & TNBC). The crystal structure of CatD at 2.5 Å resolution (PDB: 1LYB), which was complexed with Pepstatin A, was selected for computer-aided molecular modeling. The methods used in our study were pharmacophore modeling and molecular docking. Virtual screening was performed to identify small molecules from an in-house database and a large commercial chemical library. Cytotoxicity studies were performed on human normal cell line HEK293T and growth inhibition studies on breast adenocarcinoma cell lines, namely MCF-7, MDA-MB-231, SK-BR-3, and MDA-MB-468. Furthermore, RT-PCR analysis, in vitro enzyme assay, and cell cycle analysis ascertained the validity of the selected molecules. A set of 28 molecules was subjected to an in vitro fluorescence-based inhibitory activity assay, and among them six molecules exhibited \(>\)50 % inhibition at \(25\,\upmu \hbox {M}\). These molecules also exhibited good growth inhibition against TPBC and TNBC cancer types. Among them, molecules 1 and 17 showed single-digit micromolar \(\hbox {GI}_{50}\) values against MCF-7 and MDA-MB-231 cell lines.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Solid tumors, particularly in breast cancer, metastasize into different organs of the body in the advanced stages of the disease. Cancer invasion and metastasis are challenging areas of treatment, and one of the important reasons for invasion is the proteolysis of the extracellular matrix (ECM) [1]. The ECM is composed of collagen, laminin, and fibronectin along with many enzymes, such as matrix metalloproteases, serine proteases, cathepsins, etc. [2]. Among all, cathepsins are found to have a major role in growth factor-binding and ECM degradation, which emphasize the importance of cathepsins in human cancer development. The lysosomal compartment contains about 40 different kinds of acid hydrolases composed of proteases, nucleases, lipases, glycosidases, and sulfatases. Among the mentioned hydrolases, proteases are divided into three subgroups on their active site amino acid: Cysteine (B, C, H, F, K, L, O, S, V, X, and W), Aspartate (D and E), and Serine (A and G) proteases [3–5].
Our target of interest, CatD, is the most explored lysosomal aspartic protease as a tumor marker due to its elevated concentrations in the cytosol of breast cancer cells in patients diagnosed during clinical trials [6]. CatD is initially synthesized as an inactive pre-proenzyme and undergoes further post-transcriptional modifications, such as glycosylation to form mature enzyme [7]. Maturation of CatD is an ATP-driven acidification of lysosomes in relation with cysteine proteases [8]. The enzyme is active in acid milieu and cleaves the components of the extracellular matrix increasing cancer invasion by releasing growth factors into circulation [9].
There are many functionalities of CatD in normal cells, namely, the metabolic degradation of proteins and peptides, antigen processing, activation, and degradation of growth factors [10]. CatD is endocytosed into the lysosomal compartment by mannose-6-phosphate receptors and other unidentified receptors present on the surface of lysosomes [11, 12]. In the case of breast cancer (estrogen-positive), there are an increased number of large acid vesicles bearing more amounts of CatD causing cancer invasion [13]. The high level of CatD in primary tumors is indicative of local recurrence or distant metastasis [14]. CatD is also reported as an important biological marker among triple-negative breast cancer patients along with a high Ki-67 index [15, 16]. CatD is currently a target of interest due to its high expression in advanced stages of solid tumors, especially mammary tumors, resulting in aggressive metastasis and worsening of the condition of the individual [17].
Pepstatin A was identified as a specific inhibitor of CatD with an \(\hbox {IC}_{50}\) of 0.1 nM, and, to date, there are very few scaffolds reported for CatD inhibition including hydroxyl ethyl isosteres with cyclic tertiary amines, synthetic oligopeptides, non-peptidic acylguanidine, and sulfamoyl benzamide derivatives [18, 19]. In designing CatD substrates, several chemokines including interleukin-8 (CXCL8), SDF-1 (CXCL12), and SLC were studied to understand the pattern of CatD metastatic mechanism [20]. CatD has two lobes connected by a cleft accommodating eight amino acid residues in its active pocket as found in other aspartic proteases (e.g., HIV protease, pepsin, renin, cathepsin E). Furthermore, CatD inhibition is also reported as a possible therapy in other ailments due to its structural similarities with \(\beta \)-secretase, plasmepsins, and HIV proteases, in degenerative diseases, malaria, and HIV, respectively [21]. To date, CatD inhibitors have not reached clinical trials, and there remains more to explore about their use and development in cancer chemotherapy [22].
Our goal in this research area is to discover novel nonpeptidic small molecules as CatD inhibitors for the treatment of breast cancer. The reported CatD inhibitors, to date, have been estimated to be clinically useful and are further studied for their role in cancers. We took a step forward in discovering new moieties for CatD inhibition and further analyzing their role in cell cycle regulation. We performed an energy-based pharmacophore virtual screening and molecular docking studies using the ASINEX database (www.asinex.com) and BITS, an in-house database to discover novel CatD inhibitors. The chemical structures of reported CatD inhibitors (84 molecules) are considered as bioactives and a set of 1000 inactive, drug-like molecules, provided by Schrodinger are used as decoys. A single dataset file was made including both decoys and bioactives. This dataset file consisting of 1084 molecules is used to assess the screening performance of generated e-pharmacophores. The screening performance is directly proportional to the number of bioactives screened, which was further calculated as mentioned in the “Methods” section below. The virtual screening method helped us discover new molecules on protein–ligand interactions and binding affinities. After molecular docking, the selected molecules were tested for CatD inhibitory activity using a fluorescence-based assay. Furthermore, we have conducted cell-based assays consisting of a panel of cell lines with varied levels of gene expression for estrogen receptor (ER), progesterone receptor (PR), and Her2/neu in correlation with CatD expression.
Materials and methods
Computational study
Software
The pharmacophore models were generated using PHASE 3.4 implemented in Maestro 9.3 software package [23]. All docking studies were performed using GLIDE (Grid Ligand Docking with Energetics) 5.8 (Maestro 9.3), and all ADME (Absorption, Distribution, Metabolism, and Excretion) properties were calculated using QikProp 3.5 (Maestro 9.3) [23].
Energy-based Pharmacophore generation and validation
Molecular docking studies were performed using an e-pharmacophore approach which helps in the rapid in silico screening of chemical databases. ‘Scripts’ option in Maestro was used to generate pharmacophores, on the energies and features of the co-crystallized ligand. In addition, PHASE was used to identify the features in a molecule that match with the features in the generated hypothesis. The available pharmacophoric features provided by PHASE during the alignment were hydrogen bond acceptor (A), hydrogen bond donor (D), hydrophobic group (H), negatively ionizable group (N), positively ionizable group (P), and aromatic ring (R). Furthermore, different pharmacophore models were generated with minimum to maximum number of features to screen all the molecules from the ASINEX and BITS databases. Datasets were prepared using reported CatD inhibitors in the literature together with a decoy set. The decoy set, provided by Schrodinger, is a collection of 1000 ligands which exhibit “drug-like” properties, chosen from 1 million compound library [23]. A total of 84 reported non-peptidic CatD inhibitors with different scaffolds (e.g., acylguanidine [22], hydroxyl ethylamine [24]) were drawn using Maestro’s 2D sketcher and were energetically minimized using LigPrep [23]. Stereo isomers were also generated by adding sufficient hydrogen molecules at neutral pH (pH 7) using LigPrep.
On the basis of the 10 featured hypotheses generated using Pepstatin A, many 3–7 featured hypotheses were generated considering the positions of donors, acceptors and aromatic rings (Fig S1). PHASE generated scores were considered to evaluate the hypothesis ability in differentiating reported CatD inhibitors from the decoy set. Enrichment Factor (EF) and Goodness of Hit (GH) were calculated using Eqs. 1 and 2 [25]:
where ‘Ht’ is the total set of molecules used from the hit list; ‘Ha’ is the set of actives from the hit list; ‘A’ denotes the actives from the decoy set; and ‘D’ is the number of molecules from the decoy set. The best pharmacophores were further used to select molecules from the databases following molecular docking protocols.
Grid generation
The best pharmacophore models were used to screen diverse scaffolds from the ASINEX and BITS databases (Figs. S2, S3). GLIDE was used for the virtual screening of compound libraries. The crystal structure of CatD (PDB ID: 1LYB) was minimized using the protein preparation wizard (OPLS_2005 force field) [23]. The co-crystallized ligand, Pepstatin A, was selected during the receptor grid generation to define x, y, and z axes of the grid. The sites for molecular docking were defined during the grid generation which allowed us to filter molecules from the selected active site.
Virtual screening by pharmacophore model and molecular docking
The best pharmacophores with good EF and GH scores were selected and run through PHASE to find matches against the ASINEX and BITS databases. Molecules that matched the respective features of the selected pharmacophore were further considered for docking studies using GLIDE. Initially, the favorable interactions between screened ligand molecules (from pharmacophore screening) and CatD, in flexible mode of docking, were examined by means of GLIDE. The three different levels of docking precisions provided by GLIDE were utilized for virtual screening methodology, namely HTVS (high throughput virtual screening), SP (standard precision), and XP (extra precision). HTVS was first used to identify molecules binding to CatD followed by SP and then XP docking for refinement. Ligand interactions in relation to Pepstatin A, along with XP GLIDE scores, number of hydrogen bonds, alignment of the molecule inside the active site pocket, and ADME properties were considered to select the final set of compounds for further in vitro studies.
ADME profile screening
The QikProp module [23] was used to calculate the ADME properties of the selected molecules. This provided an understanding of the solubility and absorption levels of the selected set of molecules from the database. Caco-permeability with a value of \(<\)25 was considered poor and \(>\)500 as high permeability. QP log Po/w (octanol/water partition coefficient ratio) was calculated from a range of \(-2\) to 6.5; QP log S (aqueous solubility) was considered good if the value was between \(-6.5\) and 0.5. Furthermore, the percentage of human absorption was considered high if it was \(>\)80 % and poor if it was \(<\)25 %.
Experimental approaches
Chemicals used
Phosphate-buffered saline (PBS), Minimum Essential Medium (MEM), Roswell Park Memorial Institute (RPMI) 1640 medium, and fetal bovine serum (FBS) were obtained from Hi Media Laboratories Pvt. Ltd. MTT (3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyl tetrazolium bromide), trypan blue, and CatD human primers were procured from Sigma–Aldrich (Bangalore, India).
In vitro enzyme assay study
CatD enzyme inhibition studies were carried out using Cathepsin D Inhibitor Screening Kit (Fluorometric) (BioVision, Inc., Milpitas, CA). This is a fluorescence-based assay which makes use of the CatD substrate sequence GKPILFFRLK (DNP)-D-RNH2 labeled with methyl coumarin (MCA). CatD acts by cleaving the synthetic substrate to release the quenched fluorescent group MCA, which will then be measured by a fluorescence plate reader at Ex/Em of 328/460 nm. The relative efficacy of test inhibitors was compared to the positive control inhibitor, Pepstatin A \((\hbox {IC}_{50} < 0.1\,\hbox {nM})\). The assay procedure implemented was followed as described by the manufacturer’s protocol. The inhibitor reference control along with positive and background controls were prepared adding the reaction buffer and substrate in common, with and without enzyme as per the protocol. An incubation period of 1–2 h at \(37\,^{\circ }\hbox {C}\) was followed by measuring the fluorescence using a micro-plate reader. The readings were further measured as per the standardized protocol:
RNA Extraction and quantitative RT-PCR analyses
RT-PCR studies were carried out to bring out a logical selection of a set of breast cancer cell lines depending on their differential CatD gene expressions. The consideration was on the presence or the absence of differential gene expressions, e.g., estrogen receptor (ER+/\(-\)), progesterone receptor (PR+/\(-\)), and human epidermal growth factor receptor (HER/neu) which helped us study the activities of molecules considering both TNBC and TPBC types. Total mRNA was quantified from \(5\times 10^{6}\) cells MCF-7 (triple-positive), MDA-MB-231 (triple-negative), MDA-MB-468 (triple-negative), and SK-BR-3 (ER-, Her2 amplified). RNA was extracted using the TRI reagent according to the manufacturer’s protocol (Sigma). PCR amplification, for converting mRNA to cDNA, was run using Verso cDNA kit, with optimized protocol for 45 cycles. Each cycle had a 15-second denaturation time at \(95\,^{\circ }\hbox {C}\), annealing for 1 min at \(52\,^{\circ }\hbox {C}\)-\(55\,^{\circ }\hbox {C}\), and extension for 1 min at \(72\,^{\circ }\hbox {C}\). The mRNA quantities were amplified using a PCR master mix to optimize the annealing temperature of the designed CatD human primers. The cDNA entities formed were further confirmed by electrophoresis, running through 1 % agarose gel at 100 volts for 30 min.
RT-PCR was performed following the manufacturer’s instructions using a fluorescent dye, SYBR green (Kappa Bio-systems), helpful in the detection of PCR products. We used CatD (human) primers procured from Sigma, i.e., forward primer 5\(^{\prime }\) CAT TGT GGA CAC AGG CAC TTC 3\(^{\prime }\) and reverse primer 5\(^{\prime }\) GAC ACC TTG AGC GTG TAGTCC 3\(^{\prime }\) to determine DNA fragment [26]. The RT-PCR was initiated with denaturation at \(94\,^{\circ }\hbox {C}\) for 3 minutes, followed by 45 cycles of denaturation at \(95\,^{\circ }\hbox {C}\) for 20 s, annealing at \(61\,^{\circ }\hbox {C}\) for 10 s, and elongation \(72\,^{\circ }\hbox {C}\) for 2 min. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the house-keeping gene to normalize the gene expressions. Normalized relative levels of CatD expressions in the selected breast cancer cell lines were the ratios of CatD and GAPDH gene expressions.
Cell-based growth inhibition and viability assays
Based on the RTPCR analysis, a set of breast cancer cell lines was selected to evaluate the anti-proliferative activities of the selected molecules. The cell lines, namely MCF-7, MDA-MB-231, MDA-MB-468, and SK-BR-3 of American Type Culture Collection origin, were kindly provided by Incozen Therapeutics Pvt. Ltd. The cell lines were maintained in MEM and RPMI media supplemented with 10% fetal bovine serum, \(100\,\mu \hbox {g/mL}\) streptomycin, and 100 U/mL penicillin under conditions of 5 % \(\hbox {CO}_{2}\), 80 % relative humidity (RH) at \(37\,^{\circ }\hbox {C}\). Cell viability/growth inhibition was determined by MTT assay. Cells were harvested and seeded into 96-well tissue culture plates with 5,000 cells per well and allowed to attach and gain morphology overnight. Cells were treated with test compounds at concentrations ranging from \(100\, \mu \hbox {M}\) to 1 nM in triplicates for a drug-incubation period of 48–72 h. The incubation time varied depending upon the doubling time of individual cell lines. A stock solution of a test compound (10 mM) was prepared using DMSO and all the other concentrations were diluted with medium to achieve \(<\)1 % DMSO while exposing to cells. A DMSO control was also placed to measure its toxic effects, if any, on the cancer cells. A zero reading was taken to note the absorbance of day 1 cells plated. After test compound incubation, MTT was added at a concentration of 5 mg/mL, and plates were incubated for 3–4 hrs at \(37\,^{\circ }\hbox {C}\). Cell viability was quantified by dissolving the formed formazan crystals in DMSO and measuring the absorbance spectrophotometrically at 550–600 nm using a Vector X3 2030 multi-label reader (Perkin Elmer).
Cell cycle analysis
Selected molecules were further evaluated for their effect on cancer cell cycle. The DNA content was different in various phases of cell cycle and propidium iodide, which stains DNA, helped analyze the effect of a test compound on cell cycle regulation. Breast cancer cells (\(1\times 10^{6}\) cells/well) were seeded in six-well plates and treated with a test compound for 24 h. Cells were first harvested and washed, followed by fixation in 70 % ice-cold ethanol at \(4\,^{\circ }\hbox {C}\) for 2 h. The cells were further centrifuged, washed well with cold PBS, and recentrifuged. Finally, cells were evenly resuspended in \(250\,\mu \hbox {L}\) PBS, and stained with \(10\,\mu \hbox {L}\) propidium iodide (PI; 1 mg/mL) and \(10\,\mu \hbox {L}\) RNase A (10 mg/mL) for 30 min at room temperature (RT). The samples were loaded to measure the forward scatter (FS) and side scatter (SS) to identify single cells. DNA content was observed using flow sight (Amnis, Millipore), and IDEAS analysis software was used to calculate percentage of cells in G0/G1, S and G2/M phases.
Survival in acidic tumor microenvironment
Tumor cells are known to adapt in acidic microenvironment by preserving ATP levels. As a part of assessing lysosomal CatD activity in acidic conditions of tumor microenvironment, the metastatic breast cancer cell lines were plated using RPMI 1640 and MEM media with 10% FCS at pH 7.4 or 5.5, and the pH 5.5 was achieved by adding 2 N HCl to the culture medium. After growing the culture for 5 days, cells were stained with 0.4% trypan blue and analyzed by flow cytometry [27].
Results
Selection and validation of pharmacophore model
Using the co-crystallized ligand, Pepstatin A, we generated the initial hypotheses as a ten-featured e-pharmacophore through docking post-processing (Table S1). There were six acceptors and four donor groups in the generated ten-featured e-pharmacophore. On the basis of energies between the hydrogen bond acceptor (A), hydrogen bond donor (D), hydrophobic group (H), negatively ionizable (N), positively ionizable (P), and aromatic ring (R) moieties, we generated 3–7-featured pharmacophore models through combinations. The combinations were made by selecting different features each time generating respective pharmacophores. This further helped us attain best-fit small molecules filtered on the basis of specific pharmacophore features. A set of 1084 compounds were screened against all the generated pharmacophores using PHASE, and the pharmacophore efficiencies were further calculated as discussed in the “Methods” section. Out of many hypotheses generated, we selected the five best hypotheses on EF and GH scores. Pharmacophores AAADD1, AADDD, AAADD2, AADD, and AAD displayed good EF and GH scores (Table 1), and so were further selected for molecular docking to identify best hits from the databases.
Grid generation and virtual screening
The docking grid was generated by selecting the co-crystallized Pepstatin A ligand in the protein’s crystal structure. The generated grid was used to screen the ASINEX database (500,000 molecules) and the in-house BITS database (930 molecules). The docking calculations were performed in accordance with the molecular docking protocols and interactions between the molecules and target protein. A HTVS docking was performed with both the in-house BITS and ASINEX databases to identify molecules with good binding affinity. Molecules were ranked on the basis of docking score, interactions with amino acid residues, and numbers of hydrogen bonds in comparison with crystal ligand Pepstatin A and were further selected for SP and XP docking. The active site of CatD was found to be interacting with Pepstatin A with residues, namely Asp33, Asp231, Gly35, Gly79, Gly233, Ser80, and Ser235. The active site contains two aspartate residues, Asp33 and Asp 231, each of which plays a vital role in the catalytic function of CatD. The final XP docking resulted in 523, 139, and 124 molecules from 5-, 4-, and 3-featured e-pharmacophores, respectively. The active site has many areas that can accommodate bulkier substituents from the databases used. In addition, the docking score, ADME properties, and the alignment of the molecule in the active site were considered to select a total of 28 molecules for in vitro activity studies (Fig.1).
ADME predictions
A total of 13 molecules from the ASINEX database and 15 molecules from the BITS database were selected and examined for pharmaceutically relevant drug-like properties. The QikProp module was used for validating pharmaceutically relevant properties of the molecules in relation to Absorption, Distribution, Metabolism, and Excretion (ADME). Most of the hits had good calculated partition coefficients (QPlogPo/w), QPlogS, and QPPCaco, which are the key measures of solubility, absorption, and cell permeability. Lipinski’s rule of five displayed zero violations of the molecules. The expected human oral absorption for the molecules ranged from 57 to 100 %.
A total of 28 molecules were selected as best hits and their parameters such as predicted GLIDE score, amino acid interaction residues, and measures of their ADME properties are listed in (Tables 2, 3) and their chemical structures are shown in (Figs. 2, 3).
In vitro enzyme assay
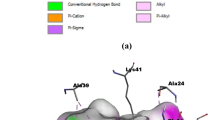
Initially, Pepstatin standard graph was plotted considering concentrations from \(100\,\mu \hbox {M}\) to 0.1 nM as per the manufacturer’s protocol (Fig. 4). Furthermore, 28 molecules were selected for in vitro CatD inhibition using a flourometric assay (BIOVISION, U.S.A). The initial screening at \(25\,\upmu \hbox {M}\) revealed that six out of 28 molecules displayed \(>\)50 % inhibition (Fig. 5). These molecules were further screened at \(10\,\upmu \hbox {M}\), and fluorescence measurements (Ex/Em of 328/460 nm) showed an inhibition range from 30 to 40 %. Active molecules were correlated with their docking calculations. Molecules 1, 3, and 10 from the ASINEX database and molecules 17, 19, and 23 from the BITS database showed strong docking interactions and docking poses inside the active pocket compared to all other database molecules. Among these, the docking poses of molecules 1, 10, and 17 with different orientations inside the active site pocket are shown in (Fig. 6) [28]. The active site pocket of CatD has two lobes and is connected by a cleft. The residues Gly79 and Ser80 forming the cleft are not displayed in the figure to provide a clear view of the alignment of molecules inside the active site pocket. These deselected residues are colored green, as shown in Fig. 6.
Binding poses displaying interactions a Pepstatin A, crystal ligand (1LYB), b most active Lead 1 from docking, c second-most active lead 17 from docking. d Active compound with good alignment in to the groove lead 10. All active site-forming crucial residues are shown in spring green and sites S1, S2, S3, S4, S2\(^{\prime }\), and S3\(^{\prime }\) are in yellow text
Gene expression studies
All the four breast cancer cell lines were analyzed for gene expression of CatD using primers mentioned in the “Methods” section. The mRNA quantifications of MCF-7, MDA-MB-231, MDA-MB-468, and SK-BR-3 were measured by reading absorbance at 260 nm \((\hbox {A}_{260})\). The relative CatD and GAPDH expression levels in all cell lines are shown in Fig. 7. CatD gene expression was higher in MCF-7 followed by triple-negative breast cancer cell lines MDA-MB-231 and MDA-MB-468. Among TNBCs, MDA-MB-231 is a highly metastatic tumor cell line [29, 30] and showed high levels of CatD gene expression. Thus, CatD expression was determined in the entire cell line panel selected and used to validate the inhibitory activities of the selected molecules.
Anti-proliferative activity studies
CatD is reported to degrade ECM and release growth factors into the blood stream causing invasion and metastasis of mammary tumors [31, 32]. Breast cancer cell lines expressing CatD gene were selected to perform this study. All the 28 molecules were evaluated for their anti-proliferative effect on MCF-7, MDA-MB-231, MDA-MB-468 and SK-BR-3. Among the selected 28 molecules, 1,3 (ASINEX), and 17,23 (BITS database) showed promising growth inhibition \((\hbox {GI}_{50})\) on MCF-7 exhibiting inhibition values of \(2.27\,\pm \, 1.13\), \(3.70\,\pm \,0.05\), \(5.7\,\pm \,0.005\),), and and \(5.6\,\pm \,2.34\,\upmu \hbox {M}\), respectively. The molecules were further tested for their cytotoxicity in normal cells, i.e., human embryonic kidney cells (HEK-293T), and \(>\)70 % of them were found to be non-toxic. The growth inhibitory values of the 28 molecules along with normal cell cytotoxicity results are shown in Tables 4 and 5. The molecules were found to inhibit the cell proliferation of MCF-7 and MDA-MB-231 adenocarcinoma compared to the other 2 cell lines.
CatD inhibition led to cell cycle arrest
One of the crucial features to examine the final arbiter of cell fate was cell cycle arrest. The effect of cell cycle arrest during CatD inhibition was determined. Breast cancer cells were incubated with \(\hbox {GI}_{50}\) concentration of molecules 1 and 17 for 24–48 h and were run through 488 channels of flow sight (AMNIS, Millipore). The imaging system showed PI fluorescence differentiating various phases of cells with their DNA content. The samples were further analyzed by IDEAS analysis software, and a histogram was plotted as Intensity in channel 4 (Propidium iodide) on X-axis and Normalized frequency on Y-axis. We observed a decrease in G0/1 phase and an eventual increase in G2/M DNA content displaying cell cycle arrest (Fig. 8).
Lysosomal volume in acidic microenvironment
The cell lines were studied for their survival in acidic microenvironment as per the protocol mentioned in “Methods” section. The media at pH 7.4 showed considerable equal cell viability when compared to media at pH 5.4 indicating adaptation of lysosomal volume at pH 7.4 analyzed through flow sight (AMNIS, Millipore). The graph (Fig. 9) was analyzed on the basis of intensity, an indicative of percentage of cell survival [27].
Conclusion
Overall, 28 molecules from two databases were identified on pharmacophore and molecular docking studies. These studies identified a set of molecules causing CatD inhibition and growth inhibition on breast cancer cells. The most potent molecules 1 and 17 displayed growth inhibition on all the cell lines tested in correlation to their respective levels of CatD gene expressions. These molecules are CatD inhibitors with novel structural features, e.g., oxoacetamide (molecule 1) and hydrazinylidine (molecule 17) moieties, when compared to reported CatD inhibitors in the literature. Thus, we anticipate that our study would accelerate drug discovery and research for the discovery and development of new CatD inhibitors as therapeutic potential agents for breast cancer treatment.
References
Oskarsson T (2013) Extracellular matrix components in breast cancer progression and metastasis. Breast 2:S66–72. doi:10.1016/j.breast.2013.07.012
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002) The extracellular matrix of animals, 4th edn. Molecular biology of the cell,. Garland Science, New York
Sakkiah S, Thangapandian S, Lee KW (2012) Ligand-based virtual screening and molecular docking studies to identify the critical chemical features of potent cathepsin D inhibitors. Chem Biol Drug Des 80:64–80. doi:10.1111/j.1747-0285.2012.01339.x
Gocheva V, Joyce JA (2007) Cysteine cathepsins and the cutting edge of cancer invasion. Cell Cycle 6:60–64. doi:10.4161/cc.6.1.3669
Dian D, Vrekoussis T, Shabani N, Mylonas I, Kuhn C, Schindlbeck C, Navrozoglou I, Friese K, Makrigiannakis A, Jeschke U (2012) Expression of cathepsin-D in primary breast cancer and corresponding local recurrence or metastasis: an immunohistochemical study. Anticancer Res 32:901–905
Jacobson-Raber G, Lazarev I, Novack V, Mermershtein W, Baumfeld Y, Geffen DB, Sion-Vardy N, Ariad S (2011) The prognostic importance of cathepsin D and E-cadherin in early breast cancer: a single-institution experience. Oncol Lett 2:1183–1190. doi:10.3892/ol.2011.393
Baldwin ET, Bhat TN, Gulnik S, Hosur MV, Sowder RC, Cachau RE, Collins J, Silva AM, Erickson JW (1993) Crystal structures of native and inhibited forms of human cathepsin D: implications for lysosomal targeting and drug design. Proc Natl Acad Sci USA 90:6796–6800. doi:10.1073/pnas.90.14.6796
Wang F, Samudio I, Safe S (2001) Transcriptional activation of cathepsin D gene expression by 17beta-estradiol: mechanism of aryl hydrocarbon receptor-mediated inhibition. Mol Cell Endocrinol 172:91–103. doi:10.1016/S0303-7207(00)00379-8
Abbott DE, Margaryan NV, Jeruss JS, Khan S, Kaklamani V, Winchester DJ, Hansen N, Rademaker A, Khalkhali-Ellis Z, Hendrix MJ (2010) Reevaluating cathepsin D as a biomarker for breast cancer: serum activity levels versus histopathology. Cancer Biol Ther 9:23–30. doi:10.4161/cbt.9.1.10378
Benes P, Vetvicka V, Fusek M (2008) Cathepsin D-many functions of one aspartic protease. Crit Rev Oncol Hemat 68:12–28. doi:10.1016/j.critrevonc.2008.02.008
Laurent-Matha V, Farnoud MR, Lucas A, Rougeot C, Garcia M, Rochefort H (1998) Endocytosis of pro-cathepsin D into breast cancer cells is mostly independent of mannose-6-phosphate receptors. J Cell Sci 111:2539–2549
Gieselmann V, Hasilik A, Von Figura K (1985) Processing of human cathepsin D in lysosomes in vitro. J Biol Chem 260:3215–3220. http://www.jbc.org/content/260/5/3215.full.pdf
Ahmed W, Jabeen U, Khaliq S (2014) New inhibitors of proteolytic enzymes cathepsin D and plasmepsin II. Pak J Biochem Mol Biol 47:129–132
Fehrenbacher N, Jäättelä M (2005) Lysosomes as targets for cancer therapy. Cancer Res 65:2993–2995. doi:10.1158/0008-5472.CAN-05-0476
Huang L, Liu Z, Chen S, Liu Y, Shao Z (2013) A prognostic model for triple-negative breast cancer patients based on node status, cathepsin-D and Ki-67 index. PLoS One 8:e83081. doi:10.1371/journal.pone.0083081
Wolf M, Clark-Lewis I, Buri C, Langen H, Lis M, Mazzucchelli L (2003) Cathepsin D specifically cleaves the chemokines macrophage inflammatory protein-1 alpha, macrophage inflammatory protein-1 beta, and SLC that are expressed in human breast cancer. Am J Pathol 162:1183–1190. doi:10.1016/S0002-9440(10)63914-4
Khalkhali-Ellis Z, Goossens W, Margaryan NV, Hendrix MJ (2014) Cleavage of Histone 3 by cathepsin D in the involuting mammary gland. PLoS One 9:e103230. doi:10.1371/journal.pone.0103230
Lin T, Williams HR (1979) Inhibition of cathepsin D by synthetic oligopeptides. J Biol Chem 254:11875–11883
Ahmed W, Khan IA, Arshad MN, Siddiqui WA, Haleem MA, Azim MK (2013) Identification of sulfamoylbenzamide derivatives as selective cathepsin D inhibitors. Pak J Pharm Sci 26:687–690
Laurent-Matha V, Derocq D, Prébois C, Katunuma N, Liaudet-Coopman E (2006) Processing of human cathepsin D is independent of its catalytic function and auto-activation: involvement of cathepsins L and B. J Biochem 139:363–371. doi:10.1093/jb/mvj037
Fusek M, Vetvicka V (2005) Dual role of cathepsin D: ligand and protease. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 149:43–50
Grädler U, Czodrowski P, Tsaklakidis C, Klein M, Werkmann D, Lindemann S, Maskos K, Leuthner B (2014) Structure-based optimization of non-peptidic cathepsin D inhibitors. Bioorg Med Chem Lett 24:4141–4150. doi:10.1016/j.bmcl.2014.07.054
Schrodinger, LLC, New York. http://www.schrodinger.com/
McConnell RM, Green AW, Trana CJ, McConnell MS, Lindley JF, Sayyar K, Godwin WE, Hatfield SE (2006) New cathepsin d inhibitors with hydroxyethylamine isosteres: preparation and characterization. Med Chem 2:27–38. doi:10.2174/157340606775197705
Sakkiah S, Meganathan C, Sohn YS, Namadevan S, Lee KW (2012) Identification of important chemical features of \(11\beta \)-hydroxysteroid dehydrogenase type1 inhibitors: application of ligand based virtual screening and density functional theory. Int J Mol Sci 13:5138–5162. doi:10.3390/ijms13045138
Fukuda ME, Iwadate Y, Machida T, Hiwasa T, Nimura Y, Nagai Y, Takiguchi M, Tanzawa H, Yamaura A, Seki N (2005) Cathepsin D is a potential serum marker for poor prognosis in glioma patients. Cancer Res 65:5190–5194. doi:10.1158/0008-5472.CAN-04-4134
Matarrese P, Ascione B, Ciarlo L, Vona R, Leonetti C, Scarsella M, Mileo AM, Catricala C, Paggi MG, Malomi W et al (2010) Cathepsin B inhibition interferes with metastatic potential of human melanoma: an in vitro and in vivo study. Mol Cancer 9:207–220. doi:10.1186/1476-4598-9-207
Carew JS, Espitia CM, Esquivel JA, Mahalingam D, Kelly KR, Reddy G, Giles FJ, Nawrocki ST (2011) Lucanthone is a novel inhibitor of autophagy that induces cathepsin D-mediated apoptosis. J Biol Chem 8:6602–6613. doi:10.1074/jbc.M110.151324
Montcourrier P, Mangeat PH, Salazar G, Morisset M, Sahuquet A, Rochefort H (1990) Cathepsin D in breast cancer cells can digest extracellular matrix in large acidic vesicles. Cancer Res 50:6045–6054
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the \(2^{-\Delta \Delta {\rm c}}\text{ T }\) Method. Methods 25:402–408
Knopfova L, Benes P, Pekarcikova L, Hermanova M, Masarik M, Pernicova Z, Soucek K, Smarda J (2012) c-Myb regulates matrix metalloproteinases 1/9, and cathepsin D: implications for matrix-dependent breast cancer cell invasion and metastasis. Mol Cancer 11:15. doi:10.1186/1476-4598-11-15
Dali B, Keita M, Megnassan E, Frecer V, Miertus S (2012) Insight into selectivity of peptidomimetic inhibitors with modified statine core for plasmepsin II of Plasmodium falciparum over human cathepsin D. Chem Biol Drug Des 79:411–430. doi:10.1111/j.1747-0285.2011.01276.x
Nakayama S, Torikoshi Y, Takahashi T, Yoshida T, Sudo T, Matsushima T, Kawasaki Y, Katayama A, Gohda K, Hortobagyi GN, Noguchi S, Sakai T, Ishihara H, Ueno NT et al (2009) Prediction of paclitaxel sensitivity by CDK1 and CDK2 activity in human breast cancer cells. Breast Cancer Res 11:1–10. doi:10.1186/bcr2231
Acknowledgments
The authors gratefully acknowledge the Birla Institute of Technology and Sciences, Hyderabad for infrastructural facilities and DST for the INSPIRE fellowship.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Anantaraju, H.S., Battu, M.B., Viswanadha, S. et al. Cathepsin D inhibitors as potential therapeutics for breast cancer treatment: Molecular docking and bioevaluation against triple-negative and triple-positive breast cancers. Mol Divers 20, 521–535 (2016). https://doi.org/10.1007/s11030-015-9645-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-015-9645-8