Abstract
Resistance to radiotherapy is a major limitation for the successful treatment of colorectal cancer (CRC). Recently, accumulating evidence supports a critical role of epigenetic regulation in tumor cell survival upon irradiation. Lysine Demethylase 4B (KDM4B) is a histone demethylase involved in the oncogenesis of multiple human cancers but the underlying mechanisms have not been fully elucidated. Here we show that KDM4B is overexpressed in human colorectal cancer (CRC) tumors and cell lines. In CRC cells, KDM4B silencing induces spontaneous double-strand breaks (DSBs) formation and potently sensitizes tumor cells to irradiation. A putative mechanism involved suppression of Signal Transducer and Activator of Transcription 3 (STAT3) signaling pathway, which is essential for efficient repair of damaged DNA. Overexpression of STAT3 in KMD4B knockdown cells largely attenuates DNA damage triggered by KDM4B silencing and increases cell survival upon irradiation. Moreover, we find evidence that transcription factor CAMP Responsive Element Binding Protein (CREB) is a key regulator of KMD4B expression by directly binding to a conserved region in KMD4B promoter. Together, our findings illustrate the significance of CREB–KDM4B–STAT3 signaling cascade in DNA damage response, and highlight that KDM4B may potentially be a novel oncotarget for CRC radiotherapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is a leading cause of cancer deaths worldwide. Despite recent advances in diagnosis and therapy, the overall prognosis of CRC remains poor [1, 2]. Ionizing radiation (IR) is commonly used in cancer therapy and is an important component of CRC treatment, however, the efficacy is not optimistic. One reason is that acquired radioresistance often occurs shortly after the treatment and subsequently leads to tumor recurrence. Another reason is that radiotherapy might cause serious gastrointestinal tissue damage and result in symptoms such as intestinal inflammation, electrolyte imbalance, diarrhea, fibrosis, and even death during the therapeutic procedure [3]. Accordingly, developing novel therapeutic approaches to improve CRC cell radiation response and overcome radioresistance is critical to improving the therapeutic efficacy and minimizing the side effects of radiotherapy.
Covalent histone modifications play important roles in regulating chromatin structure, transcription, replication, and DNA damage repair. It has been well-established that deregulation of histone modifications is causally linked to tumorigenesis of multiple human cancers [2, 4,5,6]. The Lysine Demethylase 4 (KDM4) subfamily presents a group of histone demethylases containing the Jumonji C (JmJC) domain and plays a specific critical role in the dynamic regulation of H3K9 and H3K36 tri-/di-methylation during differentiation and proliferation [7,8,9]. Aberrant expression of KDM4 proteins has been observed in several human tumor types and forced expression of KDM4 confers oncogenic pressure in vitro [10, 11]. Previous studies showed that the H3K9me3 demethylase KDM4B is overexpressed in several human cancers and plays crucial roles in maintaining genome stability and promoting tumor growth. It has been shown that EGFP-tagged KDM4D was rapidly recruited to the loci of DNA damage in living cells induced by laser irradiation (IR) [10, 11]. Forced overexpression of KDM4D was associated with decreased numbers of DNA damage foci and enhanced cell survival following irradiation [10]. Loss of KDM4B led to cell death, cell cycle arrest, and impaired DNA damage response in vitro as well as suppressed tumor xenograft growth in vivo [8, 11,12,13]. In the current study, we find evidence that KDM4B is overexpressed in CRC tumor tissues and CRC cell lines, and higher KDM4B expression positively correlates with cellular tolerance to gamma irradiation. We thus hypothesized that reducing expression of KDM4B might sensitize CRC cells to radiation treatment. To this end, we established KDM4B knockdown cell lines by stable small hairpin RNA (shRNA) transfection. Our results showed that KDM4B is involved in DNA damage response via regulating STAT3 signaling.
Results
Overexpression of KDM4B in CRC tumor tissues and cell lines
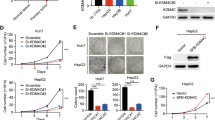
As shown in Fig. 1a, using immunohistochemical staining, we examined expression of KDM4B in a CRC tumor tissue (tumor) and the peritumoral tissue (cntl). Our results showed that KDM4B staining was predominantly observed in the nucleus, and the signal was significantly higher in tumor versus cntl, indicating overexpression of KDM4B in CRC tumors. Furthermore, we used real-time PCR to examine expression of KDM4B in 20 pairs of CRC tumor tissues and peritumoral tissues (Fig. 1b). In line with the immunohistochemistry data, our results suggested that KDM4B mRNA expression was significantly higher in CRC tumors compared to cntls (p = 0.008, paired Student’s t test). Examining KDM4B expression in two human colon epithelial cell lines (CRL-1831 and NCM-460) as well as two CRC cell lines (SW620 and HT29) using western blot revealed overexpression of KDM4B in CRC cell lines, with the highest expression in HT29 cells (Fig. 1c).
Overexpression of KDM4B in CRC tumor tissues and cell lines. a Representative immunohistochemical staining of KDM4B in CRC tumors and peritumoral tissues (scale bar 200 µm). b KDM4B mRNA expression in 20 CRC tumors and peritumoral tissues determined by real-time PCR. Data represent the mean ± s.e.m. **p < 0.01, Student’s t test. c KDM4B protein expression in human CRC cell lines and colon epithelial cell lines as determined by western blot. d Cells were irradiated with various doses of ionizing radiation and incubated for 7 days to allow colony formation. The number of colonies per dish was counted and the surviving fractions were calculated as the ratio between treated and untreated cells. *p < 0.05, **p < 0.01, paired Student’s t test
Given the putative role of KDM4B in DNA damage repair and radioresistance [10], we asked whether CRC cells and colon epithelial cells exhibit different sensitivity to IR treatment. To this end, different epithelial cells and CRC cells were irradiated with 0–8 Gy single doses of IR and cell survival was analyzed after 7 days. As shown in Fig. 1d, cell survival upon IR treatment was significantly higher in CRC cells versus epithelial cells (p = 0.009, paired Student’s t test), while HT29 cells exhibited stronger IR resistance than SW620 cells (p = 0.04, paired Student’s t test), supporting the notion that KDM4B being important in DNA damage repair. We thus suggested that targeting KDM4B might represent an important approach to improve the radiotherapy efficacy for CRC treatment.
KDM4B silencing induces DNA double-strand breaks
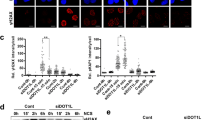
We then used HT29 cells with high KDM4B expression as a model to investigate the function of KDM4B in CRC DNA damage response. To this end, using a lentiviral shRNA-based approach, we established two stable KDM4B knockdown HT29 cell lines (shKDM4B-1 and -2) with efficient reduction of KDM4B expression compared to control knockdown (shCNTL) (Fig. 2a). We didn’t observe significant cell death or cell proliferation arrest in either cell lines with KDM4B knockdown (Supplementary Fig. 1). Consistent with previous findings [13], we could detect expression of phosphorylated H2AX (γH2AX), a characteristic marker of DSBs, in both KDM4B knockdown cell lines, but not in control knockdown cells using western blot (Fig. 2a). Consistently, immunofluorescence assays revealed spontaneous formation of γH2AX nuclear foci in KDM4B knockdown cells but not in control knockdown cells (Fig. 2b). We further used flow cytometry to quantify γH2AX signal intensities in KDM4B knockdown and control knockdown cells. As shown in Fig. 2c, KDM4B knockdown cells with elevated levels of γH2AX were clearly distinguishable from control knockdown cells with baseline levels of γH2AX in the signal intensity histograms. Notably, a distinct small cell population with high γH2AX signals (between 1.1 and 1.8, log10 transformed) was observed in both knockdown cell lines, indicating that loss of KDM4B might eventually result in accumulation of high levels of DSBs and genome instability. Overall, γH2AX signal intensities were significantly higher in both KDM4B knockdown cells versus control knockdown cells (p < 1.0 × 10−10, Student’s t test), indicating that KDM4B silencing induced spontaneous DSBs in these cells.
KDM4B silencing induces DNA double-strand breaks. a Western blot of KDM4B and γH2AX in control or KDM4B knockdown HT29 cells. b Immunofluorescence assay of γH2AX nuclear foci in control or KDM4B knockdown cells. c Overlay of histograms representing control knockdown (blue) and KDM4B knockdown (gray and orange) flow cytometry data for γH2AX signals. Data represent the mean ± s.e.m. for three independent experiments. ***p < 0.001, Student’s t test. d Cell were irradiated with various doses of ionizing radiation and incubated for 7 days and the surviving fractions were calculated as the ratio between treated and untreated cells. **p < 0.01, paired Student’s t test. (Color figure online)
H2AX phosphorylation is a critical event in the series of early responses to DNA damage upon different stress stimuli [14]. Appearance of γH2AX in KDM4B knockdown cells without apparent stimuli thus suggested defective DNA damage repair and decreased genome instability in these cells. To directly investigate the implication of KDM4B in mediating radioresistance, KDM4B knockdown and control knockdown cells were treated with different doses of IR and analyzed for cell survival. Our results showed that KDM4B knockdown significantly enhanced radiation sensitivity of HT29 cells versus control knockdown (Fig. 2d, p = 0.003, paired Student’s t test). Together, these data support a critical role of KDM4B in DNA damage repair and maintenance of genome stability. We thus suggest that combination of radiation therapy with KDM4B targeting might provide a promising approach to overcome radioresistance and enhance the overall radiation response in CRC patients.
KDM4B silencing suppresses activity of STAT3 signaling
It has been shown that loss of KDM4B is associated with disruption of STAT3 signaling pathway, which has been implicated in sensing DSBs and repair of damaged DNA [15, 16]. To examine the putative implication of STAT3 signaling in KDM4B silencing-induced DNA damage, we examined STAT3 and its upstream regulators JAK1 and JAK2 for total protein expression and activated phosphorylation (Fig. 3a). Our results showed that p-STAT3 levels were significantly downregulated in KDM4B knockdown cells compared to control knockdown cells, while total STAT3 levels were largely unaffected. In contrast, we observed no obvious disruption in JAK1/2 expression or phosphorylation, indicating that KDM4B knockdown specifically disrupted STAT3 activation. To test the influence of STAT3 inactivation on γH2AX accumulation, we treated HT29 cells with AG490, which is a specific inhibitor of STAT3 phosphorylation and activity. As shown in Fig. 3b, treatment with AG490 abolished STAT3 phosphorylation and resulted in the accumulation of γH2AX. Consistently, flow cytometry assay revealed that γH2AX signal is significantly enhanced in HT29 cells treated with AG490 versus control (Fig. 3c, p < 1.0 × 10−10, Student’s t test), supporting a role of STAT3 in DNA damage repair and removing γH2AX.
KDM4B silencing suppresses activity of STAT3 signaling. a Western blot of JAK/STAT3 signaling proteins and their phosphorylation in control or KDM4B knockdown HT29 cells. b Western blot of STAT3, p-STAT3, and γH2AX in HT29 cells treated with 10 µM STAT3 phosphorylation inhibitor AG490. c Overlay of histograms representing DMSO (blue) and AG490 (orange) flow cytometry data for γH2AX signals. Data represent the mean ± s.e.m. for three independent experiments. ***p < 0.001, Student’s t test. d Western blot of STAT3, p-STAT3, and γH2AX in KDM4B knockdown cells transfected with empty vector (EV) or STAT3. e γH2AX signals in cells described in d. Data represent the mean ± s.e.m. for three independent experiments. ***p < 0.001, Student’s t test. f Cell were irradiated with various doses of ionizing radiation and incubated for 7 days and the surviving fractions were calculated as the ratio between treated and untreated cells. **p < 0.01, paired Student’s t test. (Color figure online)
To investigate whether STAT3 inactivation is causally linked to DSBs accumulation upon KDM4B silencing, we overexpressed STAT3 in KDM4B knockdown cells via transient transfection. Our results showed that overexpression efficiently increased KDM4B protein expression as well as phosphorylation in these cells (Fig. 3d), leading to the removal of γH2AX induced by KDM4B silencing (Fig. 3d, e). Furthermore, overexpression of STAT3 in KDM4B knockdown cells largely enhanced cell survival in response to IR treatment (Fig. 3f, p = 0.03, paired Student’s t test). Together, these data indicate that loss of KDM4B triggered DNA damage in CRC cells via suppression of STAT3 activation.
KDM4B is a direct transcriptional target of CREB
Overexpression of KDM4B has been observed in multiple cancers and has been implicated in tumor cell survival, proliferation, and resistance to chemo/radiotherapies [12, 17]. However, little is known about the regulatory mechanisms controlling KDM4B expression in cancer cells. Focusing on transcriptional regulation, we scanned KDM4B promoter region − 1 kb upstream transcription start site (TSS) for putative transcription factor (TF) binding sites using ENCODE datasets and UCSC genome browser, which contain experimentally identified TF binding sites. As shown in the snapshot in Fig. 4a, we observed a CREB binding sites within − 300 bp region upstream TSS. Notably, compared with the flanking regions, this putative CREB binding site is highly conserved across vertebrates (Fig. 4a), indicating the functional importance.
KDM4B is a direct transcriptional target of CREB. a UCSC genome browser tracks for CREB binding site in KDM4B promoter. b After chromatin immunoprecipitation, DNA fragments crosslinked with CREB amplified by PCR for CREB binding region in KDM4B promoter as well as two negative control regions in KDM4B introns 1 and 2 containing no CREB binding sites. c Western blot of KDM4B and CREB expression following CREB knockdown. d Western blot of KDM4B and CREB expression following CREB inhibition using H89. e Schematic of luciferase reporter vectors for the CREB binding site in KDM4B promoter region (− 566 to + 27) (WT) and the antisense sequences (MT). f Luciferase activities of reporters described in e with no treatment, DMSO, or H89 treatment. Data represent the mean ± s.e.m. for three independent experiments. **p < 0.01, Student’s t test
We then performed chromatin immunoprecipitation (ChIP) assays to test whether CREB could directly bind to KDM4B promoter in HT29 cells (Fig. 4b). DNA fragments crosslinked with CREB were collected by an antibody against CREB and amplified by PCR for CREB binding region in KDM4B promoter as well as two negative control regions in KDM4B introns 1 and 2, which contain no CREB binding sites. Our results showed that CREB could specifically bind to the predicted binding site, but not the negative control sites (Fig. 4b). Moreover, both CREB knockdown (Fig. 4c) or inactivation using specific inhibitor H89 (Fig. 4d) resulted in potent reduction in KDM4B expression, suggesting that KDM4B is a direct transcriptional target of CREB.
We further transfected HT29 cells with luciferase reporters containing the KDM4B promoter (− 566 to + 27) (WT) or the reverse complementary sequence (MT) (Fig. 4e). Interestingly, luciferase activity of the WT reporter is significantly higher than that of the MT vector (p = 0.0035, Student’s t test) following transfection, indicating that the CREB binding site is essential to KDM4B transcription. Treatment with H89 largely attenuated luciferase activity of WT vector (p = 0.0017, Student’s t test), but not that of MT vector (p = 0.25, Student’s t test) (Fig. 4f), consistent with direct CREB binding to the KDM4B promoter being important in promoting transcription. In multiple human cancers, CREB overexpression is frequently associated with worse prognoses and relapse following radiotherapies. Our results thus raise the possibility that high radiotherapy resistance can result from increased DNA damage repair activity via the CREB–KDM4B signaling transduction.
Discussion
Efficient repair of damaged DNA is essential to facilitate tumor cell survival upon radiotherapy [18, 19]. Here we show that KDM4B is overexpressed in CRC cells and correlates with resistance to irradiation. KDM4B-deficient CRC cells exhibit enhanced DNA damage and are found to be more sensitive to irradiation stimuli, suggesting that KDM4B is required for optimum DNA damage response. Mechanistically, KDM4B silencing suppresses STAT3 activity, resulting in the accumulation of spontaneous DSBs and reduced capacity to repair damaged DNA. Moreover, our results suggest that CREB directly binds to KDM4B promoter and play a critical role in enhancing KDM4B expression, providing a putative mechanism underlying KDM4B overexpression in CRC. Taken together, these data suggest that overexpression of KDM4B in CRC may contribute to the failure of radiotherapy and possibly other DSB-inducing chemotherapeutic drugs.
Radiation therapy causes DNA damage, resulting in the accumulation of DSBs and other forms of DNA lesions in the cell [19]. Phosphorylation of H2AX is a key event in sensing damaged DNA, a crucial early step in DNA damage response, leading to the activation of STAT3 signaling cascade and recruitment of repair factors to DSBs [14, 16, 20]. Failure to repair DSBs might induce cell cycle arrest and cell death by apoptosis and necrosis, and sensitize tumor cells to irradiation stimuli. Previous studies showed that histone modifications might play a critical role in mediating DNA damage response via affecting chromatin organization [4,5,6]. In line with this notion, alternations in histone methylation levels as well as expression of methyltransferase and demethyltransferase are predictive of patient outcomes in multiple human cancer types [17]. Specifically, it has been shown that H3K9me3 presents a barrier to the DSB repair machinery and demethylation of H3K9 is required for efficient DNA damage repair [21]. It has been shown that the demethylase catalytic domain of KDM4B was required for KDM4B accumulation on irradiation-induced DNA damage tracks, supporting the mechanistic link between demethylation and DNA damage repair [22]. Given the critical role of KDM4B in H3K9me3 demethylation, failure to remove H3K9me3 in KDM4B silencing cells might block signal transduction triggered by DSB and compromise STAT3 activation and damage repair. In line with this, our results showed that overexpression of STAT3 in KDM4B knockdown cells largely restored the repair of DSBs, as monitored by γH2AX formation. Accordingly, overexpression of KDM4B in CRC patient samples might thus confer a survival advantage upon irradiation stimuli and lead to resistance to radiotherapy.
Although overexpression of KDM4B has been reported in different human cancers, little is known about the mechanism underlying regulation of KDM4B expression. In the current study, we provide evidence supporting the vital role of CREB in regulating KDM4B expression in CRC cells. CREB is a member of the CREB/ATF family nuclear transcription factor which directly regulates expression of a number of critical genes involved in differentiation, proliferation, and survival. Aberrant expression and/or activation of CREB has been observed in multiple cancers, and previous studies highlighted its implications in tumor initiation, progression, metastasis. Particularly, accumulating evidence suggests that CREB plays a critical role in cellular resistance to irradiation, indicating a mechanistic link between CREB target genes and DNA damage response [23]. Our results established KDM4B as a direct target of CREB and thus provide a putative explanation for CREB-dependent irradiation resistance in CRC cells. Interestingly, it has been shown that the molecular chaperon Hsp90 plays a critical role in stabilizing KDM4B protein via inhibition of ubiquitin-dependent proteasomal degradation [24, 25], suggesting that the abundance and activity of this demethylase are regulated by distinct mechanisms. It is possible that CREB and HSP90 might play a synergic role in regulating KDM4B expression via regulating transcription and protein stability. Notably, recent advances in small organic molecules have identified several chemical inhibitors targeting CREB-mediated gene transcription and proteasomal-induced protein turnover. It would be interesting to test whether a proper dose of the inhibitors could be achieved in certain CRC cell lines or subset of patients that the residual KDM4B is insufficient to support high resistance to irradiation. Further studies are also required to investigate the correlation between KDM4B expression and CRC radioresistance in large cohorts of CRC patients and to provide putative information for patient selection. Whether CREB and/or HSP90 expression are independent determinants of KDM4B expression also wait further investigation. Nevertheless, our results suggest that manipulation of CREB or KDM4B expression/activity might be a promising new strategy to enhance the efficacy of radiation therapeutic intervention in CRC patients.
Materials and methods
Chemicals and antibodies
AG490 and H89 were purchased from Sigma-Aldrich (St. Louis, MO, USA). Primary antibodies against KDM4B, p-STAT3, STAT3, p-JAK1, JAK1, and p-JAK2 were purchased from Cell Signaling (Danvers, MA, USA). Antibodies against JAK2, γH2AX, and α-tubulin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Alexa Fluor 594 Goat Anti-Rabbit IgG (H + L) Antibody and DAPI dye were purchased from Thermo Fisher (Waltham, MA, USA).
Cell culture
The human CRC cell lines HT29 and SW620 (ATCC, Manassas, VA, USA) were cultured at McCoy’s 5A or RPMI-1640 media, respectively, supplemented with 10% fetal bovine serum and 1% Penicillin/Streptomycin (Gibco, Gaithersburg, MD, USA) in a 37 °C incubator with 5% CO2. Primary colon epithelial cells CRL-1831 (ATCC, Manassas, VA, USA) and NCM-460 (INCELL, San Antonio, TX, USA) were cultured in DMEM:F12 Medium (Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum and 1% Penicillin/Streptomycin (Gibco, Gaithersburg, MD, USA) in a 37 °C incubator with 5% CO2.
CRC patient samples
A total of 20 patients who were diagnosed as stage II and III CRC and received colon resection surgery in First Affiliated Hospital of Nanchang University from 2016 to 2017 were included in this study. CRC tumor tissue and peritumoral tissue pairs were formalin-fixed for immunohistochemistry, or snap-frozen in liquid nitrogen and stored at − 80 °C until used for RNA extraction. The project protocol was approved by the Institutional Review Board of First Affiliated Hospital of Nanchang University. Informed consent for clinical research was obtained from each patient and all experiments were performed in accordance with relevant guidelines.
Immunohistochemistry
CRC tumor tissues and matched peritumoral samples were formalin-fixed and paraffin-embedded in 4 µm tissue sections. The labeled antigen was detected by the EnVision™ Detection Systems Peroxidase/DAB detection kit (Agilent, Santa Clara, CA, USA). The slides were counterstained with hematoxylin and the tissue image was captured with a digital camera.
Real-time PCR
Total RNA from CRC tumor tissue and peritumoral tissue were extracted with Trizol (Thermo Fisher Scientific, Waltham, MA, USA). The quantity and quality of RNA were determined by NanoDrop UV–vis spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). 1 µg of total RNA was used as a template for cDNA synthesis with the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA) using random primers for cDNA synthesis. 10 ng of the synthesized cDNA was used as a template for real-time PCR using SYBR® Green Real-Time PCR Master Mixes (Thermo Fisher Scientific, Waltham, MA, USA) and primers KDM4B forward: 5′-GGACTGACGGCAACCTCTAC-3′; KDM4B reverse: 5′-CGTCCTCAAACTCCACCTG-3′; GAPDH forward: 5′-AGCCACATCGCTCAGACAC-3′; GAPDH reverse: 5′-GCCCAATACGACCAAATCC-3′.
Cell survival upon ionizing radiation treatment
HT29 cells were seeded in 12-well plates and irradiated with various doses of ionizing radiation using a 137Cs irradiation unit with a dose rate of 1 Gy min−1. Cells were then incubated for 7 days to allow colony formation. Cells were washed twice with PBS, fixed with 100% ethanol, and were stained with crystal violet (Sigma-Aldrich, St. Louis, MO, USA). The number of colonies per dish was counted and the surviving fractions were calculated as the ratio between treated and untreated cells.
Chromatin immunoprecipitation assay
Chromatin Immunoprecipitation assay was performed using the Chromatin Immunoprecipitation (ChIP) Assay Kit (Millipore, Bedford, MA, USA). Briefly, HT29 cells were crosslinked with 1.0% formaldehyde (Sigma, St. Louis, MO) and 5 µg sonicated DNA was incubated with 2 µg antibody linked to Magnetic Beads (Thermo Fisher Scientific, Waltham, MA, USA). Isolated DNA was purified using a PCR Clean-up kit (Qiagen, Hilden, Germany) before PCR amplification using KDM4B primers promoter forward: 5′-CTGAGGCTCCCTTGTCAATCCA-3′; promoter reverse: 5′-CCCGCTGACCGGCTCTGTT-3′; Intron 1 forward: 5′-CTGACTTGCCCTTTGCTG-3′; Intron 1 reverse: 5′- CCTCCTAAGGCTCACAACC-3′; Intron 2 forward: 5′-TCGGCTGTTGGCTTATT-3′; Intron 2 reverse: 5′-TGGTATGACAGGCGTGAG-3′. PCR products were separated on a 1.2% agarose gel.
Knockdown and overexpression
For stable KDM4B knockdown, HT29 cells were transduced with lentiviral-based pLKO.1-shRNA vectors (shKDM4B-1: TTTGGAGGGTAATGTGGCCGG; shKDM4B-1: TACTTCTTCAGGATGATGGGC) and selected in puromycin (Sigma-Aldrich, St. Louis, MO, USA). pLKO.1 empty vector was used as control. For transient CREB knockdown, cells were transfected with 50 nM of siRNA (AM16708, Thermo Fisher Scientific, Waltham, MA, USA) using lipofectamine RNAiMAX (Thermo Fisher Scientific, Waltham, MA, USA). pCDNA3-STAT3 was a gift from Jie Chen (Addgene plasmid # 74433).
Luciferase reporter assay
KDM4B promoter region (− 566 to + 27) containing CREB binding site was amplified from genomic DNA using primers forward: 5′-GGGAACGCTAAGCCCACATT-3′ and reverse: 5′-TCGGTTGCTGGCGACCGA-3′. Both sense (WT) and antisense (WT) fragments were subcloned into pGL3 plasmid to generate luciferase reporters. HT29 cells were transfected with the pGL3 constructs along with Renilla luciferase vector (pRL-TK) using Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA). 36 h post-transfection cells were harvested using Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA).
References
Ross W, Lynch P, Raju G, Rodriguez A, Burke T, Hafemeister L, Hawk E, Wu X, Dubois RN, Mishra L (2012) Biomarkers, bundled payments, and colorectal cancer care. Genes Cancer 3:16–22. https://doi.org/10.1177/1947601912448958
Migliore L, Migheli F, Spisni R, Coppede F (2011) Genetics, cytogenetics, and epigenetics of colorectal cancer. J Biomed Biotechnol 2011:792362. https://doi.org/10.1155/2011/792362
Shadad AK, Sullivan FJ, Martin JD, Egan LJ (2013) Gastrointestinal radiation injury: symptoms, risk factors and mechanisms. World J Gastroenterol 19:185–198. https://doi.org/10.3748/wjg.v19.i2.185
Lao VV, Grady WM (2011) Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol 8:686–700. https://doi.org/10.1038/nrgastro.2011.173
Crea F, Nobili S, Paolicchi E, Perrone G, Napoli C, Landini I, Danesi R, Mini E (2011) Epigenetics and chemoresistance in colorectal cancer: an opportunity for treatment tailoring and novel therapeutic strategies. Drug Resist Updat 14:280–296. https://doi.org/10.1016/j.drup.2011.08.001
van Engeland M, Derks S, Smits KM, Meijer GA, Herman JG (2011) Colorectal cancer epigenetics: complex simplicity. J Clin Oncol 29:1382–1391. https://doi.org/10.1200/JCO.2010.28.2319
Fodor BD, Kubicek S, Yonezawa M, O’Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K, Schotta G, Jenuwein T (2006) Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev 20:1557–1562. https://doi.org/10.1101/gad.388206
Yang J, Jubb AM, Pike L, Buffa FM, Turley H, Baban D, Leek R, Gatter KC, Ragoussis J, Harris AL (2010) The histone demethylase JMJD2B is regulated by estrogen receptor alpha and hypoxia, and is a key mediator of estrogen induced growth. Cancer Res 70:6456–6466. https://doi.org/10.1158/0008-5472.CAN-10-0413
Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y (2006) Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125:467–481. https://doi.org/10.1016/j.cell.2006.03.028
Young LC, McDonald DW, Hendzel MJ (2013) Kdm4b histone demethylase is a DNA damage response protein and confers a survival advantage following gamma-irradiation. J Biol Chem 288:21376–21388. https://doi.org/10.1074/jbc.M113.491514
Khoury-Haddad H, Guttmann-Raviv N, Ipenberg I, Huggins D, Jeyasekharan AD, Ayoub N (2014) PARP1-dependent recruitment of KDM4D histone demethylase to DNA damage sites promotes double-strand break repair. Proc Natl Acad Sci USA 111:E728–E737. https://doi.org/10.1073/pnas.1317585111
Berry WL, Janknecht R (2013) KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res 73:2936–2942. https://doi.org/10.1158/0008-5472.CAN-12-4300
Chen L, Fu L, Kong X, Xu J, Wang Z, Ma X, Akiyama Y, Chen Y, Fang J (2014) Jumonji domain-containing protein 2B silencing induces DNA damage response via STAT3 pathway in colorectal cancer. Br J Cancer 110:1014–1026. https://doi.org/10.1038/bjc.2013.808
Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE (2006) H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc Natl Acad Sci USA 103:9891–9896. https://doi.org/10.1073/pnas.0603779103
Xiong H, Zhang ZG, Tian XQ, Sun DF, Liang QC, Zhang YJ, Lu R, Chen YX, Fang JY (2008) Inhibition of JAK1, 2/STAT3 signaling induces apoptosis, cell cycle arrest, and reduces tumor cell invasion in colorectal cancer cells. Neoplasia 10:287–297
Barry SP, Townsend PA, Knight RA, Scarabelli TM, Latchman DS, Stephanou A (2010) STAT3 modulates the DNA damage response pathway. Int J Exp Pathol 91:506–514. https://doi.org/10.1111/j.1365-2613.2010.00734.x
Young LC, Hendzel MJ (2013) The oncogenic potential of Jumonji D2 (JMJD2/KDM4) histone demethylase overexpression. Biochem Cell Biol 91:369–377. https://doi.org/10.1139/bcb-2012-0054
Yang Q, Zhu Q, Lu X, Du Y, Cao L, Shen C, Hou T, Li M, Li Z, Liu C, Wu D, Xu X, Wang L, Wang H, Zhao Y, Yang Y, Zhu WG (2017) G9a coordinates with the RPA complex to promote DNA damage repair and cell survival. Proc Natl Acad Sci USA 114:E6054–E6063. https://doi.org/10.1073/pnas.1700694114
Roos WP, Thomas AD, Kaina B (2016) DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer 16:20–33. https://doi.org/10.1038/nrc.2015.2
Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD (2012) Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol Cell 48:723–733. https://doi.org/10.1016/j.molcel.2012.09.026
Price BD, D’Andrea AD (2013) Chromatin remodeling at DNA double-strand breaks. Cell 152:1344–1354. https://doi.org/10.1016/j.cell.2013.02.011
Zheng H, Chen L, Pledger WJ, Fang J, Chen J (2014) p53 promotes repair of heterochromatin DNA by regulating JMJD2b and SUV39H1 expression. Oncogene 33:734–744. https://doi.org/10.1038/onc.2013.6
Pregi N, Belluscio LM, Berardino BG, Castillo DS, Canepa ET (2017) Oxidative stress-induced CREB upregulation promotes DNA damage repair prior to neuronal cell death protection. Mol Cell Biochem 425:9–24. https://doi.org/10.1007/s11010-016-2858-z
Ipenberg I, Guttmann-Raviv N, Khoury HP, Kupershmit I, Ayoub N (2013) Heat shock protein 90 (Hsp90) selectively regulates the stability of KDM4B/JMJD2B histone demethylase. J Biol Chem 288:14681–14687. https://doi.org/10.1074/jbc.C113.462770
Fu L, Chen L, Yang J, Ye T, Chen Y, Fang J (2012) HIF-1alpha-induced histone demethylase JMJD2B contributes to the malignant phenotype of colorectal cancer cells via an epigenetic mechanism. Carcinogenesis 33:1664–1673. https://doi.org/10.1093/carcin/bgs217
Acknowledgements
This study was supported by Natural Science Foundation of China (NSFC) 81660096.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11010_2018_3345_MOESM1_ESM.eps
Supplementary Figure 1. KDM4B silencing doesn’t induce cell death or proliferation arrest. Growth curves of KDM4B knockdown or control knockdown cells. 10,000 cells were cultured for up to 72 h and the number of cells overtime were calculated. Data represent the mean ± s.e.m. for three independent experiments. No significant difference was observed between groups. Supplementary material 1 (EPS 1910 KB)
Rights and permissions
About this article
Cite this article
Deng, WW., Hu, Q., Liu, ZR. et al. KDM4B promotes DNA damage response via STAT3 signaling and is a target of CREB in colorectal cancer cells. Mol Cell Biochem 449, 81–90 (2018). https://doi.org/10.1007/s11010-018-3345-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-018-3345-5